Head Kidney Transcriptome Analysis and Characterization for the Sub-Antarctic Notothenioid Fish Eleginops maclovinus
Abstract
:1. Introduction
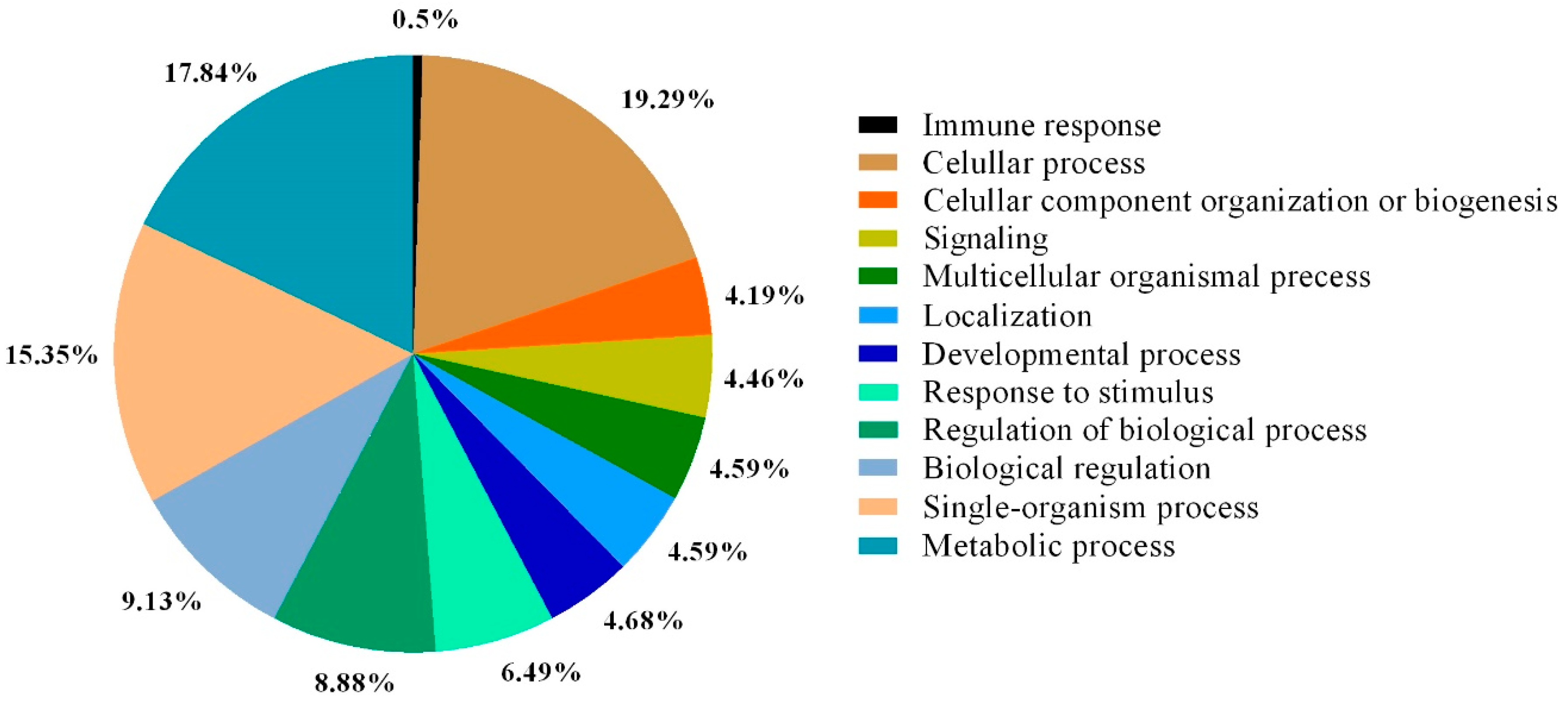
2. Results
3. Materials and Methods
3.1. Animal Specimens
3.2. RNA Isolation, Library Construction, and Sequencing
3.3. De Novo Transcriptome Assembly, Identification of Protein Coding Region, and Annotation
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Near, T.J. Estimating divergence times of notothenioid fishes using a fossil-calibrated molecular clock. Antarct. Sci. 2004, 16, 37–44. [Google Scholar] [CrossRef]
- Papetti, C.; Windisch, H.S.; La Mesa, M.; Lucassen, M.; Marshall, C.; Lamare, M.D. Non-Antarctic notothenioids: Past phylogenetic history and contemporary phylogeographic implications in the face of environmental changes. Mar. Genom. 2016, 25, 1–9. [Google Scholar] [CrossRef] [PubMed]
- La Mesa, M.; Caputo, V.; Eastman, J.T. Some reproductive traits of the Tristan klipfish, Bovichtus diacanthus (Carmichael 1819) (Notothenioidei: Bovichtidae) from Tristan da Cunha (South Atlantic). Polar Biol. 2010, 33, 337–346. [Google Scholar] [CrossRef]
- Hofmann, G.E.; Lund, S.G.; Place, S.P.; Whitmer, A.C. Some like it hot, some like it cold: The heat shock response is found in New Zealand but not Antarctic notothenioid fishes. J. Exp. Mar. Bio. Ecol. 2005, 316, 79–89. [Google Scholar] [CrossRef]
- Beers, J.M.; Jayasundara, N. Antarctic notothenioid fish: what are the future consequences of “losses” and “gains” acquired during long-term evolution at cold and stable temperatures? J. Exp. Biol. 2015, 218, 1834–1845. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cheng, C.-H.C.; Zhang, J.; Cao, L.; Chen, L.; Zhou, L.; Jin, Y.; Ye, H.; Deng, C.; Dai, Z.; et al. Transcriptomic and genomic evolution under constant cold in Antarctic notothenioid fish. Proc. Natl. Acad. Sci. USA 2008, 105, 12944–12949. [Google Scholar] [CrossRef] [PubMed]
- Thorne, M.A.S.; Burns, G.; Fraser, K.P.P.; Hillyard, G.; Clark, M.S. Transcription profiling of acute temperature stress in the Antarctic plunderfish Harpagifer antarcticus. Mar. Genom. 2010, 3, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Coppe, A.; Agostini, C.; Marino, I.A.M.; Zane, L.; Bargelloni, L.; Bortoluzzi, S.; Patarnello, T. Genome evolution in the cold: Antarctic icefish muscle transcriptome reveals selective duplications increasing mitochondrial function. Genome Biol. Evol. 2013, 5, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Huth, T.J.; Place, S.P. De novo assembly and characterization of tissue specific transcriptomes in the emerald notothen, Trematomus bernacchii. BMC Genom. 2013, 14, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gerdol, M.; Buonocore, F.; Scapigliati, G.; Pallavicini, A. Analysis and characterization of the head kidney transcriptome from the Antarctic fish Trematomus bernacchii (Teleostea, Notothenioidea): A source for immune relevant genes. Mar. Genom. 2015, 20, 13–15. [Google Scholar] [CrossRef] [PubMed]
- Bilyk, K.T.; Cheng, C.H.C. Model of gene expression in extreme cold—Reference transcriptome for the high-Antarctic cryopelagic notothenioid fish Pagothenia borchgrevinki. BMC Genom. 2013, 14, 634. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.C.; Kim, S.J.; Lee, J.K.; Ahn, D.H.; Kim, M.G.; Lee, H.; Lee, J.; Kim, B.K.; Park, H. Transcriptomics and comparative analysis of three Antarctic Notothenioid fishes. PLoS ONE 2012, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.C.; Ahn, D.H.; Kim, S.J.; Pyo, C.W.; Lee, H.; Kim, M.K.; Lee, J.; Lee, J.E.; Detrich, H.W.; Postlethwait, J.H.; et al. The genome sequence of the Antarctic bullhead notothen reveals evolutionary adaptations to a cold environment. Genome Biol. 2014, 15, 468. [Google Scholar] [CrossRef] [PubMed]
- Pequeño, G. The geographical distribution and taxonomic arrangement of south american notothenhd fishes (Osteichthyes, Notothenhdae). Bol. Soc. Biol. Concepc. 1989, 183–200. [Google Scholar]
- Brickle, P.; Laptikhovsky, V.; Arkhipkin, A. Reproductive strategy of a primitive temperate notothenioid Eleginops maclovinus. J. Fish Biol. 2005, 66, 1044–1059. [Google Scholar] [CrossRef]
- Contreras-Lynch, S.; Olmos, P.; Vargas, A.; Figueroa, J.; González-Stegmaier, R.; Enríquez, R.; Romero, A. Identification and genetic characterization of Piscirickettsia salmonis in native fish from southern Chile. Dis. Aquat. Organ. 2015, 115, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Soulliere, C.; Dixon, B. Immune System Organs of Bony Fishes; Elsevier Ltd.: Amsterdam, The Netherlands, 2017; ISBN 9780128096338. [Google Scholar]
- Gallo, V.P.; Civinini, A. Survey of the Adrenal Homolog in Teleosts. Int. Rev. Cytol. 2003, 230, 89–187. [Google Scholar] [PubMed]
- Vargas-Chacoff, L.; Moneva, F.; Oyarzún, R.; Martínez, D.; Muñoz, J.L.P.; Bertrán, C.; Mancera, J.M. Environmental salinity-modified osmoregulatory response in the sub-Antarctic notothenioid fish Eleginops maclovinus. Polar Biol. 2014, 37, 1235–1245. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
| GO Category | Description |
|---|---|
| GO:0002376 | H-2 class ii histocompatibility antigen gamma chain-like |
| GO:0045087 | Tyrosine-protein kinase fgr isoform ×1 |
| GO:0002755 | Ubiquitin-40s ribosomal protein s27a |
| GO:0045087 | Af280815_1calmodulin partial |
| GO:0006955 | Major histocompatibility class i receptor |
| GO:0006955 | Tumor necrosis factor ligand superfamily member 13b |
| GO:0006054 | Macrophage colony-stimulating factor 1 receptor 2-like |
| GO:0006055 | Ectodysplasin splice variant-8 partial |
| GO:0002474 | Mhc class i alpha partial |
| GO:0001916 | Mhc class i antigen |
| GO:0006955 | Invariant chain-like protein 14-1 |
| GO:0002504 | Mhc class ii antigen beta chain |
| GO:0006355 | Paired amphipathic helix protein sin3a |
| GO:0002376 | G-protein coupled receptor 183-like |
| GO:0045087 | Actin-related protein 3 |
| GO:0045087 | Cell division control protein 42 homolog |
| GO:0006954 | Toll-like receptor 1 |
| GO:0045087 | Catenin beta-1-like |
| GO:0006954 | Toll-like receptor 8 |
| A. Number of Obtained Readings | |
| Raw Wells | 291,469 |
| Key Pass Wells | 281,128 |
| Passed Filter Wells | 208,809 |
| Total Bases | 95,309,200 |
| B. Sizes of the Obtained Sequences | |
| Length Average | 456 |
| Longest Read Length | 1173 |
| Shortest Read Length | 40 |
| Median Read Length | 500 |
| Mode Read Length | 542 |
| Assembled Transcripts | Unassembled Reads | |
|---|---|---|
| Number of transcripts | 5088 | 6119 |
| Base pairs | 2,398,375 | 1,780,104 |
| Annotated with “nr” | 2000 | 1602 |
| Not annotated with “nr” | 3088 | 4517 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez, D.; Pontigo, J.P.; Morera, F.J.; Yañéz, A.; Vargas-Chacoff, L. Head Kidney Transcriptome Analysis and Characterization for the Sub-Antarctic Notothenioid Fish Eleginops maclovinus. Fishes 2018, 3, 8. https://doi.org/10.3390/fishes3010008
Martínez D, Pontigo JP, Morera FJ, Yañéz A, Vargas-Chacoff L. Head Kidney Transcriptome Analysis and Characterization for the Sub-Antarctic Notothenioid Fish Eleginops maclovinus. Fishes. 2018; 3(1):8. https://doi.org/10.3390/fishes3010008
Chicago/Turabian StyleMartínez, Danixa, Juan Pablo Pontigo, Francisco J. Morera, Alejandro. Yañéz, and Luis Vargas-Chacoff. 2018. "Head Kidney Transcriptome Analysis and Characterization for the Sub-Antarctic Notothenioid Fish Eleginops maclovinus" Fishes 3, no. 1: 8. https://doi.org/10.3390/fishes3010008






