PDMS-Based Microfluidic Devices for Cell Culture
Abstract
:1. Introduction
- pH—Most normal mammalian cell lines grow well at pH 7.4. However, some transformed cell lines have been shown to grow better in slightly more acidic environments (pH 7.0–7.4), and some normal fibroblast cell lines prefer slightly more basic environments (pH 7.4–7.7).
- Dissolved Oxygen (DO)—The dissolved oxygen concentration in culture medium in standard culture conditions is approximately 20–50%.
- Dissolved Carbon Dioxide—A dissolved carbon dioxide level less than 200 mmHg (1% CO2 = 7.2 mmHg) should be generally maintained in cell cultures.
- Temperature—There are optimal temperature values associated with different cell lines. Most human and mammalian cell lines are maintained at 36–37 °C for optimal growth. Insect cells are cultured at 27 °C since they grow more slowly at lower temperatures and at temperatures between 27–30 °C; above 30 °C, the viability of insect cells decreases and the cells do not recover even after they are returned to 27 °C. Cell lines derived from cold-blooded animals (e.g., amphibians, cold-water fish) tolerate a wide temperature range between 15 and 26 °C.
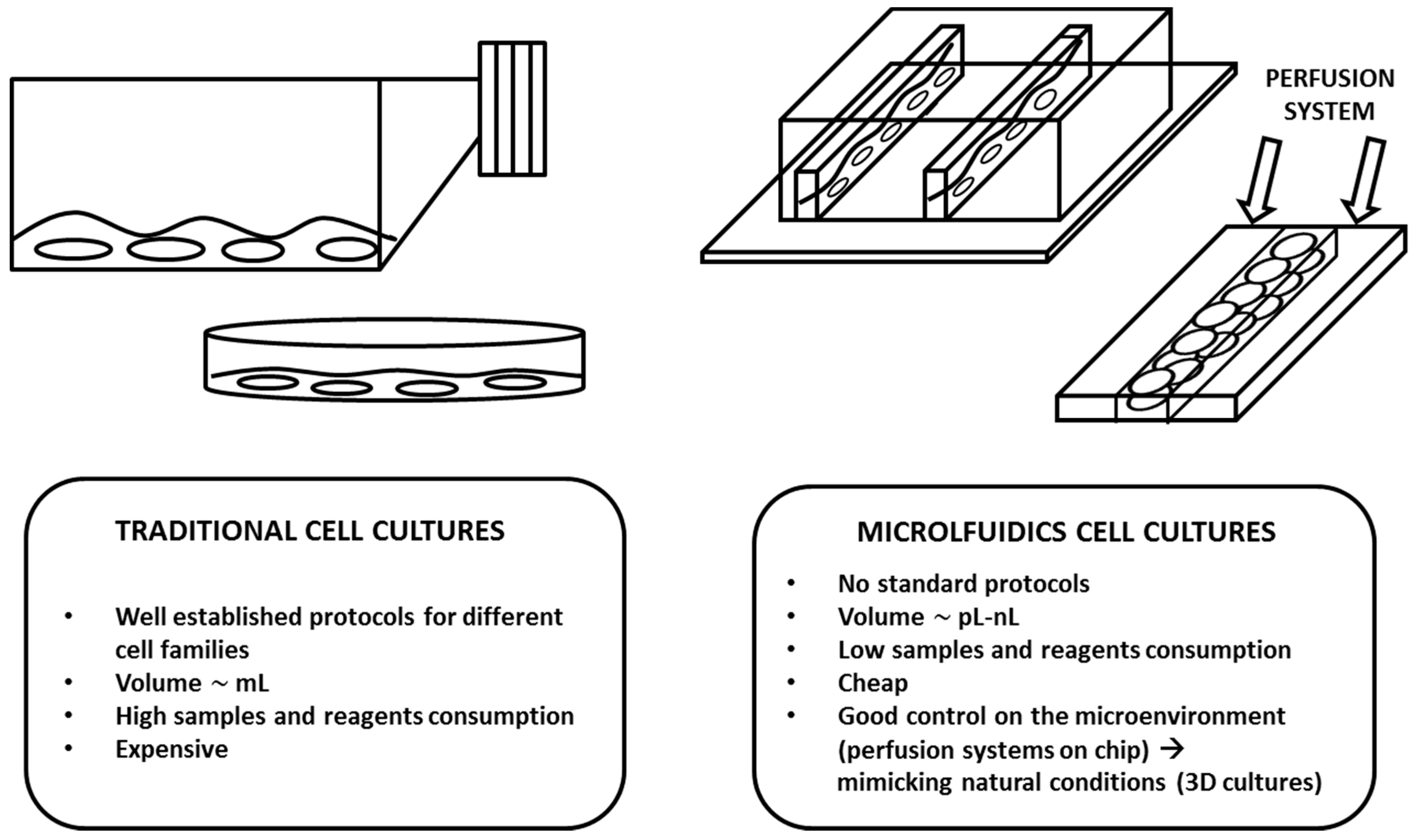
2. Design and Theory
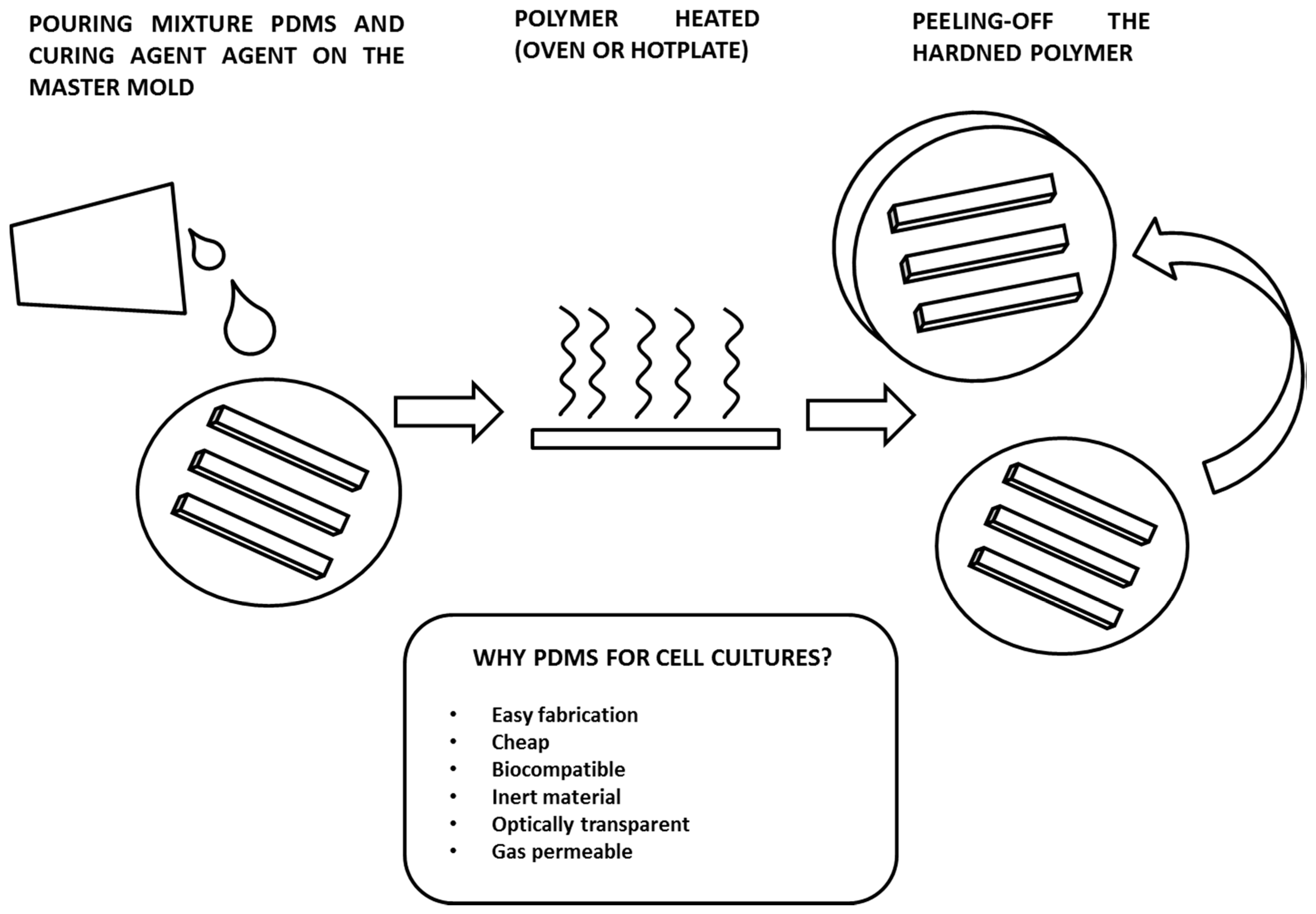
3. Fabrication of Microfluidic Culture Chambers by PDMS Soft-Lithography
- Optical transparency. PDMS is optically transparent from a 240–1100 nm wavelength and is therefore compatible with many optical detection methods. Although PDMS is optically transparent, it has an intrinsic fluorescence, with intensity values smaller than the ones given by other polymers, that is considerably lower than the signals from fluorescence measurements performed on cells. If the PDMS microculture chamber is sealed with a glass slide using an inverted microscope with acquisition in reflection, the fluorescence from the polymer will be a weak signal on the background. However, for some applications, diffraction and reflection of light waves through the material could be a problem, for example, when the detection of fluorescent reporters within the growth chamber should be correlated with gene expression or molecular binding events [60].
- Gas permeability. PDMS is gas permeable and therefore a perfect material for cell cultures, since it allows the control of the amount of gas through the exchange across the polymer matrix [61]. The concentration of O2 and CO2 can be kept in the required range for good cell viability. This is a critical issue especially when these microsystems are to be used for establishing long-term cultures (i.e., days or weeks). On the other hand, the gas permeability of PDMS can introduce some issues, such as the evaporation of the media. This aspect strongly affects cell cultures in microchambers due the small quantity of liquid samples (in the order of few microliters). Evaporation leads to bubble formation, which consequently lyses the cells in the chamber. Several solutions have been proposed to overcome this issue, such as: insuring a sufficient humidity level in the incubator in which the microculture chambers are placed [62]; using a large-volume reservoir filled with the culture media to compensate the rate of evaporation [63]; and coating the PDMS channel with a polymer layer, usually parylene, to prevent evaporation [64].
- Inert material. Inert surfaces are of great interest since they allow the spatial patterning of proteins or cells. The most commonly used technique for making these patterned substrates is microcontact printing [65]. An elastomeric PDMS stamp is first inked with a solution containing the patterning component, and then the stamp is brought into physical contact with the target surface. This can be another PDMS layer or a substrate of a different material, such as a metal or glass. With this technique, cells are well confined to a specific region of a substrate, allowing the precise control of the size and shape of the cell population. This approach is quite widely used in applications for tissue engineering or bio-sensing [66,67].
- Biocompatibility. This is the most important requirement for a material that interfaces with biological samples. It can be defined as the property by which the material does not produce undesirable effects on a biological organism [68]. In medicine and biology, there had been a tendency to use natural polymers or modified natural polymers. However, nowadays, there are many synthetic polymers that are widely used for bio-applications, such as polylactic acid (PLA) and polyglycolic acid (PGA), which are the most used materials for scaffold fabrication in tissue engineering. PDMS is highly biocompatible. Several studies have demonstrated the success of using the polymer for the fabrication of microdevices for cell cultures, for organ-on-chip models, and for making implantable devices or parts of them. In some cases, issues related to the compatibility of devices made of other materials, i.e., having metal parts, have been solved by covering them with a PDMS film [69].
4. Discussions and Conclusions
Conflicts of Interest
References
- Bergmann, S.; Steinert, M. Chapter One—From Single Cells to Engineered and Explanted Tissues: New Perspectives in Bacterial Infection Biology. Int. Rev. Cell Mol. Biol. 2015, 319, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Mather, J.P. (Ed.) Cell Culture Studies Using Extracts of Extracellular Matrix to Study Growth and Differentiation in Mammalian Cells. In Mammalian Cell Culture; Springer: Boston, MA, USA, 1984; ISBN 978-1-4615-9361-4. [Google Scholar]
- Michael Conn, P. (Ed.) Laboratory Methods in Cell Biology: Biochemistry and Cell Culture, 11st ed.; Elsevier Academic Press: Cambridge, UK, 2012; Volume 112, ISBN 9780124055490. [Google Scholar]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Beebe, D.J.; Mensing, A.J.; Walker, G.M. Physics and applications of microfluidics in biology. Annu. Rev. Biomed. Eng. 2002, 4, 261–286. [Google Scholar] [CrossRef] [PubMed]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Abgrall, P.; Gué, A.M. Lab-on-chip technologies: Making a microfluidic network and coupling it into a complete microsystem—A review. J. Micromech. Microeng. 2007, 17, R15–R49. [Google Scholar] [CrossRef]
- Halldorsson, S.; Lucumi, E.; Gómez-Sjöberg, R.; Fleming, R.M.T. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens. Bioelectron. 2015, 63, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Young, E.W.K.; Beebe, D.J. Fundamentals of microfluidic cell culture in controlled microenvironments. Chem. Soc. Rev. 2010, 39, 1036–1048. [Google Scholar] [CrossRef] [PubMed]
- Mehling, M.; Tay, S. Microfluidic Cell Culture. Curr. Opin. Biotechnol. 2014, 25, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Luni, C.; Giulitti, S.; Serena, E.; Ferrari, L.; Zambon, A.; Gagliano, O.; Giobbe, G.G.; Michielin, F.; Knöbel, S.; Bosio, A.; et al. high-efficiency cellular reprogramming with microfluidics. Nat. Methods 2016, 13, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Novo, P.; Dell’Aica, M.; Janasek, D.; Zahedi, R.P. High spatial and temporal resolution cell manipulation techniques in microchannels. Analyst 2016, 141, 1888–1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, B.G.; Flanagan, L.A.; Rhee, S.W.; Schwartz, P.H.; Lee, A.P.; Monuki, E.S.; Jeon, N.L. Human neural stem cell growth and differentiation in a gradient-generating microfluidic device. Lab Chip 2005, 5, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Sjöberg, R.; Leyrat, A.A.; Pirone, D.M.; Chen, C.S.; Quake, S.R. Versatile, fully automated, microfluidic cell culture system. Anal. Chem. 2007, 79, 8557–8563. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Ghassemi, P.; Babahosseini, H.; Strobl, J.S.; Agah, M. Single-Cell Mechanical Characteristics Analyzed by Multiconstriction Microfluidic Channels. ACS Sens. 2017, 2, 290–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tam, J.; Cordier, G.A.; Balint, S.; Sandoval Alvarez, A.; Borbely, J.S.; Lakadamyali, M. A Microfluidic Platform for Correlative Live-Cell and Super-Resolution Microscopy. PLoS ONE 2014, 9, e115512. [Google Scholar] [CrossRef] [PubMed]
- Martewicz, S.; Gabrel, G.; Campesan, M.; Canton, M.; di Lisa, F.; Elvassore, N. Live Cell Imaging in Microfluidic Device Proves Resistance to Oxygen/Glucose Deprivation in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Anal. Chem. 2018, 90, 5687–5695. [Google Scholar] [CrossRef] [PubMed]
- Jovic, A.; Howell, B.; Takayama, S. Timing is everything: Using fluidics to understand the role of temporal dynamics in cellular systems. Microfluid. Nanofluid. 2009, 6, 717–729. [Google Scholar] [CrossRef]
- Kamei, K.; Guo, S.; Yu, Z.T.F.; Takahashi, H.; Gschweng, E.; Suh, C.; Wang, X.; Tang, J.; McLaughlin, J.; Witte, O.N.; et al. An integrated microfluidic culture device for quantitative analysis of human embryonic stem cells. Lab Chip 2009, 9, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Khademhosseini, A.; Langer, R. A decade of progress in tissue engineering. Nat. Protoc. 2016, 11, 1775–1781. [Google Scholar] [CrossRef] [PubMed]
- Lanz, H.L.; Saleh, A.; Kramer, B.; Cairns, J.; Ng, C.P.; Yu, J.; Trietsch, S.J.; Hankemeier, T.; Joore, J.; Vulto, P.; et al. Therapy response testing of breast cancer in a 3D high-throughput perfused microfluidic platform. BMC Cancer 2017, 17, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, P.S.; Manz, A. Lab-on-a-chip: Microfluidics in drug discovery. Nat. Rev. Drug Discov. 2006, 5, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Mattern, K.; Beißner, N.; Reichl, S.; Dietzel, A. DynaMiTES—A dynamic cell culture platform for in vitro drug testing PART 1—Engineering of microfluidic system and technical simulations. Eur. J. Pharm. Biopharm. 2018, 126, 159–165. [Google Scholar] [CrossRef] [PubMed]
- van Duinen, V.; Trietsch, S.J.; Joore, J.; Vulto, P.; Hankemeier, T. Microfluidic 3D cell culture: From tools to tissue models. Curr. Opin. Biotechnol. 2015, 35, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Sung, K.E.; Beebe, D.J. Microfluidic 3D models of cancer. Adv. Drug Deliv. Rev. 2014, 79–80, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chen, Q.; Liu, W.; He, Z.; Lin, J.M. Recent advances in microfluidic 3D cellular scaffolds for drug assays. TrAC Trends Anal. Chem. 2017, 87, 19–31. [Google Scholar] [CrossRef]
- Park, D.H.; Jeon, H.J.; Kim, M.J.; Nguyen, X.D.; Morten, K.; Go, J.S. Development of a microfluidic perfusion 3D cell culture system. J. Micromech. Microeng. 2018, 28, 045001. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.P.; Ma, Y.; Lou, Q.; Zhu, H.; Yang, B.; Fang, Q. Three-Dimensional Cell Culture and Drug Testing in a Microfluidic Sidewall-Attached Droplet Array. Anal. Chem. 2017, 89, 10153–10157. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Vickerman, V.; Blundo, J.; Chung, S.; Kamm, R. Design, fabrication and implementation of a novel multi-parameter control microfluidic platform for three-dimensional cell culture and real-time imaging. Lab Chip 2008, 8, 1468–1477. [Google Scholar] [CrossRef] [PubMed]
- Huh, D. A Human Breathing Lung-on-a-Chip. Ann. Am. Thorac. Soc. 2015, 12, S42–S44. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Ingber, D.E. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. 2013, 5, 1130–1140. [Google Scholar] [CrossRef] [PubMed]
- Bhise, N.S.; Manoharan, V.; Massa, S.; Tamayol, A.; Ghaderi, M.; Miscuglio, M.; Lang, Q.; Zhang, Y.S.; Shin, S.R.; Calzone, G.; et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication 2016, 8, 014101. [Google Scholar] [CrossRef] [PubMed]
- Weltin, A.; Slotwinski, K.; Kieninger, J.; Moser, I.; Jobst, G.; Wego, M.; Ehret, R.; Urban, G.A. Cell culture monitoring for drug screening and cancer research: A transparent, microfluidic, multi-sensor microsystem. Lab Chip 2014, 14, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Jeevarajan, A.S.; Vani, S.; Taylor, T.D.; Anderson, M.M. Continuous pH monitoring in a perfused bioreactor system using an optical pH sensor. Biotechnol. Bioeng. 2002, 78, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.G.; Jeevarajan, A.S.; Anderson, M.M. Long-term continuous monitoring of dissolved oxygen in cell culture medium for perfused bioreactors using optical oxygen sensors. Biotechnol. Bioeng. 2004, 86, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Pattison, R.N.; Swamy, J.; Mendenhall, B.; Hwang, C.; Frohlich, B.T. Measurement and control of dissolved carbon dioxide in mammalian cell culture processes using an in situ fiber optic chemical sensor. Biotechnol. Prog. 2000, 16, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Naciri, M.; Kuystermans, D.; Al-Rubeai, M. Monitoring pH and dissolved oxygen in mammalian cell culture using optical sensors. Cytotechnology 2008, 57, 245–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabeling, P. Introduction to Microfluidics, 6th ed.; Oxford University Press: New York, NY, USA, 2005. [Google Scholar]
- Bruus, H. Theoretical Microfluidics; Oxford University Press Inc.: New York, NY, USA, 2008. [Google Scholar]
- Nguyen, N.T.; Wereley, S.T. Fundamental and Applications of Microfluidics, 2nd ed.; Artech House: Norwood, MA, USA, 2002. [Google Scholar]
- Kim, L.; Toh, Y.C.; Voldman, J.; Yu, H. A practical guide to microfluidic perfusion culture of adherent mammalian cells. Lab Chip 2007, 7, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.; Zhou, J.; Wu, H. Materials for Microfluidic Chip Fabrication. Acc. Chem. Res. 2013, 46, 2396–2406. [Google Scholar] [CrossRef] [PubMed]
- Becker, H.; Gärtner, C. Polymer microfabrication technologies for microfluidic systems. Anal. Bioanal. Chem. 2008, 390, 89–111. [Google Scholar] [CrossRef] [PubMed]
- Hong, T.F.; Ju, W.J.; Wu, M.C.; Tai, C.H.; Tsai, C.H.; Fu, L.M. Rapid prototyping of PMMA microfluidic chips utilizing a CO2 laser. Microfluid. Nanofluid. 2010, 9, 1125–1133. [Google Scholar] [CrossRef]
- Shadpour, H.; Hupert, M.L.; Patterson, D.; Liu, C.; Galloway, M.; Stryjewski, W.; Goettert, J.; Soper, S.A. Multichannel Microchip Electrophoresis Device Fabricated in Polycarbonate with an Integrated Contact Conductivity Sensor Array. Anal. Chem. 2007, 79, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Pugmire, D.L.; Waddell, E.A.; Haasch, R.; Tarlov, M.J.; Locascio, L.E. Surface Characterization of Laser-Ablated Polymers Used for Microfluidics. Anal. Chem. 2002, 74, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Metz, S.; Holzer, R.; Renaud, P. Polyimide-based microfluidic devices. Lab Chip 2001, 1, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Steigert, J.; Haeberle, S.; Brenner, T.; Muller, C.; Steinert, C.P.; Koltay, P.; Gottschlich, N.; Reinecke, H.; Ruhe, J.; Zengerle, R.; et al. Rapid prototyping of microfluidic chips in COC. J. Micromech. Microeng. 2007, 17, 333–341. [Google Scholar] [CrossRef]
- Young, E.W.K.; Berthier, E.; Guckenberger, D.J.; Sackmann, E.; Lamers, C.; Meyvantsson, I.; Huttenlocher, A.; Beebe, D.J. Rapid Prototyping of Arrayed Microfluidic Systems in Polystyrene for Cell-Based Assays. Anal. Chem. 2011, 83, 1408–1417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, J.C.; Duffy, D.C.; Anderson, J.R.; Chiu, D.T.; Wu, H.; Schueller, O.J.; Whitesides, G.M. Fabrication of microfluidic systems in poly(dimethylsiloxane). Electrophoresis 2000, 21, 27–40. [Google Scholar] [CrossRef]
- Fujii, T. PDMS-based microfluidic devices for biomedical applications. Microelectron. Eng. 2002, 61–62, 907–914. [Google Scholar] [CrossRef]
- Hongbin, Y.; Guangya, Z.; Siong, C.F.; Shouhua, W.; Feiwen, L. Novel polydimethylsiloxane (PDMS) based microchannel fabrication method for lab-on-a-chip application. Sens. Actuator B-Chem. 2009, 137, 754–761. [Google Scholar] [CrossRef]
- Xia, Y.; Whitesides, G.M. Soft lithography. Annu. Rev. Mater. Sci. 1998, 28, 153–184. [Google Scholar] [CrossRef]
- Mack, C. Introduction to Semiconductor Lithography. In Fundamental Principles of Optical Lithography: The Science of Microfabrication; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2007; ISBN 978-0-470-01893-4. [Google Scholar]
- Mohanty, S.; Larsen, L.B.; Trifol, J.; Szabo, P.; Burri, H.V.R.; Canali, C.; Dufva, M.; Emnéus, J.; Wolff, A. Fabrication of scalable and structured tissue engineering scaffolds using water dissolvable sacrificial 3D printed moulds. Mater. Sci. Eng. C 2015, 55, 569–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mata, A.; Fleischman, A.J.; Roy, S. Characterization of Polydimethylsiloxane (PDMS) Properties for Biomedical Micro/Nanosystems. Biomed. Microdevices 2005, 7, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Ng, W. Transparent PET optical window for microfluidic cell culture device. PeerJ Prepr. 2017, 5, e3245v1. [Google Scholar] [CrossRef]
- Merkel, T.C.; Bondar, V.I.; Nagai, K.; Freeman, B.D.; Pinnau, I. Gas sorption, diffusion, and permeation in poly(dimethylsiloxane). J. Polym. Sci. B Polym. Phys. 2000, 38, 415–434. [Google Scholar] [CrossRef] [Green Version]
- Christen, J.B.; Andreou, A.G. Design, Fabrication, and Testing of a Hybrid CMOS/PDMS Microsystem for Cell Culture and Incubation. IEEE Trans. Biomed. Circuits Syst. 2007, 1, 3–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Chu, L.Y.; Chueh, B.; Shen, M.; Hazarika, B.; Phadke, N.; Takayama, S. Arrays of horizontally-oriented mini-reservoirs generate steady microfluidic flows for continuous perfusion cell culture and gradient generation. Analyst 2004, 129, 1026–1031. [Google Scholar] [CrossRef] [PubMed]
- Flueckiger, J.; Bazargan, V.; Stoeber, B.; Cheung, K.C. Characterization of postfabricated parylene C coatings inside PDMS microdevices. Sens. Actuators B Chem. 2011, 160, 864–874. [Google Scholar] [CrossRef]
- Ruiz, S.A.; Chen, C.S. Microcontact printing: A tool to pattern. Soft Matter 2007, 3, 168–177. [Google Scholar] [CrossRef] [Green Version]
- Bernard, A.; Delamarche, E.; Schmid, H.; Michel, B.; Bosshard, H.R.; Biebuyck, H. Printing patterns of proteins. Langmuir 1998, 14, 2225–2229. [Google Scholar] [CrossRef]
- Andersson, H.; van den Berg, A. Microfabrication and microfluidics for tissue engineering: State of the art and future opportunities. Lab Chip 2004, 4, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.F. On the mechanisms of biocompatibility. Biomaterials 2008, 29, 2941–2953. [Google Scholar] [CrossRef] [PubMed]
- Bélanger, M.C.; Marois, Y. Hemocompatibility, biocompatibility, inflammatory and in vivo studies of primary reference materials low-density polyethylene and polydimethylsiloxane: A review. J. Biomed. Mater. Res. Appl. Biomater. 2001, 58, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, E.; Sakai, Y.; Fujii, T. Cell Culture in 3-Dimensional Microfluidic Structure of PDMS (polydimethylsiloxane). Biomed. Microdevices 2003, 5, 109–114. [Google Scholar] [CrossRef]
- Regehr, K.J.; Domenech, M.; Koepsel, J.T.; Carver, K.C.; Ellison-Zelski, S.J.; Murphy, W.L.; Schuler, L.A.; Alarid, E.T.; Beebe, D.J. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip 2009, 9, 2132–2139. [Google Scholar] [CrossRef] [PubMed]
- Toepke, M.W.; Beebe, D.J. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip 2006, 6, 1484–1486. [Google Scholar] [CrossRef] [PubMed]
- Modena, M.M.; Chawla, K.; Misun, P.M.; Hierlemann, A. Smart Cell Culture Systems: Integration of Sensors and Actuators into Microphysiological Systems. ACS Chem. Biol. 2018, 13, 1767–1784. [Google Scholar] [CrossRef] [PubMed]
- Mannino, R.G.; Qiu, Y.; Lam, W.A. Endothelial cell culture in microfluidic devices for investigating microvascular processes. Biomicrofluidics 2018, 12, 042203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bein, A.; Shin, W.; Jalili-Firoozinezhad, S.; Park, M.H.; Sontheimer-Phelps, A.; Tovaglieri, A.; Chalkiadaki, A.; Kim, H.J.; Ingber, D.E. Microfluidic Organ-on-a-Chip Models of Human Intestine. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, G.; Leijten, J.; Khademhosseini, A. Concise Review: Organ Engineering: Design, Technology, and Integration. Stem Cells 2016, 35, 51–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torino, S.; Corrado, B.; Iodice, M.; Coppola, G. PDMS-Based Microfluidic Devices for Cell Culture. Inventions 2018, 3, 65. https://doi.org/10.3390/inventions3030065
Torino S, Corrado B, Iodice M, Coppola G. PDMS-Based Microfluidic Devices for Cell Culture. Inventions. 2018; 3(3):65. https://doi.org/10.3390/inventions3030065
Chicago/Turabian StyleTorino, Stefania, Brunella Corrado, Mario Iodice, and Giuseppe Coppola. 2018. "PDMS-Based Microfluidic Devices for Cell Culture" Inventions 3, no. 3: 65. https://doi.org/10.3390/inventions3030065