Inference of Genome-Scale Gene Regulatory Networks: Are There Differences in Biological and Clinical Validations?
Abstract
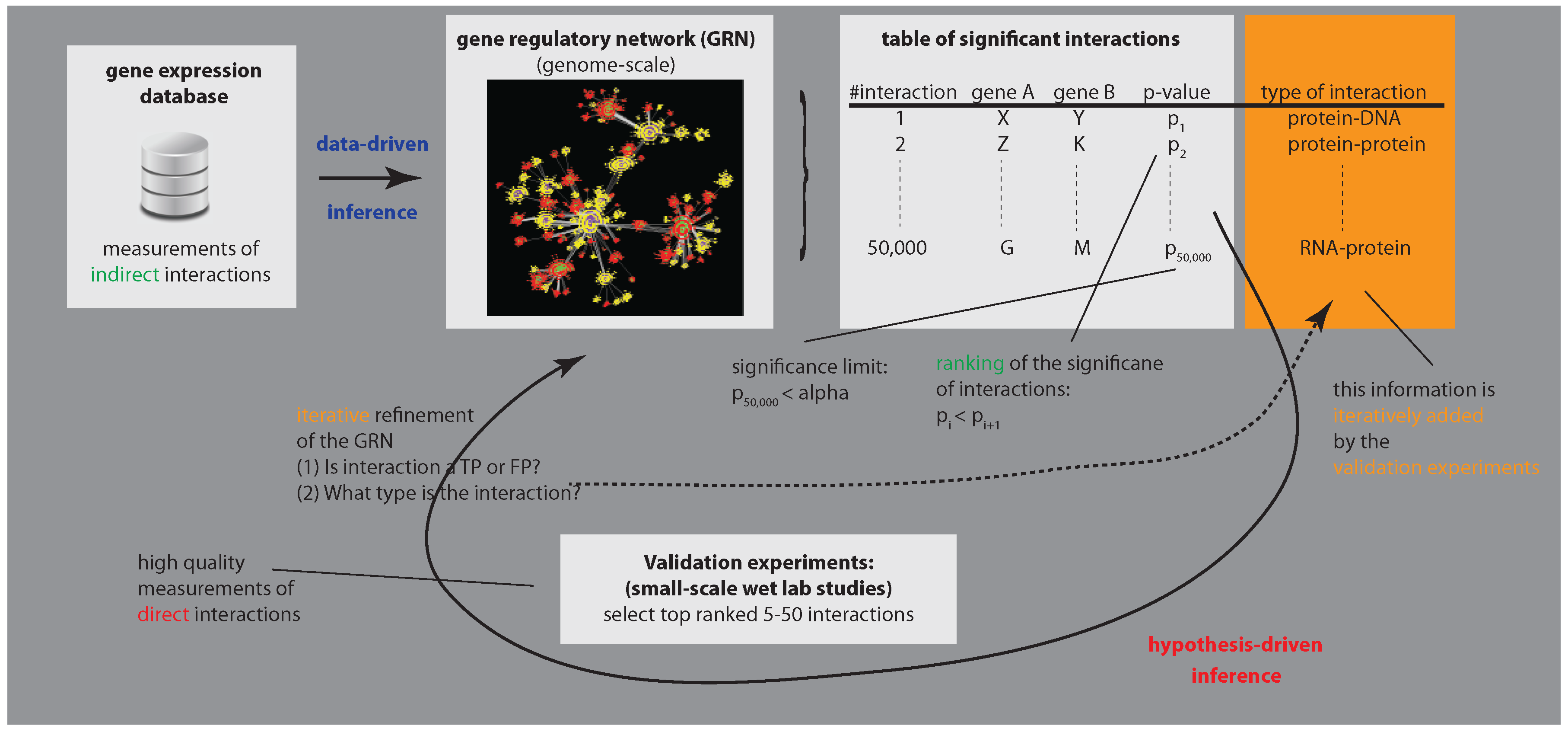
:1. Introduction
2. How Large Are Gene Regulatory Networks?
3. Biological Validation of GRNs
3.1. Enhancing Experimental Assays by Perturbing the System
- A single gene knockdown experiment was selected from the collection that included all replicates as a validation set.
- The genes whose expressions are significantly affected by the perturbation were identified.
- The capacity of the network model to predict which genes are affected by the perturbation by focusing on connections local to the gene being perturbed was assessed.
- Steps 1–3 were repeated to assess the predictive power of the network model until all perturbations had been tested.
3.2. Existing Data Repositories
4. General Considerations about Validation
- A validation is necessary because the entities to be validated are generated by a statistical model corresponding to its predictions.
- The entities to be predicted are scientifically meaningful for particular research fields.
5. Clinical Validation of GRNs
Higher-Level Networks
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ghahramani, Z. Probabilistic machine learning and artificial intelligence. Nature 2015, 521, 452. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436. [Google Scholar] [CrossRef] [PubMed]
- Witten, D.M.; Friedman, J.H.; Simon, N. New Insights and Faster Computations for the Graphical Lasso. J. Comput. Graph. Stat. 2011, 20, 892–900. [Google Scholar] [CrossRef]
- De Matos Simoes, R.; Emmert-Streib, F. Bagging statistical network inference from large-scale gene expression data. PLoS ONE 2012, 7, e33624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opgen-Rhein, R.; Strimmer, K. Learning causal networks from systems biology time course data: An effective model selection procedure for the vector autoregressive process. BMC Bioinform. 2007, 8, S3. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mardis, E.R. Next-Generation DNA Sequencing Methods. Annu. Rev. Genom. Hum. Genet. 2008, 9, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, J.; Wyrwicz, L.S. Integrating genomics, proteomics and bioinformatics in translational studies of molecular medicine. Expert Rev. Mol. Diagn. 2009, 9, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Barabási, A.L.; Oltvai, Z.N. Network Biology: Understanding the Cell’s Functional Organization. Nat. Rev. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Chuang, H.Y.; Lee, E.; Liu, Y.T.; Ideker, T. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 2007, 3, 140. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, A.; Dehmer, M.; Jurisica, I. Knowledge discovery and interactive data mining in bioinformatics-state-of-the-art, future challenges and research directions. BMC Bioinform. 2014, 15, I1. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Hidalgo, D.; Domínguez-Cejudo, M.A.; Amore, G.; Brockmann, A.; Lemos, M.C.; Córdoba, A.; Casares, F. A Hh-driven gene network controls specification, pattern and size of the Drosophila simple eyes. Development 2012, 082172. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Hidalgo, D.; Zurita, A.C.; Fernández, M.C.L. Complex networks evolutionary dynamics using genetic algorithms. Int. J. Bifurc. Chaos 2012, 22, 1250156. [Google Scholar] [CrossRef]
- Aguilar-Hidalgo, D.; Lemos, M.C.; Córdoba, A. Evolutionary dynamics in gene networks and inference algorithms. Computation 2015, 3, 99–113. [Google Scholar] [CrossRef]
- Ideker, T.; Krogan, N.J. Differential network biology. Mol. Syst. Biol. 2012, 8, 565. [Google Scholar] [CrossRef] [PubMed]
- Shen-Orr, S.; Milo, R.; Mangan, S.; Alon, U. Network motifs in the transcriptional regulatory network of Escherichia coli. Nat. Genet. 2002, 31, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Roukos, D.H. Genome network medicine: Innovation to overcome huge challenges in cancer therapy. Wiley Interdiscip. Rev. Syst. Biol. Med. 2014, 6, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Yachie-Kinoshita, A.; Onishi, K.; Ostblom, J.; Langley, M.A.; Posfai, E.; Rossant, J.; Zandstra, P.W. Modeling signaling-dependent pluripotency with Boolean logic to predict cell fate transitions. Mol. Syst. Biol. 2018, 14, e7952. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, E.R. Validation of inference procedures for gene regulatory networks. Curr. Genom. 2007, 8, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, E.R. Validation of gene regulatory networks: Scientific and inferential. Brief. Bioinform. 2011, 12, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Walhout, A. What does biologically meaningful mean? A perspective on gene regulatory network validation. Genome Biol. 2011, 12, 109. [Google Scholar] [CrossRef] [PubMed]
- Schaffter, T.; Marbach, D.; Floreano, D. GeneNetWeaver: In silico benchmark generation and performance profiling of network inference methods. Bioinformatics 2011, 27, 2263–2270. [Google Scholar] [CrossRef] [PubMed]
- Van den Bulcke, T.; Van Leemput, K.; Naudts, B.; van Remortel, P.; Ma, H.; Verschoren, A.; De Moor, B.; Marchal, K. SynTReN: A generator of synthetic gene expression data for design and analysis of structure learning algorithms. BMC Bioinform. 2006, 7, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emmert-Streib, F.; Glazko, G.; Altay, G.; de Matos Simoes, R. Statistical inference and reverse engineering of gene regulatory networks from observational expression data. Front. Genet. 2012, 3, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearl, J. Causality: Models, Reasoning, and Inference; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2000. [Google Scholar]
- Spirtes, P.; Glymour, C.; Scheines, R. Causation, Prediction, and Search; Springer: New York, NY, USA, 1993. [Google Scholar]
- Emmert-Streib, F. The Chronic Fatigue Syndrome: A Comparative Pathway Analysis. J. Comput. Biol. 2007, 14, 961–972. [Google Scholar] [CrossRef] [PubMed]
- De Matos Simoes, R.; Dehmer, M.; Emmert-Streib, F. Interfacing cellular networks of S. cerevisiae and E. coli: Connecting dynamic and genetic information. BMC Genom. 2013, 14, 324. [Google Scholar] [CrossRef] [PubMed]
- Basso, K.; Margolin, A.A.; Stolovitzky, G.; Klein, U.; Dalla-Favera, R.; Califano, A. Reverse Engineering of Regulatory Networks in Human B Cells. Nat. Genet. 2005, 37, 382–390. [Google Scholar] [CrossRef] [PubMed]
- De Matos Simoes, R.; Dehmer, M.; Emmert-Streib, F. B-cell lymphoma gene regulatory networks: Biological consistency among inference methods. Front. Genet. 2013, 4, 281. [Google Scholar] [CrossRef] [PubMed]
- Emmert-Streib, F.; de Matos Simoes, R.; Mullan, P.; Haibe-Kains, B.; Dehmer, M. The gene regulatory network for breast cancer: Integrated regulatory landscape of cancer hallmarks. Front. Genet. 2014, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Emmert-Streib, F.; de Matos Simoes, R.; Glazko, G.; McDade, S.; Haibe-Kains, B.; Holzinger, A.; Dehmer, M.; Campbell, F. Functional and genetic analysis of the colon cancer network. BMC Bioinform. 2014, 15, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godfrey-Smith, P. Theory and Reality: An Introduction to the Philosophy of Science; Science and Its Conceptual Foundations Series; University of Chicago Press: Chicago, IL, USA, 2003. [Google Scholar]
- Kell, D.B.; Oliver, S.G. Here is the evidence, now what is the hypothesis? The complementary roles of inductive and hypothesis-driven science in the post-genomic era. BioEssays 2004, 26, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Bruce, J. Chemical Cross-Linking for Protein? Protein Interaction Studies. In Mass Spectrometry of Proteins and Peptides; Methods in Molecular Biology; Lipton, M., Pasa-Tolic, L., Eds.; Humana Press: New York, NY, USA, 2009; Volume 492, pp. 283–293. [Google Scholar]
- Liu, Q.; Dinu, I.; Adewale, A.; Potter, J.; Yasui, Y. Comparative evaluation of gene-set analysis methods. BMC Bioinform. 2007, 8, 431. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, D.G.; Roth, S.Y. Current Protocols in Molecular Biology; Chapter Identification of Protein Interactions by Far Western Analysis; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2001. [Google Scholar]
- Kerppola, T.K. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 2006, 1, 1278–1286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aranda, B.; Achuthan, P.; Alam-Faruque, Y.; Armean, I.; Bridge, A.; Derow, C.; Feuermann, M.; Ghanbarian, A.T.; Kerrien, S.; Khadake, J.; et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2009, 38, D525–D531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buck, M.; Lieb, J. ChIP-chip: Considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 2004, 83, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Kidder, B.L.; Hu, G.; Zhao, K. ChIP-Seq: Technical considerations for obtaining high-quality data. Nat. Immunol. 2011, 12, 918–922. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Seeholzer, S.; Ohh, M. Identification of associated proteins by coimmunoprecipitation. In Molecular Cloning—A Laboratory Manual; Sambrook, J., Russell, D.W., Eds.; CSHL Press: New York, NY, USA, 2002; Volume 3, pp. 18–60. [Google Scholar]
- Joung, J.K.; Ramm, E.I.; Pabo, C.O. A bacterial two-hybrid selection system for studying protein–DNA and protein–protein interactions. Proc. Natl. Acad. Sci. USA 2000, 97, 7382–7387. [Google Scholar] [CrossRef] [PubMed]
- Konig, J.; Zarnack, K.; Rot, G.; Curk, T.; Kayikci, M.; Zupan, B.; Turner, D.J.; Luscombe, N.M.; Ule, J. iCLIP–transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J. Vis. Exp. 2011, 50, 2638. [Google Scholar] [CrossRef] [PubMed]
- Hamosh, A.; Scott, A.F.; Amberger, J.; Valle, D.; McKusick, V.A. Online Mendelian inheritance in man (OMIM). Hum. Mutat. 2000, 15, 57–61. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Parker, J.; Karginova, O.; Fan, C.; Livasy, C.; Herschkowitz, J.; He, X.; Perou, C. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010, 12, R68. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Haibe-Kains, B.; Desmedt, C.; Loi, S.; Culhane, A.C.; Bontempi, G.; Quackenbush, J.; Sotiriou, C. A Three-Gene Model to Robustly Identify Breast Cancer Molecular Subtypes. J. Natl. Cancer Inst. 2012, 104, 311–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [PubMed]
- Olsen, C.; Fleming, K.; Prendergast, N.; Rubio, R.; Emmert-Streib, F.; Bontempi, G.; Haibe-Kains, B.; Quackenbush, J. Inference and validation of predictive gene networks from biomedical literature and gene expression data. Genomics 2014, 103, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Cho, J.W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2017, 46, D380–D386. [Google Scholar] [CrossRef] [PubMed]
- Ben-Hamo, R.; Efroni, S. Gene expression and network-based analysis reveals a novel role for hsa-miR-9 and drug control over the p38 network in glioblastoma multiforme progression. Genome Med. 2011, 3, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dehmer, M.; Mueller, L.; Emmert-Streib, F. Quantitative Network Measures as Biomarkers for Classifying Prostate Cancer Disease States: A Systems Approach to Diagnostic Biomarkers. PLoS ONE 2013, 8, e77602. [Google Scholar] [CrossRef] [PubMed]
- Cun, Y.; Frohlich, H. Network and Data Integration for Biomarker Signature Discovery via Network Smoothed T-Statistics. PLoS ONE 2013, 8, e73074. [Google Scholar] [CrossRef] [PubMed]
- Frohlich, H. Including network knowledge into Cox regression models for biomarker signature discovery. Biom. J. 2014, 56, 287–306. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, G.; Chen, L. Prediction and early diagnosis of complex diseases by edge-network. Bioinformatics 2014, 30, 852–859. [Google Scholar] [CrossRef] [PubMed]
- Zeng, T.; Zhang, W.; Yu, X.; Liu, X.; Li, M.; Liu, R.; Chen, L. Edge biomarkers for classification and prediction of phenotypes. Sci. China Life Sci. 2014, 57, 1103–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, T.; Sun, S.y.; Wang, Y.; Zhu, H.; Chen, L. Network biomarkers reveal dysfunctional gene regulations during disease progression. FEBS J. 2013, 280, 5682–5695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.Q.; Tingle, S.R.; Filipski, K.K.; Khoury, M.J.; Lam, T.K.; Schully, S.D.; Ioannidis, J.P. An overview of recommendations and translational milestones for genomic tests in cancer. Genet. Med. 2014, 17, 431. [Google Scholar] [CrossRef] [PubMed]
- Van De Vijver, M.J.; He, Y.D.; van’t Veer, L.J.; Dai, H.; Hart, A.A.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J.; et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002, 347, 1999–2009. [Google Scholar] [CrossRef] [PubMed]
- Cava, C.; Bertoli, G.; Castiglioni, I. In silico identification of drug target pathways in breast cancer subtypes using pathway cross-talk inhibition. J. Transl. Med. 2018, 16, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaeger, S.; Igea, A.; Arroyo, R.; Alcalde, V.; Canovas, B.; Orozco, M.; Nebreda, A.R.; Aloy, P. Quantification of pathway cross-talk reveals novel synergistic drug combinations for breast cancer. Cancer Res. 2016. [Google Scholar] [CrossRef] [PubMed]
| Organism (Network Type) | # of Interactions | References |
|---|---|---|
| E. coli (GRN) | 21,820 | [28] |
| S. cerevisiae (GRN) | 27,493 | [4] |
| B-cell lymphoma (GRN) | 129,000; 57,905 | [29,30] |
| Breast cancer (GRN) | 180,171 | [31] |
| Colon cancer (GRN) | 135,194 | [32] |
| S. cerevisiae (TRN) | 12,873 | [28] |
| S. cerevisiae (PIN) | 112,562 | [28] |
| Human (TRN) | 51,871 | [30] |
| Human (PIN) | 185,433 | [30] |
| Experimental Technique | Type of Interaction | Reference |
|---|---|---|
| ChIP-chip/ ChIP-seq | protein–DNA interaction | [40,41] |
| Co-immunoprecipitation | protein–protein interaction | [42] |
| Yeast two-hybrid | protein–protein interaction | [43] |
| Crosslinking Protein Interaction Analysis | protein–protein interaction | [35] |
| Label Transfer Chemistry | protein–protein interaction | [36] |
| Far-Western Blot Analysis | protein–protein interaction | [37] |
| BiFCassays | protein–protein interaction | [38] |
| iCLIP | protein–RNA interaction | [44] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Emmert-Streib, F.; Dehmer, M. Inference of Genome-Scale Gene Regulatory Networks: Are There Differences in Biological and Clinical Validations? Mach. Learn. Knowl. Extr. 2019, 1, 138-148. https://doi.org/10.3390/make1010008
Emmert-Streib F, Dehmer M. Inference of Genome-Scale Gene Regulatory Networks: Are There Differences in Biological and Clinical Validations? Machine Learning and Knowledge Extraction. 2019; 1(1):138-148. https://doi.org/10.3390/make1010008
Chicago/Turabian StyleEmmert-Streib, Frank, and Matthias Dehmer. 2019. "Inference of Genome-Scale Gene Regulatory Networks: Are There Differences in Biological and Clinical Validations?" Machine Learning and Knowledge Extraction 1, no. 1: 138-148. https://doi.org/10.3390/make1010008





