Practical Method for Isolation of Phage Deletion Mutants
Abstract
:1. Introduction
2. Experimental Design
2.1. Materials
- S. aureus Sa9 [11], isolated from a mastitic milk sample was used as the host strain of the phage.
- Phage phiH5, isolated from raw milk was used to select “clear lysis plaques” deletion mutants [11]
- TSB, Tryptic Soy Broth (Scharlau, Barcelona, Spain; Cat. no.: 02-200-500).
- Bacteriological agar (Cat. no.: A6686), Tris HCl (Cat. no.: 10812846001), MgSO4 (Cat. no.: M2643), CaCl2 (Cat. no.: C5670), NaCl (Cat. no.: S5886), KCl (Cat. no.: P9541), Na2HPO4 (Cat. no.: RES20908-A7), KH2PO4 (Cat. no.: NIST200B), Sodium pyrophosphate (Cat. no.: P8135), PEG 8000 (Cat. no.: 89510), CsCl (Cat. no.: 203025) and RNAse (Cat. no.: R6513) where purchased at Sigma-Aldrich, now Merck, Darmstadt, Germany.
2.2. Equipment
- Incubator shaker model Excella E24 (New-Brunswick Scientific UK Ltd, Cambridge, UK).
- Sanyo MIR-153 refrigerated Incubator (Sanyo electric, Ora-Gun, Japan).
- Spectofotometer BioPhotometer Eppendorf (Eppendorf Ibérica S.L.U., Madrid, Spain).
- Laminar Airflow Cabinet FASTER TWO 30 Cabinet (Manson Technology, Dublin, Ireland).
- STUART Mini Gyro Rocker SSM3 (Cole-Parmer, Stone, UK).
- Microcentrifuge 5415 R Epperdorf (Eppendorf Ibérica S.L.U., Madrid, Spain).
- Beckman Optima MAX Ultracentrifuge (Beckman Coulter, Brea, CA, USA).
- Centrifuge Thermo Scientific™ Sorvall™ RC 6 Plus Centrifuge (Fisher Scientific, Loughborough, UK).
- Vortex ZX3 (VELP SCIENTIFICA, Usmate Velate, Italy).
- Vacuum pump Diaphragm pumps, VACUUBRAND (VACUUBRAND, GMBH, Wertheim, Germany). 0.45 µm cellulose acetate filter (Whatman™, GE Healthcare, Amersham, UK; Cat. no.: 462100).
- 0.2 µm polyethersulphone filter (VWR, Radnor, USA; Cat. no.: 28145-501).
- 0.025 µm VSWP MF-Millipore™ Membrane Filters (Merck, Millipore, Cork, Ireland; Cat. no.: VSWP02500).
- 1 L, 0.45 µm pore size, Corning® bottle-top vacuum filter system (Sigma-Aldrich, now Merck, Darmstadt, Germany; Cat. no.: CLS430516).
- 1.5–3 mL VWR® Two-Sided Disposable Plastic Cuvette (VWR, Radnor, USA; Cat. no.: 97000-586).
- 90 mm Polystyrene Petri dishes (Labbox, Labware S.L., Barcelona, Spain; Cat. no.: PDIP-09N-500).
- Tube, thin wall, Ultra-clear, 5 mL, 13 × 51 mm ultracentrifuge tubes (Beckman Coulter, Brea, CA, USA; Cat. no.: 344057).
- PIPETMAN L Pipettes: P20L, P200L, and P1000L (Gilson, Dunstable, UK).
- Sterile aerosol filter pipet tips (VWR, Radnor, PA, USA).
3. Procedure
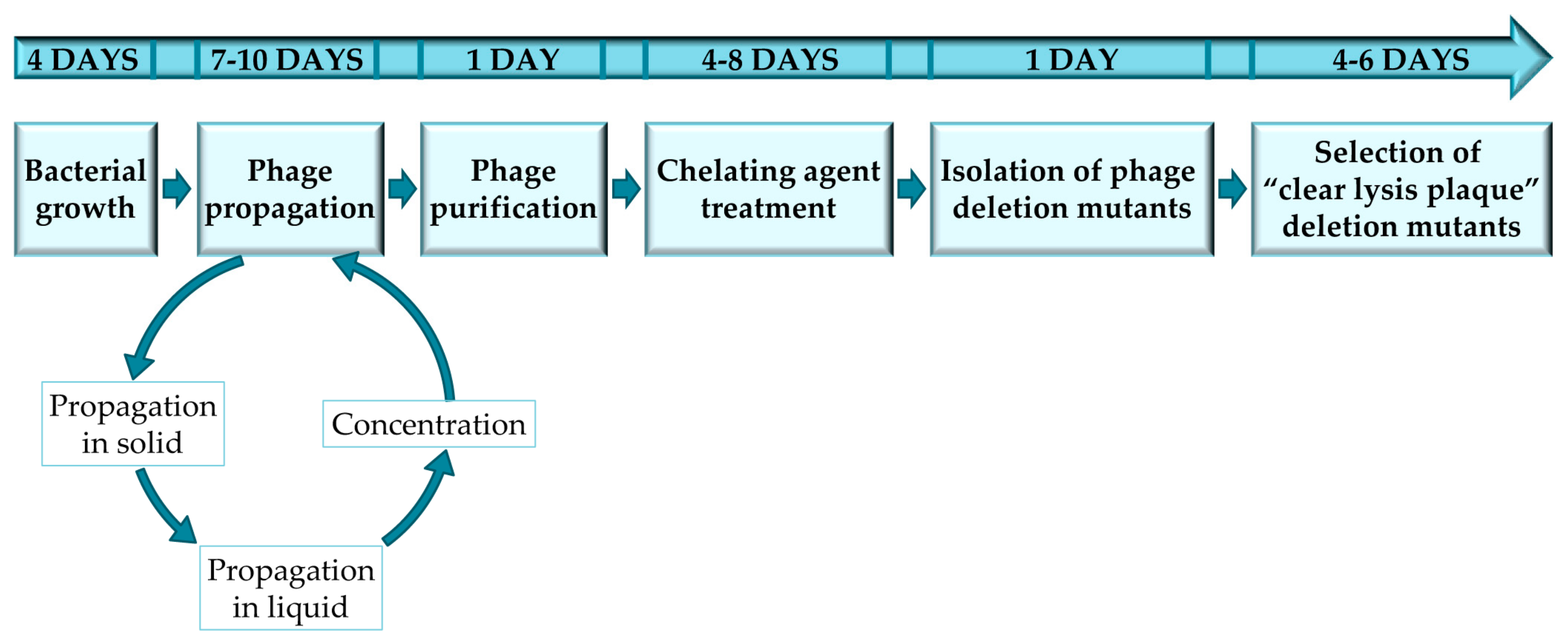
3.1. Bacterial Growth Conditions and Phage Titration. 4 Days
- Streak the S. aureus host strain onto a TSA (TSB supplemented with 2% w/v agar) plate using a sterile tip and incubate the plate at 37 °C for 18 h to obtain isolated colonies.
PAUSE STEP After the colonies are grown, plates can be stored at 4 °C for up to 1 month.
- Inoculate one S. aureus single colony from the TSA plate into a sterile 10 mL tube containing 5 mL of sterile TSB). Incubate at 37 °C with shaking (250 rpm) for 18 h to obtain an overnight (o/n) culture.OPTIONAL STEP Take 1 mL of the o/n culture and add glycerol to a final concentration of 20% in order to prepare a frozen stock of the host strain. Store this stock at −80 °C.
- In order to perform a bacterial growth curve, inoculate 50 mL of TSB with 1% (v/v) of the S. aureus o/n culture into a 100 mL sterile bottle and incubate at 37 °C with shaking.
- Take 2 mL samples at time points during incubation (0, 1, 2, 3, 4, 5, 6, 7, 8 and 24 h). Transfer 1 mL sample to a plastic cuvette and measure the optical density at an absorbance wavelength of 600 nm (OD600). At the same time, perform 1:10 serial dilutions of the sample into PBS buffer. Add 100 µL of the desired dilution to a TSA plate and spread using a sterile L-shape spreader until liquid dries. Incubate the plate at 37 °C for 18 h.
- Count the number of colonies in the plate containing 30–300 colony forming units (CFU). Calculate the viable cell counts of the culture using the equation:where N is the number of colonies counted on the plate (expressed as CFU); DF is the dilution factor and V is the sample volume poured onto the plate (expressed in mL).
- Repeat the experiment two extra times to obtain three biological replicates.
- Represent graphically the mean ± standard deviation of the CFU/mL in a logarithmic scale or the OD600 in the y-axis and the time of incubation of the culture in the x-axis.
CRITICAL STEP The number of CFU/mL should be calculated for each host strain, but generally speaking, it is very similar among strains belonging to the same species. The calculation of the CFU/mL in the exponential culture (Cb) is necessary to allow for the accurate determination of the multiplicity of infection (MOI). Calculation of this parameter is decisive in order to achieve an optimum yield in the phage propagation procedure.
3.2. Phage Titration and Propagation in Small Volume. 2–5 Days
- Inoculate one S. aureus single colony to obtain an o/n culture (see above).
- Make 1:10 serial dilutions of the initial phage stock.
- Determine the number of infective phage particles by using the double-layer plaque assay. Take 100 µL of a S. aureus o/n culture and mix with 100 µL of the desired phage dilution into a 15 mL tube. Add 3 mL of semisolid TSA (0.7% agar) and pour the mixture onto a TSA plate. Let the soft agar medium solidify, and then, incubate the plate at 37 °C for 18 h. This experiment must be performed in triplicate.
- After incubation, count the number of plaque forming units (PFU) in those plates where the number is between 30–300 PFU. Calculate the phage titer of the original phage sample by using the equation:where N is the number of plaques of lysis counted on the plate (expressed as PFU); DF is the dilution factor and V is the sample volume poured onto the plate (expressed in mL).
CRITICAL STEP If the titer of the phage suspension is ≥109 PFU/mL, then the phage propagation and concentration protocol can be followed from Section 3.3. If not, propagation in solid medium should be performed before proceeding to the next step.
- In order to carry out the propagation in solid medium, take 100 µL of diluted phage stock mix with 100 µL of a S. aureus o/n culture into a 15 mL tube. Add 3 mL of TSA 0.7% and pour the mixture onto a TSA plate. Repeat the procedure to get 5 plates. Let the medium solidify and then, incubate the plates at 37 °C for 18 h.
CRITICAL STEP After incubation, the overlaid plate should be completely transparent reflecting the occurrence of confluent lysis. The minimum concentration of phages that should be plated to ensure confluent lysis varies between 104 and 106 PFU/plate. Add 1 mL of SM buffer to each plate and incubate with gentle shaking (60 rpm) at 20 °C for 1 h. Recover the SM buffer from the five plates and transfer it to a 15 mL tube. Centrifuge 10 min at 13,600 × g. Transfer the supernatant containing the phages to a new sterile tube and perform phage titration again. If it is needed, solid propagation can be repeated again with this new phage stock in order to obtain a concentration ≥109 PFU/mL.
PAUSE STEP Phage stock solutions can be kept at 4 °C for up to three months after filtration using a 0.45 µm cellulose acetate membrane filter. For longer periods of time, a phage stock can be kept at −80 °C with 20% of glycerol.
3.3. Phage Propagation in Large Volume and Concentration. 5 Days
- Inoculate one S. aureus single colony from the TSA plate into a sterile 10 mL tube containing 5 mL of sterile TSB. Incubate at 37 °C with shaking (250 rpm) for 18 h to obtain an o/n culture.
- Inoculate one sterile 100 mL bottle containing 50 mL of sterile TSB with 1% v/v of the S. aureus o/n culture and incubate at 37 °C with shaking.OPTIONAL STEP Supplement the growth medium with 10 mM CaCl2 and 10 mM MgSO4. Addition of these cations is recommended for most phages, although it is not always strictly necessary, as some phages can be propagated efficiently without them.
- Take 1 mL sample after 1 h incubation and measure the OD600. Continue incubating the culture at 37 °C with shaking until it reaches the exponential growth phase.
CRITICAL STEP The bacterial cells should be in the exponential growing state to ensure proper phage propagation.
- Transfer the exponential growing bacteria to four sterile 50 mL tubes (10 mL in each tube). One tube will be the control and the other three tubes will be used for phage propagation with three different MOIs (0.1, 1 and 10). To calculate the volume (V0) of the phage that should be added to the different tubes follow the equation:where V1 is the final volume of the mixture (in this case 10 mL); C0 is the concentration of the previously titrated phage stock (≥109 PFU/mL), and C1 is the desired final phage concentration. For calculating C1 follow the equation:where Cb is the concentration of the bacterial culture at an exponential growing phase (expressed as CFU/mL) and MOI is the multiplicity of infection defined as PFU/CFU.
CRITICAL STEP The optimal MOI for phage propagation should be determined by testing different MOIs (MOI = 0.1, 1 and 10) since it may differ between phages.
- Incubate the tubes with shaking at 37 °C for at least 2 h. Check for lysis of the phage treated cultures by visual inspection.
CRITICAL STEP Incubation should proceed until complete lysis of the culture is observed (transparent tube compared to the turbid control tube). This can take more or less time depending on the MOI, the phage and the host strain used.
- Centrifuge the lysed cultures (13,600× g, 15 min at 4 °C) and keep the supernatant. Titer the supernatants as indicated above.
PAUSE STEP Phage suspensions can be kept at 4 °C for up to three months.
- Repeat the propagation protocol twice by scaling up the volume, and using the MOI resulting in the highest concentrated phage titer. Use the 10 mL phage stock obtained previously to perform propagation in 100 mL. Then, use the 100 mL phage stock to perform propagation in 1 L.
CRITICAL STEP The need for scaling up and the number of extra propagation rounds may vary among phages, depending on both the phage concentration obtained and the optimal MOI used for propagation.
- After carrying out propagation in 1 L volume, filter the lysate using a vacuum pump and 0.45 µm cellulose acetate vacuum filters. Concentrate the phage suspension by adding NaCl (0.5 M, final concentration) and PEG 8000 (10%, final concentration). Mix the components until they dissolve and maintain for 18 h at 4 °C. Divide the culture into four 500 mL centrifuge bottles and centrifuge at 16,000× g for 30 min at 4 °C.
- After centrifugation, decant the supernatant very carefully and suspend each phage pellet in 1 mL of SM buffer containing RNAse (40 µg/mL, final concentration). Measure the total volume of phage obtained.
PAUSE STEP This phage stock can be kept at 4 °C for at least 3 months or stored at −80 °C for more than two years after adding glycerol (20% (v/v), final concentration).
- Titer the concentrated phage stock, as described previously.
CRITICAL STEP The titer of the concentrated phage stock is usually 100 times higher than the one obtained in a liquid propagation.
- OPTIONAL STEP Perform phage purification.
3.4. Phage Purification. 1 Day (OPTIONAL STEP)
- Using the concentrated phage stock previously obtained, add CsCl to a final concentration of 0.75 g/mL, and mix vigorously.
PAUSE STEP Mixture can be kept at 4 °C for up to 1 week.
- Transfer the mixture to two Ultra-clear centrifuge tubes. Fill the tubes with filling solution (SM buffer supplemented with 0.75 g/mL CsCl). Place the tubes into buckets and centrifuge the samples in the swinging bucket rotor at 100,000× g, at 4 °C for 20 h, without brake.
- After ultracentrifugation, the purified phage can be identified as a light blue band in the middle of the tube. Pinch the tube carefully and extract the band using a 2 mL syringe and a 0.5 × 16 mm needle.
PAUSE STEP Purified phage stocks in a CsCl suspension can be kept at 4 °C for long periods of time (>5 years), as most phages are very stable in these conditions.
3.5. Sodium Pyrophosphate Treatment. 4–8 Days
- Place 150 µL of purified phage onto 0.025 µm VSWP Membrane MF-Millipore filter floating over 10 mL of SM buffer in a Petri dish. Incubate without shaking at 20 °C for 1 h. Calculate the titer of the dialyzed phage stock.
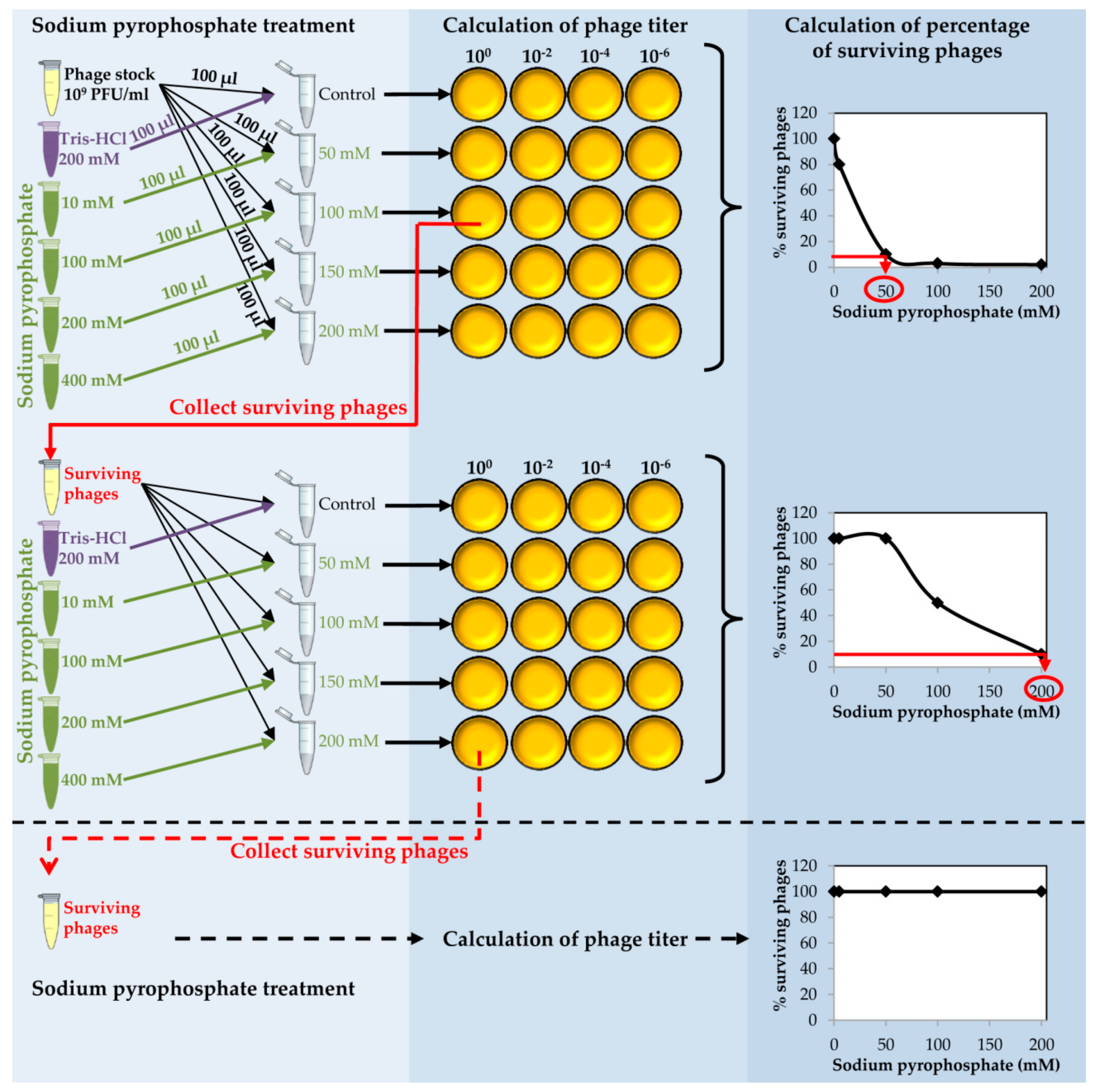
- Take an aliquot (10 µL) of dialyzed purified phage and make a 1:100 dilution into 990 µL of SM buffer (Figure 2).
CRITICAL STEP The dilution step is carried out in order to obtain a phage stock of 109 PFU/mL to proceed with the chelating agent treatment.
- Prepare a series of tubes with 100 µL of the diluted phage in each tube. Add 100 µL of each sodium pyrophosphate concentration (10–400 mM) to each tube in order to dilute the chelating agent 1:2 and get a final concentration from 5 to 200 mM in a final volume of 200 µL (Figure 2).
CRITICAL STEP For some phages, the destabilizing effect might be higher using other chelating agents such as EDTA or sodium citrate, but the procedure to follow in these cases is identical to that using sodium pyrophosphate.
- To prepare the control, mix 100 µL of purified phage with 100 µL of sterile Tris-HCl 200 mM, pH 8.
- Incubate the tubes at 37 °C for 30 min.
- Calculate the phage titer of each sample after the sodium pyrophosphate treatment making 1:100 dilutions in SM buffer and plate by the double layer technique using S. aureus as the host strain. At the same time, directly plate the undiluted sample (Figure 2).
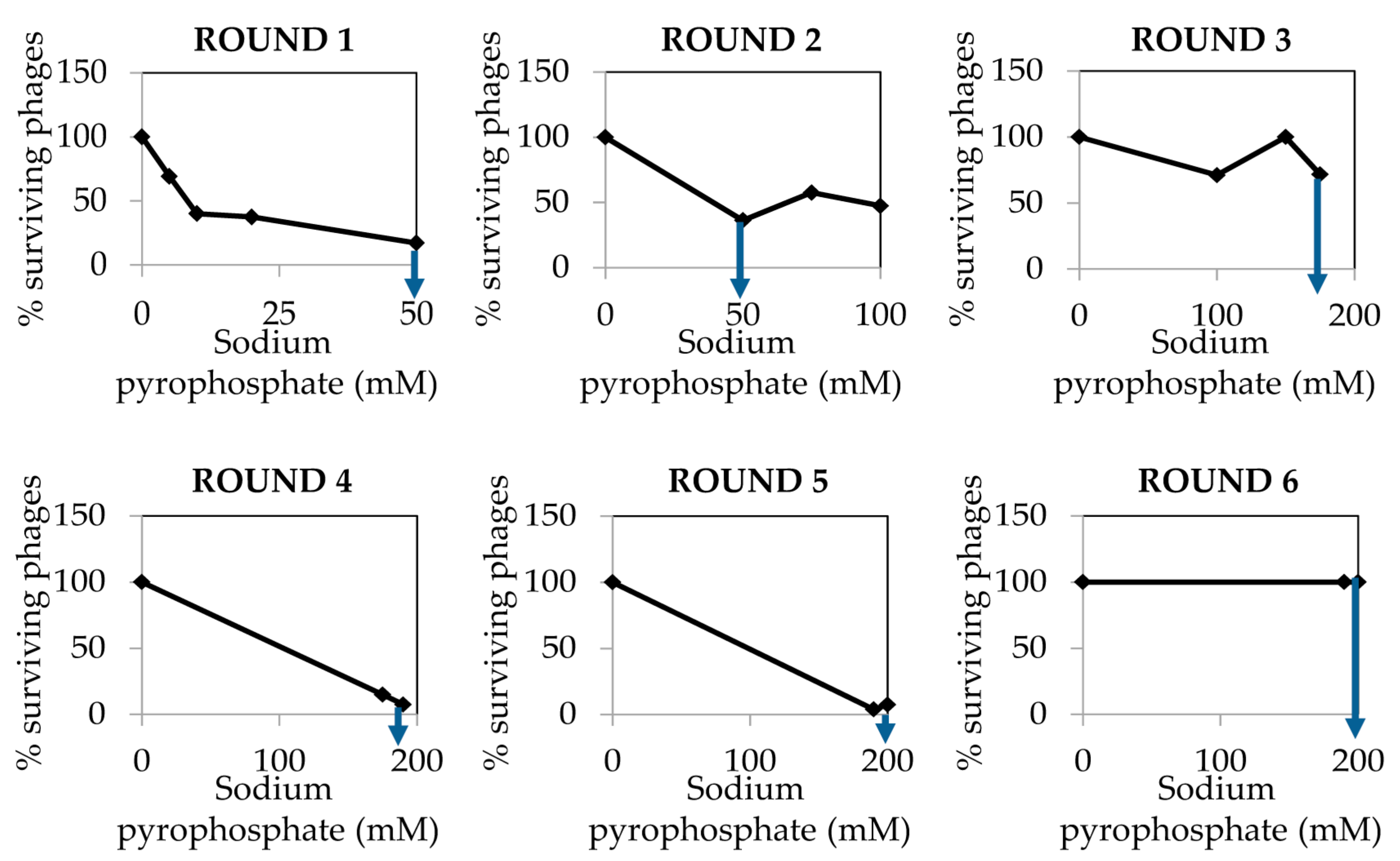
- Determine the percentage of surviving phages after the treatment compared to the control sample. Results must be depicted as a function of sodium pyrophosphate concentration (Figure 2).
CRITICAL STEP The sodium pyrophosphate treatment causes a decrease in the initial phage titer that should be around 90–99%. Some phages have a very stable particle structure; in these cases, a higher concentration of the chelating agent or an increase in the incubation temperature might be necessary.
- Collect the surviving phages from the plate in which the sodium pyrophosphate treatment led to a survival rate of about 10% of the phage population (Figure 2). To do this, add 1 mL of SM buffer to the plate corresponding to the phage dilution 10° and incubate 1 h with slight shaking (60 rpm) at room temperature. Afterwards, collect the liquid and centrifuge 10 min at 13,600× g, at 4 °C. Keep the supernatant containing the surviving phages.
PAUSE STEP The phage stock can be kept at 4 °C for 1 month before continuing with the protocol.
- Repeat the sodium pyrophosphate treatment (50–200 mM), as described above using the surviving phages obtained previously (Figure 2).
CRITICAL STEP Repeat the pyrophosphate treatment from 3 to 5 rounds until phage survival reaches a plateau of 100% even at high concentrations of the chelating agent (Figure 2).
3.6. Isolation of Phage Deletion Mutants. 1 Day
- Pick isolated lysis plaques with a sterile tip from the sample treated with the highest concentration of sodium pyrophosphate, and suspend each lysis plaque in 100 µL of SM buffer.
PAUSE STEP The phage stock can be kept at 4 °C for 1 month.
CRITICAL STEP The number of lysis plaques to be tested cannot be calculated in advance, but it is recommended to select about 10–20 lysis plaques for an initial screening for the desired mutant.
OPTIONAL STEP Propagate the putative phage deletion mutants following previous Section 3.1 and Section 3.2. - For temperate phages, the protocol can be extended to select virulent phages with “clear lysis plaque” phenotype (following section).
3.7. Selection of “Clear Lysis Plaque” Phage Deletion Mutants. 4 Days
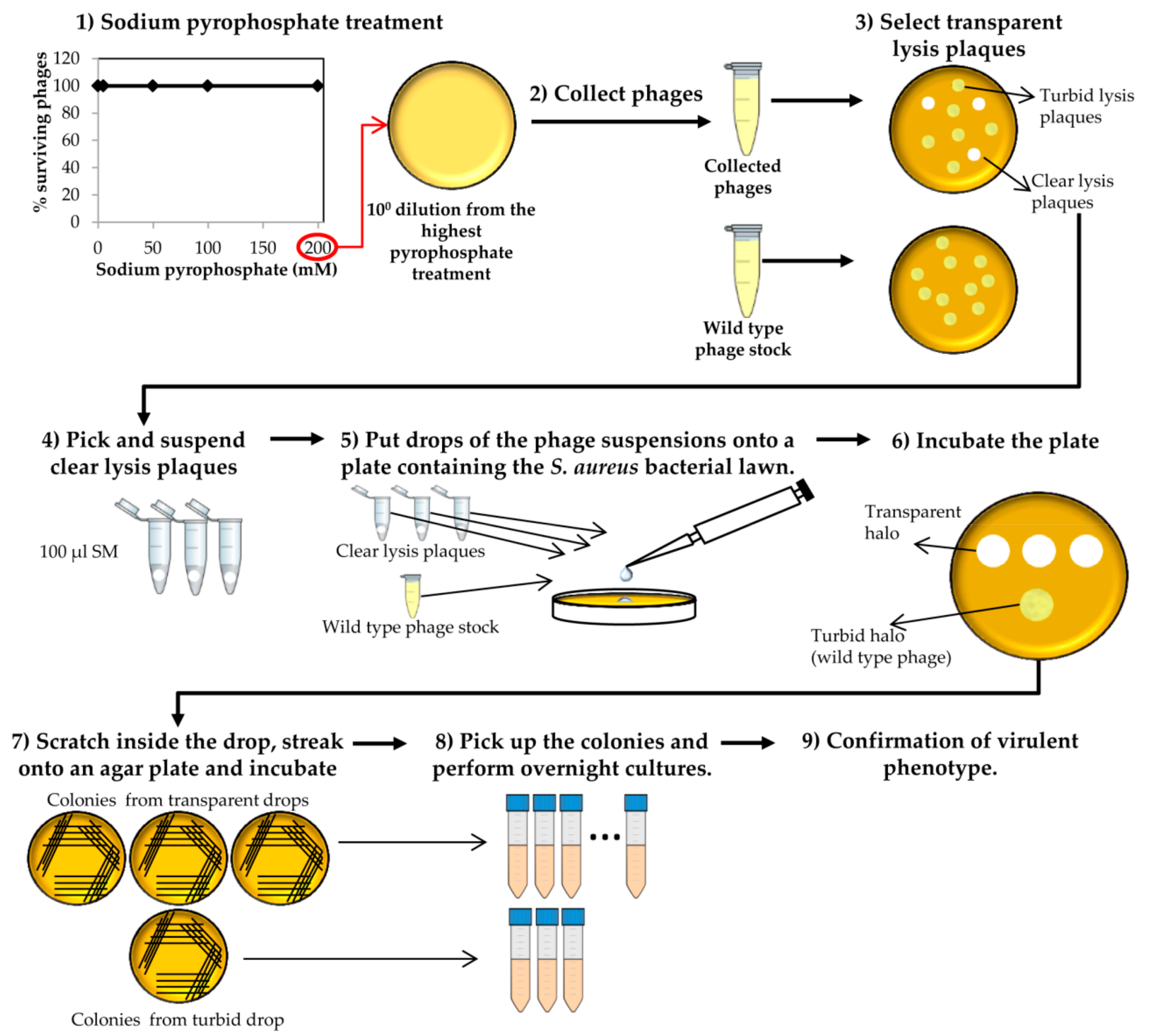
- Collect phages from the plate that resulted from the last round of treatment with the highest sodium pyrophosphate concentration, in which a plateau of 100% phage survival had been reached (Figure 3). To do this, add 1 mL of SM buffer to the plate corresponding to the phage dilution 10° and incubate 1 h with slight shaking (60 rpm) at room temperature. Afterwards, collect the liquid and centrifuge at 10,000 rpm for 10 min at 4 °C. Keep the supernatant containing the surviving phages.
- Make 1:10 dilutions of the phage suspensions in SM buffer and plate the appropriate dilutions using the double layer technique and S. aureus as host strain. For control purposes, make dilutions and plate the wild-type phage (not treated with any chelating agent). Let the medium solidify, and then incubate the plates at 37 °C for 18 h (Figure 3).At the same time, inoculate one isolated colony of S. aureus in TSB to obtain an o/n culture (see above).
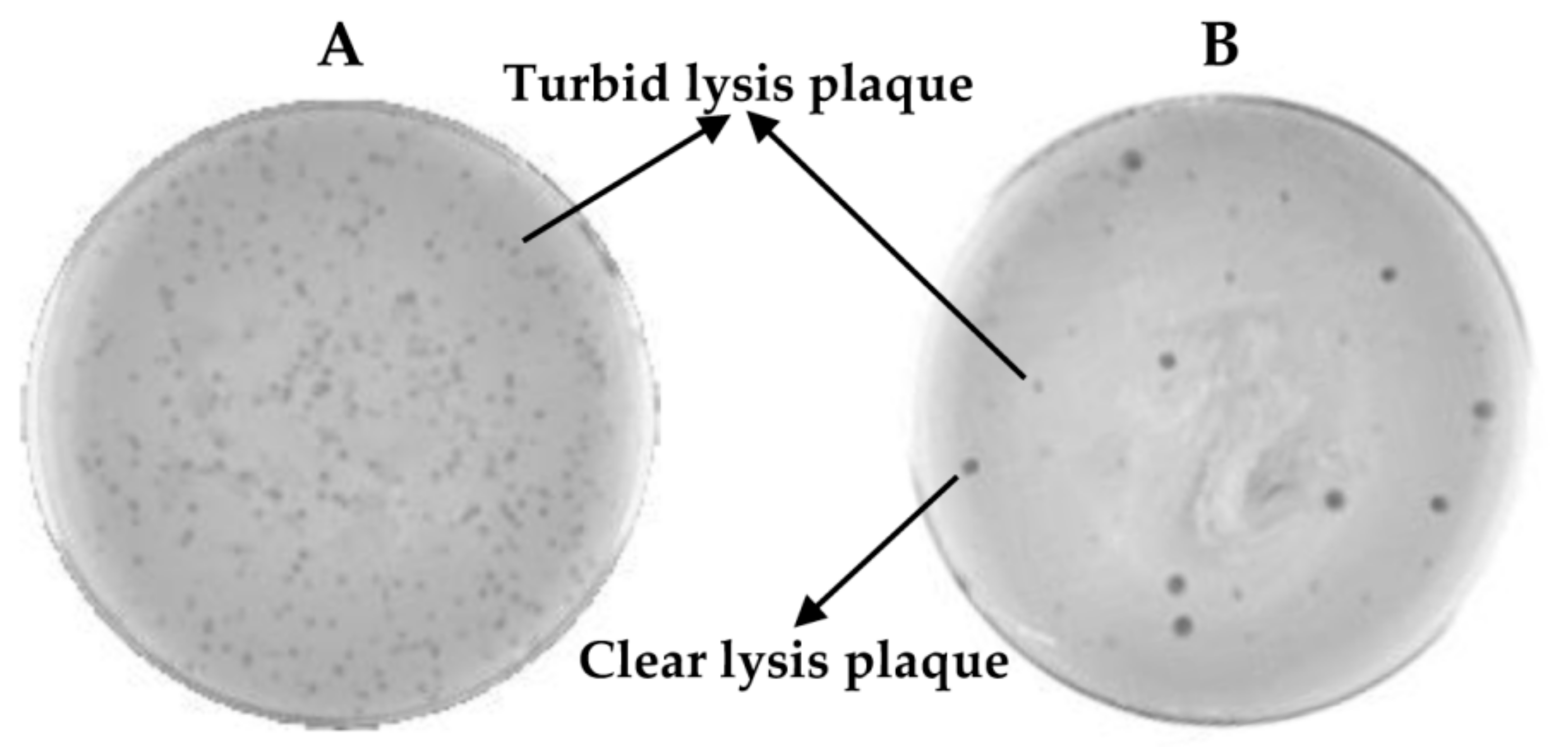
- Observe carefully the lysis plaques formed by the putative phage deletion mutants and compare with those formed by the wild-type phage. Pick up each putative “clear lysis plaque” (transparent phenotype) and suspend it in 100 µL of SM buffer (Figure 3).
CRITICAL STEP For some phages, it is necessary to incubate the plates for a longer period in order to get a more evident transparent phenotype.
- Take 100 µL of the S. aureus o/n culture and mix with 5 mL of TSA 0.7%. Pour the mixture onto a TSA 2% plate, allowing to harden. Place a 5 µL drop of each putative phage mutant suspension and a drop of the wild-type phage stock onto the plate surface. Keep the plates at room temperature until drops become dried and then incubate at 37 °C for 18 h.
- Scratch with a sterile tip inside the halos generated by the putative “clear lysis plaque” mutants (transparent halo) and streak onto a TSA 2% plate. Repeat the same with the halo generated by the wild-type phage (turbid halo). Incubate the plates at 37 °C for 18 h (Figure 3).
CRITICAL STEP The number of bacteria from the wild-type phage halo is expected to be high, while none or a low number of colonies should be present in the “clear lysis plaque” halo. Virulent phages with “clear lysis plaque” phenotype are not able to lysogenize their host; therefore, no cells are expected to survive after infection. By contrast, a high number of lysogenic cells (resistant to phage infection) will be generated inside the halo of the wild-type temperate phage.
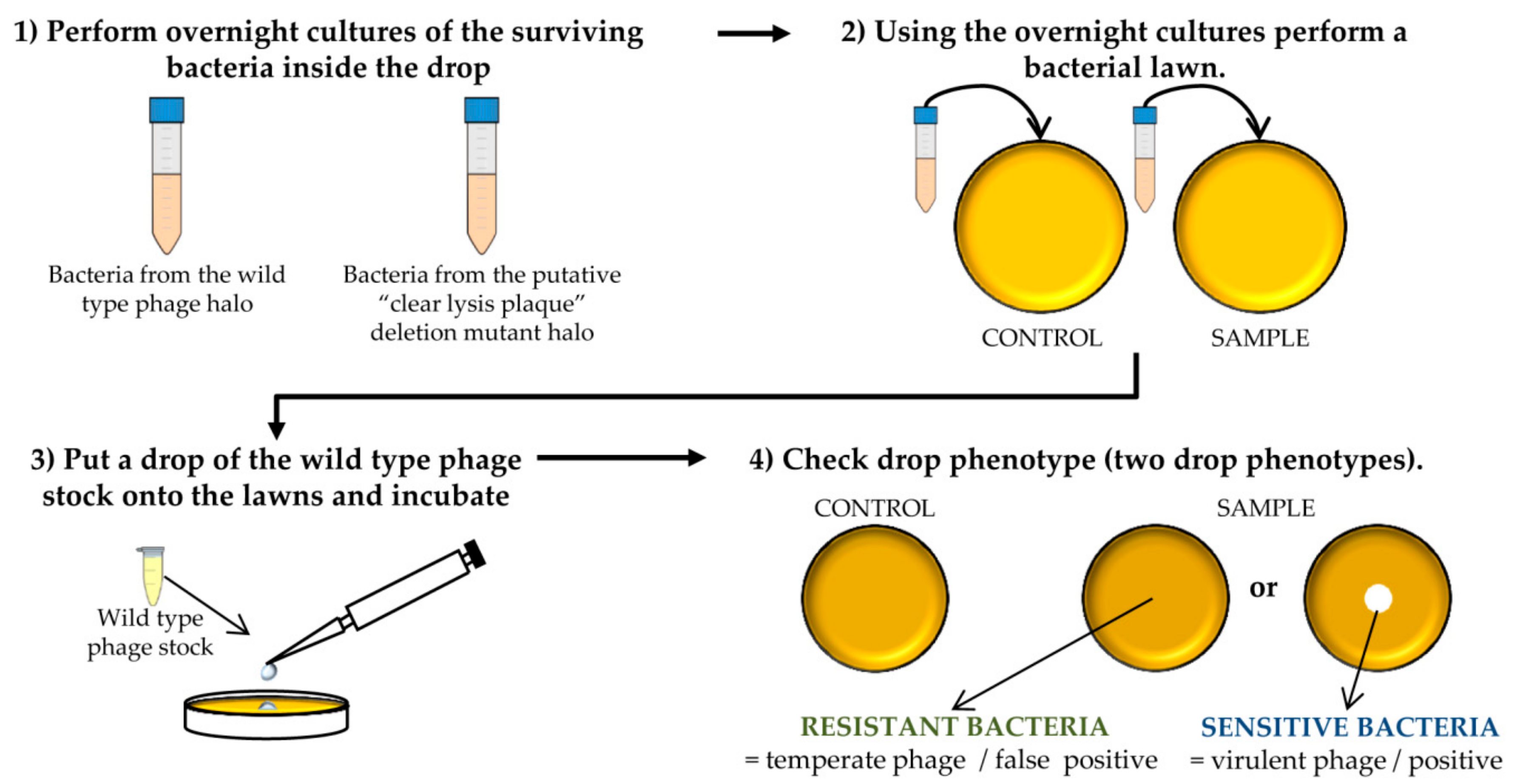
- Confirm the lytic phenotype. Pick up 3 isolated colonies from the plate that comes from the wild-type phage halo and all the colonies from the “clear lysis plaque” mutant halo. Introduce these colonies into sterile tubes containing 3 mL TSB and incubate with shaking (250 rpm) for 18 h at 37 °C (Figure 3 and Figure 4).
PAUSE STEP Prepare a stock of the colonies by adding 20% glycerol (final concentration) to the o/n cultures and freezing at −80 °C.
- Take 100 µL of each o/n culture and mix with 5 mL of TSA 0.7%. Pour the mixture onto TSA 2% plates and allow hardening. Place a 5 µL drop of the wild-type phage stock. Keep the plates at room temperature until it drops dry and then incubate them at 37 °C for 18 h (Figure 4).
CRITICAL STEP After the incubation, two phenotypes can be observed: the presence of a clear halo, which means that the strain is sensitive to the phage, or complete growth of the bacteria inside the halo which indicates resistance to the phage. The presence of a transparent halo on the surviving bacterial lawn is indicative that the putative “clear lysis plaque” phage mutant is a real virulent phage.
4. Expected Results
5. Reagents Setup
- Medium for bacterial growth: TSB; TSA (TSB supplemented with 2% w/v agar) and semisolid TSA medium (TSB supplemented with 0.7% w/v agar). TSB is prepared following the supplier’s recommendations. For the preparation of TSA, agar should be added prior to autoclaving. Sterilize by autoclaving at 121 °C for 15 min. TSA can be stored at room temperature for up to one month. Pour TSA 2% into 9 cm plates and allowed to solidify. Plates can be kept at 4 °C for up to one month. Allow TSA 0.7% to solidify at room temperature, and then it can be stored for up to one month. Before using, melt TSA-0.7% in a microwave and tempered to 50 °C in a water bath.
- SM buffer: 200 mM Tris HCl, 10 mM MgSO4, 10 mM CaCl2, and 100 mM NaCl in distilled water. Adjust pH to 7.5 with HCl. Sterilize by autoclaving at 121 °C 15 min. The buffer can be kept for up to one month at room temperature.
- PBS buffer: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 2 mM KH2PO4 in distilled water. Adjust pH to 7.4 with HCl. Sterilize by autoclaving at 121 °C 15 min. The buffer can be kept for up to one month at room temperature.
- CaCl2 solution: dissolve CaCl2 into distilled water at a final concentration of 1 M. Sterilize by autoclaving at 121 °C 15 min. The solution can be kept for up to one month at room temperature.
- MgSO4 solution: dissolve MgSO4 into distilled water at a final concentration of 1 M. Sterilize by autoclaving at 121 °C 15 min. The solution can be kept for up to one month at room temperature.
- PEG 8000 solution: dissolve PEG 8000 into distilled water to a final concentration of 30% (w/v). Sterilize by autoclaving at 121 °C 15 min. The solution can be kept for up to one month at room temperature.
- NaCl solution: dissolve NaCl into distilled water to a final concentration of 5 M. Sterilize by autoclaving at 121 °C 15 min. The solution can be kept for up to one month at room temperature.
- RNAse: dissolve RNAse into double distilled water to a final concentration of 5 mg/mL (w/v). Sterilize by filtering using a 0.2 µm PES filter. The stock can be kept at −20 °C for up to the expiration date that is provided by the supplier.
- Sodium pyrophosphate solutions: dissolve sodium pyrophosphate in Tris HCl 200 mM pH = 8 at a final concentration of 400 mM and then dilute to 10 mM, 20 mM, 40 mM 100 mM, 150 mM, 200 mM, 300 mM, and 350 mM. Sterilize by autoclaving at 121 °C 15 min. These solutions can be kept for up to one month at room temperature.
- Tris HCl 100 mM pH = 7.4: dissolve Tris HCl into distilled water to a final concentration of 100 mM. Adjust pH to 7.4 with HCl. Sterilize by autoclaving at 121 °C 15 min. The solution can be kept for up to one month at room temperature.
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Drake, J.W. Ultraviolet mutagenesis in bacteriophage T-4. I. Irradiation of extracellular phage particles. J. Bacteriol. 1966, 91, 1775–1780. [Google Scholar] [PubMed]
- Pjura, P.; Matsumura, M.; Baase, W.A.; Matthews, B.W. Development of an in vivo method to identify mutants of phage T4 lysozyme of enhanced thermostability. Protein Sci. 1993, 2, 2217–2225. [Google Scholar] [CrossRef] [PubMed]
- Marinelli, L.J.; Hatfull, G.F.; Piuri, M. Recombineering: A powerful tool for modification of bacteriophage genomes. Bacteriophage 2012, 2, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Marinelli, L.J.; Piuri, M.; Swigonova, Z.; Balachandran, A.; Oldfield, L.M.; van Kessel, J.C.; Hatfull, G.F. BRED: A simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS ONE 2008, 3, e3957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martel, B.; Moineau, S. CRISPR-Cas: An efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res. 2014, 42, 9504–9513. [Google Scholar] [CrossRef] [PubMed]
- Bari, S.M.N.; Walker, F.C.; Cater, K.; Aslan, B.; Hatoum-Aslan, A. Strategies for editing virulent Staphylococcal phages using CRISPR-Cas10. ACS Synth. Biol. 2017, 6, 2316–2325. [Google Scholar] [CrossRef] [PubMed]
- Bassel, A.; Shaw, M.; Campbell, L.L. Dissociation by chelating agents and substructure of the thermophilic bacteriophage TP84. J. Virol. 1971, 7, 663–672. [Google Scholar] [PubMed]
- Yamamoto, N.; Fraser, D.; Mahler, H.R. Chelating agent shock of bacteriophage T5. J. Virol. 1968, 2, 944–950. [Google Scholar] [PubMed]
- Parkinson, J.S.; Huskey, R.J. Deletion mutants of bacteriophage lambda. I. Isolation and initial characterization. J. Mol. Biol. 1971, 56, 369–384. [Google Scholar] [CrossRef]
- Ladero, V.; García, P.; Bascarán, V.; Herrero, M.; Álvarez, M.A.; Suárez, J.E. Identification of the repressor-encoding gene of the Lactobacillus bacteriophage A2. J. Bacteriol. 1998, 180, 3474–3476. [Google Scholar] [PubMed]
- García, P.; Madera, C.; Martínez, B.; Rodríguez, A. Biocontrol of Staphylococcus aureus in curd manufacturing processes using bacteriophages. Int. Dairy J. 2007, 17, 1232–1239. [Google Scholar] [CrossRef]
- García, P.; Martínez, B.; Obeso, J.M.; Lavigne, R.; Lurz, R.; Rodríguez, A. Functional genomic analysis of two Staphylococcus aureus phages isolated from the dairy environment. Appl. Environ. Microbiol. 2009, 75, 7663–7673. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.W.; Charlesworth, B.; Charlesworth, D.; Crow, J.F. Rates of spontaneous mutation. Genetics 1998, 148, 1667–1686. [Google Scholar] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gutiérrez, D.; Fernández, L.; Rodríguez, A.; García, P. Practical Method for Isolation of Phage Deletion Mutants. Methods Protoc. 2018, 1, 6. https://doi.org/10.3390/mps1010006
Gutiérrez D, Fernández L, Rodríguez A, García P. Practical Method for Isolation of Phage Deletion Mutants. Methods and Protocols. 2018; 1(1):6. https://doi.org/10.3390/mps1010006
Chicago/Turabian StyleGutiérrez, Diana, Lucía Fernández, Ana Rodríguez, and Pilar García. 2018. "Practical Method for Isolation of Phage Deletion Mutants" Methods and Protocols 1, no. 1: 6. https://doi.org/10.3390/mps1010006






