Protein-Mediated Biotemplating on the Nanoscale
Abstract
:1. Introduction
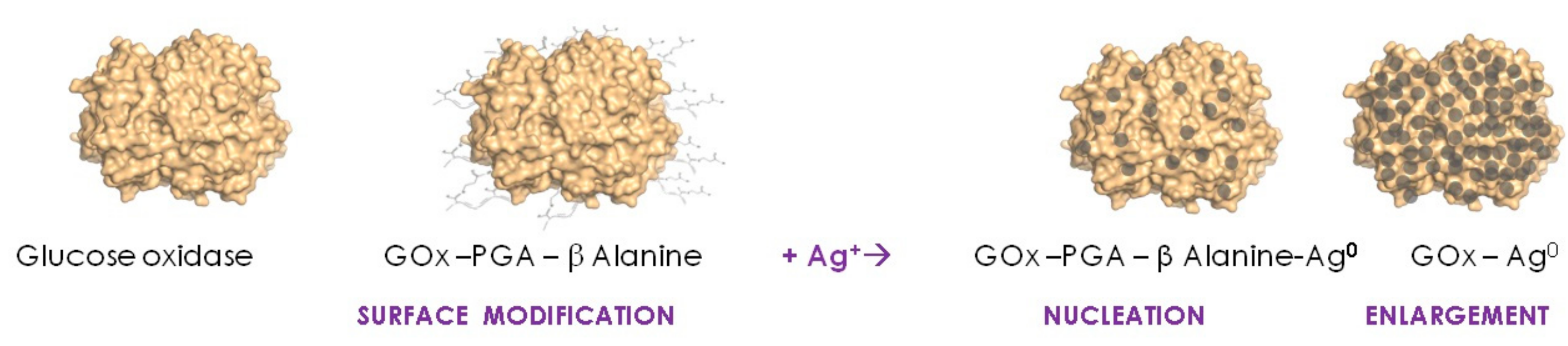
2. Single Soluble Protein Molecules Biotemplating
3. Protein “Cages” as Biotemplates
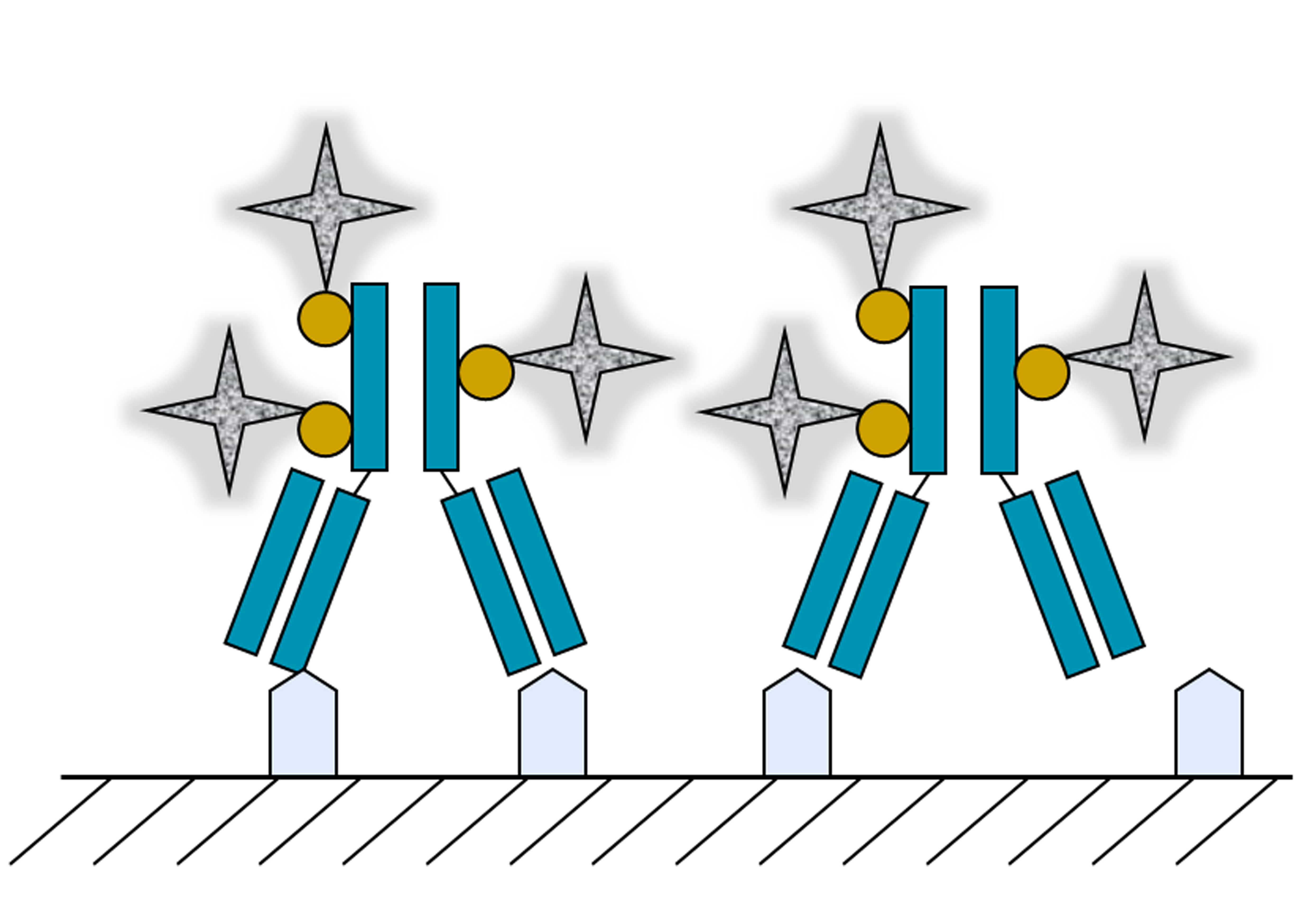
4. Two-Dimensional Protein Arrays as Biotemplates
5. Protein-Made Fibers and Tubes as Biotemplates
6. Viral Envelopes as Biotemplates
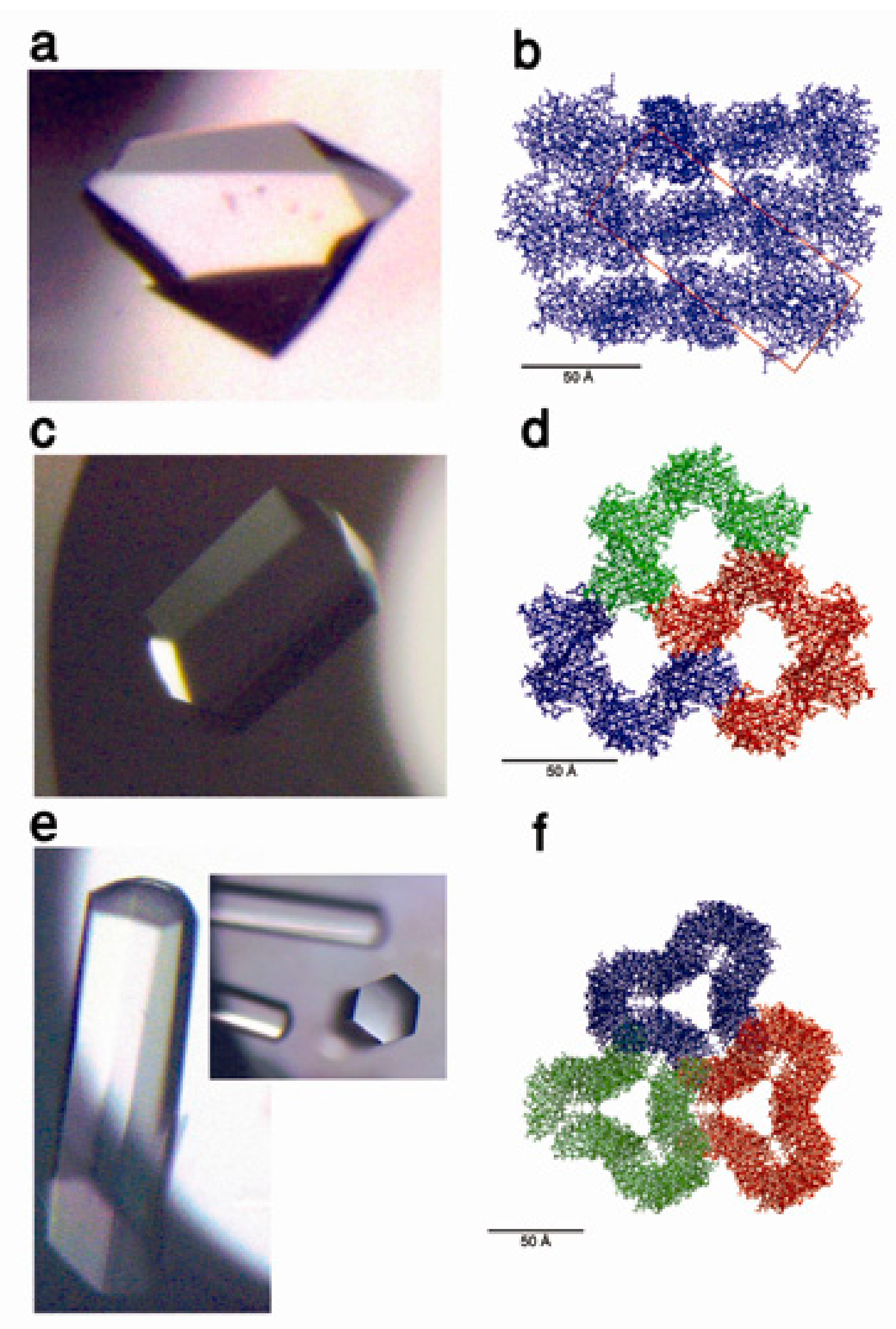
7. Protein Crystal-Mediated Biotemplating
8. Protein-Mediated Biotemplating by Cell Fragments
9. Conclusions and Future Perspectives
Conflicts of Interest
References
- Lagziel-Simis, S.; Cohen-Hadar, N.; Moscovich-Dagan, H.; Wine, Y.; Freeman, A. Protein-mediated nanoscale biotemplating. Curr. Opin. Biotechol. 2006, 17, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Sargent, E.H.; Kelley, S.O. Biotemplated nanostructures: Directed assembly of electronic and optical materials using nanoscale complementarity. J. Mater. Chem. 2008, 18, 954–964. [Google Scholar] [CrossRef]
- Behrens, S.S. Synthesis of inorganic nanomaterials mediated by protein assemblies. J. Mater. Chem. 2008, 18, 3788–3798. [Google Scholar] [CrossRef]
- Galloway, J.M.; Stainland, S.S. Protein and peptide biotemplated metal and metal oxide nanoparticles and their patterning onto surfaces. J. Mater. Chem. 2012, 22, 12423–12434. [Google Scholar] [CrossRef]
- Yang, W.; Guo, W.; Chang, J.; Zhang, B. Protein/peptide-terminated biomimetic synthesis of inorganic nanoparticles for biomedical applications. J. Mater. Chem B 2017, 5, 401–417. [Google Scholar] [CrossRef]
- Zan, G.; Wu, Q. Biomimetic and bioinspired synthesis of nanomaterials/nanostructures. Adv. Mater. 2016, 28, 2099–2147. [Google Scholar] [CrossRef] [PubMed]
- Dagan-Moscovich, H.; Cohen-Hadar, N.; Ophir, C.; Rishpon, J.; Shacham-Diamand, Y.; Freeman, A. Nanowiring of the catalytic site of novel molecular enzyme–metal hybrids to electrodes. J. Phys. Chem. C 2007, 111, 5766–5769. [Google Scholar] [CrossRef]
- Vernick, S.; Dagan-Moscovich, H.; Porat-Ophir, C.; Rishpon, J.; Freeman, A.; Shacham-Diamand, Y. Directed metallization of single-enzyme molecules with preserved enzymatic activity. IEEE Trans. Nanotech. 2009, 8, 95–99. [Google Scholar] [CrossRef]
- Mor, G.; Vernick, S.; Dagan-Moscovich, H.; Dror, Y.; Freeman, A. Novel biologically active silver-avidin hybrids. J. Phys. Chem. C 2011, 115, 22695–22700. [Google Scholar] [CrossRef]
- Freeman, A.; Dror, Y.; Ophir-Porat, C.; Hadar, N.; Shacham-Diamand, Y. Silver coated biologically active protein hybrids: Antimicrobial applications. Appl. Mechan. Mater. 2015, 749, 453–456. [Google Scholar] [CrossRef]
- Veelders, M.; Essen, L.O. Complex gadolinium-oxo clusters formed along concave protein surfaces. ChemBioChem 2012, 13, 2187–2190. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Chen, J.; Gao, F.; Li, W.; Xu, X.; Pandoli, O.; Yang, H.; Ji, J.; Cui, D. A multifunctional ribonuclease-A-conjugated CdTe quantum dot cluster nanosysytem for synchronous cancer imaging and therapy. Small 2010, 6, 2367–2373. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Kong, Y.; Ruan, J.; Wang, K.; Gao, F.; Cui, D. Protein-induced structural evolution of silver sulfide at the nanoscale: From hollow particles to solid spheres. Nanoscale 2012, 4, 4455–4458. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, L.; Huang, Y.; Jia, N. Albumin-mediated platinum nanocrystals for in vivo enhanced computed tomography imaging. J. Mater. Chem. B 2017, 5, 3498–3510. [Google Scholar] [CrossRef]
- Wang, J.; Yang, S.; Li, C.; Miao, Y.; Zhu, L.; Mao, C. Nucleation and assembly of silica into protein-based nanocomposites as effective anticancer drug carriers using self-assembled silk protein nanostructures as biotemplates. ACS Appl. Mater. Interfaces 2017, in press. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Zhang, Y.; Muneyama, J.C.; Wu, T.; Yang, Z.; Chen, H.; Qu, W.; Xiao, J. Tuning the hierarchical nanostructure of hematite mesocrystals via collagen-templated biominaralization. J. Mater. Chem. B 2017, 5, 1423–1429. [Google Scholar] [CrossRef]
- Zhang, L.; Knez, M. Spherical nanoscale templates for biomedical applications: A review on ferritin. J. Nanosci. Lett. 2012, 2, 1–13. [Google Scholar]
- Jutz, G.; Rijn, P.; Miranda, B.S.; Boker, A. Ferritin: A versatile building block for bionanotechnology. Chem. Rev. 2015, 115, 1653–1701. [Google Scholar] [CrossRef] [PubMed]
- Zang, J.; Chen, H.; Zhao, G.; Wang, F.; Ren, F. Ferritin cage for encapsulation and delivery of bioactive nutrients: From structure, property to applications. Crit. Rev. Food Sci. Nutr. 2017, 57, 3673–3683. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, S.; Xu, C.; Zhao, G. Engineering protein interfaces yields ferritin disassembly and reassembly under benign experimental conditions. Chem. Commun. 2016, 52, 7402–7405. [Google Scholar] [CrossRef] [PubMed]
- Eloi, J.C.; Ward Jones, S.E.; Poor, V.; Okuda, M.; Gwyther, J.; Schwarzacher, W. Electrochemically triggered adsorption of biotemplated nanoparticles on self-assembled organometallic diblock copolymer thin films. Adv. Funct. Mater. 2012, 22, 3273–3278. [Google Scholar] [CrossRef]
- Sano, K.I.; Yoshii, S.; Yamashita, I.; Shiba, K. In aqua structuralization of a three-dimensional configuration using biomolecules. Nano Let. 2007, 7, 3200–3202. [Google Scholar] [CrossRef] [PubMed]
- Iwahori, K.; Enomoto, T.; Furusho, H.; Miura, A.; Nishio, K.; Mishima, Y.; Yamashita, I. Cadmium sulfide nanoparticle synthesis in Dps protein from Listeria innocua. Chem. Mater. 2007, 19, 3105–3111. [Google Scholar]
- Okuda, M.; Suzumoto, Y.; Iwahori, K.; Kang, S.; Uchida, M.; Douglas, T.; Yamashita, I. Biotemplated CdSe nanoparticle synthesis in a cage shaped protein Listeria-Dps and their two dimensional ordered array self-assembly. Chem. Commun. 2010, 46, 8797–8799. [Google Scholar] [CrossRef] [PubMed]
- Huggins, K.N.L.; Schoen, A.P.; Arunagirinathan, M.A.; Heishorn, S.C. Multi-site funcionalization of protein scaffolds for bimetallic nanoparticle templating. Adv. Funct. Mater. 2014, 24, 7737–7744. [Google Scholar] [CrossRef]
- Voet, A.R.D.; Noguchi, H.; Addy, C.; Zhang, K.Y.J.; Tame, J.R.H. Biomineralization of a cadmium chloride nanocrystal by a designed symmetrical protein. Angew. Chem. Int. Ed. 2015, 54, 9857–9860. [Google Scholar] [CrossRef] [PubMed]
- Behrens, S.; Heyman, A.; Maul, R.; Essig, S.; Steigenwald, S.; Quintilla, A.; Wenzel, W.; Burck, J.; Degany, O.; Shoseyov, O. Constrained synthesis and organization of catalytically active metal nanoparticles by self-assembled protein templates. Adv. Mater. 2009, 21, 3515–3519. [Google Scholar] [CrossRef]
- Zhou, Z.; Bedwell, G.J.; Li, R.; Prevelige, P.E.; Gupta, A. Formation mechanism of chalcogenide nanocrystals confined inside genetically engineered virus-like particles. Sci. Rep. 2014, 4, 3832. [Google Scholar] [CrossRef] [PubMed]
- Maity, B.; Abe, S.; Ueno, T. Observation of gold sub-nanocluster nucleation within a crystalline protein cage. Nat. Comm. 2017. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Bedwell, J.; Li, R.; Pachoudhury, S.; Prevelige, P.E.; Gupta, A. Pathways for gold nucleation over protein cages. Langmuir 2017, 33, 5925–5931. [Google Scholar] [CrossRef] [PubMed]
- Slytr, U.B.; Messner, P.; Pum, D.; Sara, M. Crystalline bacterial cell surface layers (S layers): From supramolecular cell structure to biomimetics and nanotechnology. Angew. Chem. Int. Ed. 1999, 38, 1034–1054. [Google Scholar] [CrossRef]
- Mark, S.S.; Bergkvist, M.; Bhatnager, P.; Welch, C.; Goodyear, L.A.; Yang, X.; Angert, E.R.; Batt, C.A. Thin film processing using S-layer proteins: Biotemplating assembly of colloidal gold etch mask for fabrication of silicon nanopillar arrays. Colloid. Surf. B Biointerface 2007, 57, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Sierra-Sastre, Y.; Choi, S.; Picraux, S.T.; Batt, C.A. Vertical growth of Ge nanowires from biotemplated Au nanoparticle catalysts. J. Am. Chem. Soc. 2008, 130, 10488–10489. [Google Scholar] [CrossRef] [PubMed]
- Queitsch, U.; Hamann, C.; Schaffel, F.; Rellinghaus, B.; Schultz, L.; Bluher, A.; Mertig, M. Toward dense biotemplated magnetic nanoparticle array: Probing the particle–template interaction. J. Phys. Chem. C 2009, 113, 10471–10476. [Google Scholar] [CrossRef]
- Varga, M.; Roedel, G.; Pompe, W. Engineering of self-assembling proteins for biosensing applications. In Proceedings of the 11th IEEE International Conference on Nanotechnology, Portland, OR, USA, 15–18 August 2011; pp. 1602–1606. [Google Scholar]
- Valero, E.; Martin, M.; Galvez, N.; Sanchez, P.; Raff, J.; Merroun, M.L.; Dominguez-Vera, J.M. Nanopatterning of magnetic CrNi Prussian blue nanoparticles using a bacterial S-layer as biotemplate. Inorg. Chem. 2015, 54, 6758–6762. [Google Scholar] [CrossRef] [PubMed]
- Shindel, M.M.; Mohraz, A.; Mumm, D.R.; Wang, S.W. Modulating colloidal adsorption on a two- dimensional protein crystal. Langmuir 2009, 25, 1038–1046. [Google Scholar] [CrossRef] [PubMed]
- Shindel, M.M.; Mumm, D.R.; Wang, S.W. Biotemplating of metallic nanoparticle arrays through site specific electrostatic adsorption on strepravidin crystals. Langmuir 2010, 26, 11103–11112. [Google Scholar] [CrossRef] [PubMed]
- Shindel, M.M.; Mumm, D.R.; Wang, S.W. Manipulating energy landscapes to tune ordering in biotemplated nanoparticle arrays. Langmuir 2011, 27, 7768–7775. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.D.; Liu, H.F.; Li, C.Y.; Song, Y.Y. Biotemplated synthesis of Au nanoparticles-TiO2 nanotube junctions for enhanced direct electrochemistry of heme proteins. Chem. Commun. 2013, 49, 774–776. [Google Scholar] [CrossRef] [PubMed]
- Galloway, M.J.; Bramble, J.P.; Rawlings, A.E.; Burnell, G.; Evans, S.D.; Staniland, S.S. Biotemplated magnetic nanoparticle arrays. Small 2012, 8, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Bird, S.M.; Galloway, J.M.; Rawlings, A.E.; Bramble, J.P.; Staniland, S.S. Taking a hard line with biotemplating: Cobalt-doped magnetite magnetic nanoparticles arrays. Nanoscale 2015, 7, 7340–7351. [Google Scholar] [CrossRef] [PubMed]
- Colby, R.; Hulleman, J.; Padalkar, S.; Rochet, J.C.; Stanciu, L.A. Biotemplated synthesis of metallic nanoparticle chains on a α-synuclein fiber scaffold. J. Nanosci. Nanotech. 2008, 8, 973–978. [Google Scholar]
- Malisauskas, M.; Meskys, R.; Morozova-Roche, L.A. Ultrathin silver nanowires produced by amyloid biotemplating. Biotechnol. Prog. 2008, 24, 1166–1170. [Google Scholar] [CrossRef] [PubMed]
- Slocik, J.M.; Kim, S.N.; Whitehead, T.A.; Clark, D.S.; Naik, R.R. Biotemplated metal nanowires using hyperthermophilic protein filaments. Small 2009, 18, 2038–2042. [Google Scholar] [CrossRef] [PubMed]
- Ionov, L.; Bocharova, V.; Diez, S. Biotemplated synthesis of stimuli-responsive nanopatterned polymer brushes on microtubules. Soft Matter 2009, 5, 67–71. [Google Scholar] [CrossRef]
- Juarez, J.; Cambon, A.; Goy-Lopez, S.; Topete, A.; Taboada, P.; Mosquera, V. Obtention of metallic nanowires by protein biotemplating and their catalytic application. J. Phys. Chem. Lett. 2010, 1, 2680–2687. [Google Scholar] [CrossRef]
- Leroux, F.; Gysemans, M.; Bals, S.; Batenburg, K.J.; Snauwaert, J.; Verbiest, T.; Hasendonck, C.V.; Tendeloo, G.V. Three-dimensional characterization of helical silver nanochains mediated by protein assemblies. Adv. Mater. 2010, 22, 2193–2197. [Google Scholar] [CrossRef] [PubMed]
- Juarez, J.; Cambon, A.; Topete, A.; Taboda, P.; Mosquera, V. One-dimensional magnetic nanowires obtained by protein fibril biotemplating. Chem. Eur. J. 2011, 17, 7366–7373. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, N.; Ostermann, K.; Rodel, G. Calcium dependent formation of tubular assemblies by recombinant S-layer protein in vivo and in vitro. Nanotechnology 2011, 22, 095601. [Google Scholar] [CrossRef] [PubMed]
- Padalkar, S.; Schroeder, K.; Won, Y.H.; Jang, H.S.; Stanciu, L. Biotemplated silica and titania nanowires: Synthesis, characterization and potential applications. J. Nanosci. Nanotech. 2012, 12, 227–235. [Google Scholar] [CrossRef]
- Zhou, X.; Li, R.; Dai, B.; Zhang, Y.; Xu, P.; Zhang, Y. The fabrication and electrical characterization of protein fibril-templated one-dimensional palladium nanaostructures. Eur. Polym. J. 2013, 49, 1957–1963. [Google Scholar] [CrossRef]
- Batzli, K.M.; Love, B.J. Formation of platinum-coated templates of insulin nanowires used in reducing 4-nitrophenol. Mater. Sci. Eng. C 2015, 48, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Glover, D.J.; Giger, L.; Kim, S.S.; Naik, R.R.; Clark, D.S. Geometrical assembly of ultrastable protein templates for nanomaterials. Nat. Commun. 2016. [Google Scholar] [CrossRef] [PubMed]
- Young, M.; Willits, D.; Uchida, M.; Douglas, T. Plant viruses as biotemplates for materials and their use in nanotechnology. Ann. Rev. Phytopathol. 2008, 46, 361–384. [Google Scholar] [CrossRef] [PubMed]
- Sanghamitra, N.J.; Inaba, H.; Kitagawa, S.; Ueno, T. Inorganic design of protein assemblies as supramolecular platforms. J. Inorg. Polym. 2013, 23, 50–60. [Google Scholar] [CrossRef]
- Love, A.J.; Makarov, V.; Yaminsky, I.; Kalinina, N.O.; Taliansky, M.E. The use of tobacco mosaic virus and cowpea mosaic virus for the production of novel metal nanomaterials. Virology 2014, 449, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Steele, J.F.C.; Peyret, H.; Saunder, K.; Castells-Graells, R.; Marsian, J.; Meshcheriakova, Y.; Lomonossoff, G.P. Synthetic plant virology for nanobiotechnology and nanomedicine. WIREs Nanomed. Nanobiotechnol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, R.; Muraoka, M.; Seki, I.; Tabata, H.; Yamashita, I. Synthesis of CoPt and FePt3 nanowires using the central channel of TMV as a biotemplate. Chem. Mater. 2007, 19, 2389–2391. [Google Scholar] [CrossRef]
- Neltner, B.; Peddie, B.; Xu, A.; Donelen, W.; Durand, K.; Yun, D.S.; Speakman, S.; Peterson, A.; Belcher, A.M. Production of hydrogen using nanocrystalline protein-templated catalysts on M13 phage. ACS Nano 2010, 4, 3227–3235. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.S.; Kim, S.M.; Lee, S.Y.; Stach, E.A.; Culver, J.N.; Harris, M.T. Biotemplated aqueous-phase palladium crystallization in the absence of external reducing agents. Nano Lett. 2010, 10, 3863–3867. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Seki, M.; Tabata, H.; Watanabe, Y.; Yashimata, I. Fabrication of aligned magnetic nanoparticles using tobamoviruses. Nano Lett. 2010, 10, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Nuraje, N.; Dang, X.; Allen, M.A.; Lei, Y.; Belcher, A.M. Biotemplated synthesis of perovskite nanomaterials for solar energy conversion. Adv. Mater. 2012, 24, 2885–2889. [Google Scholar] [CrossRef] [PubMed]
- Alloyeau, D.; Stephanidis, B.; Zhao, X.; Larquet, E.; Boisset, N.; Ricolleau, C. Biotemplated synthesis of metallic nanoclusters organized in tunable two-dimensional superlattices. J. Phys. Chem. C 2011, 115, 20926–20930. [Google Scholar] [CrossRef]
- Zhou, J.C.; Soto, C.M.; Chen, M.S.; Bruckman, M.A.; Moore, M.H.; Barry, E.; Ratna, B.R.; Pehrsson, P.E.; Spies, B.R.; Confer, T.S. Biotemplating rod-like viruses for the synthesis of copper nanorods and nanowires. J. Nanobiotech. 2012, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Vera-robles, L.I.; Nhieu, G.V.T.; Laberty-Robert, C.; Livage, J.; Sanchez, C. Flexible electroactive nanomaterials biotemplated with versatile M13 phage platforms. Adv. Eng. Mater. 2013, 15, 954–960. [Google Scholar] [CrossRef]
- Cung, K.; Han, B.J.; Nguyen, T.D.; Mao, S.; Yeh, Y.W.; Xu, S.; Naik, R.R.; Poirier, G.; Purohit, P.K.; McAlpine, M.C. Biotemplated synthesis of PZT nanowires. Nano Lett. 2013, 13, 6197–6202. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.; Manivannan, S.; Kang, I.; Lee, S.W.; Kim, K. Gold dendrites co-deposited with M13 virus as a biosensor platform for nitrite ions. Biosens. Bioelectron. 2017, 94, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Adigum, O.; Retlaff-Roberts, E.L.; Novikova, G.; Wang, L.; Kim, B.S.; Ilavsky, J.; Miller, J.T.; Loesch-Fries, L.S.; Harris, M.T. BSMV as a biotemplate for palladium nanomaterial synthesis. Langmuir 2017, 33, 1716–1724. [Google Scholar] [CrossRef] [PubMed]
- Carreno-Fuentes, L.; Bahena, D.; Palomares, L.A.; Ramirez, O.T.; Jose-Yacaman, M.; Plascencia-Villa, G. Molecular docking and aberration corrected STEM of palladium nanoparticles on viral templates. Metals 2016, 6, 200. [Google Scholar] [CrossRef]
- Cohen-Hadar, N.; Wine, Y.; Lagziel-Simis, S.; Moscovich-Dagan, H.; Dror, Y.; Frolow, F.; Freeman, A. Protein crystal-mediated biotemplating. J. Porous Med. 2009, 12, 213–220. [Google Scholar] [CrossRef]
- Uraoka, Y.; Uenuma, M.; Ishikawa, Y.; Kumagai, S.; Tomita, S.; Watanabe, H.; Yamashita, I. Biotemplates and their application to electronic devices. In Intelligent Nanosystems for Energy, Information and Biological Techniques; Sone, J., Tsuji, S., Eds.; Springer: Berlin, Germany, 2016; pp. 119–143. [Google Scholar]
- Dotan, N.; Cohen, N.; Kalid, O.; Freeman, A. Supramolecular assemblies made of biological macromolecules. In Nano-Surface Chemistry; Rosoff, M., Ed.; Marcel Dekker: New York, NY, USA, 2001; pp. 461–472. [Google Scholar]
- Rica, R.D.L.; Matsui, H. Applications of peptide and protein based materials in bionanotechnology. Chem. Soc. Rev. 2010, 39, 3499–3509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sotiropoulou, S.; Sierra-Sastre, Y.; Mark, S.S.; Batt, C.A. Biotemplated nanostructured materials. Chem. Mater. 2008, 20, 821–834. [Google Scholar] [CrossRef]
- Howorka, S. Rationally engineering natural protein assemblies in nanobiotechnology. Curr. Opin. Biotechnol. 2011, 22, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Hadar, N.; Wine, Y.; Nachlieli, E.; Huppert, D.; Gutman, M.; Frolow, F.; Freeman, A. Monitoring of the stability of crosslinked protein crystals biotemplates: A feasibility study. Biotechnol. Bioeng. 2006, 94, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- Yan, E.K.; Cao, H.L.; Zhang, C.Y.; Lu, Q.Q.; Ye, Y.J.; He, J.; Huang, L.J.; Yin, D.C. Cross-linked protein crystals by glutaraldehyde and their applications. RSC Adv. 2015, 5, 26163–26174. [Google Scholar] [CrossRef]
- Wine, Y.; Cohen-Hadar, N.; Freeman, A.; Frolow, F. Elucidation of the mechanism and end products of glutaraldehyde crosslinking reaction by X-ray structure analysis. Biotechnol. Bioeng. 2007, 98, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Buch, M.; Wine, Y.; Dror, Y.; Rosenhak, S.; Lebendiker, M.; Giordano, R.; Leal, R.M.F.; Popov, A.N.; Freeman, A.; Frolow, F. Protein products obtained by site-preferred partial crosslinking in protein crystals and “liberated” by redissolution. Biotechnol. Bioeng. 2014, 111, 1296–1303. [Google Scholar] [CrossRef] [PubMed]
- Dotan, N.; Arad, D.; Frolow, F.; Freeman, A. Self-assembly of a tetrahedral lectin into predesigned diamond-like protein crystals. Angew. Chem. Int. Ed. 1999, 38, 2363–2366. [Google Scholar] [CrossRef]
- Wine, Y.; Cohen-Hadar, N.; Lamed, R.; Freeman, A.; Frolow, F. Modification of protein crystal packing by systematic mutations of surface residues: Implications on biotemplating and crystal porosity. Biotechnol. Bioeng. 2009, 104, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Wine, Y.; Cohen-Hadar, N.; Lagziel-Simis, S.; Dror, Y.; Frolow, F.; Freeman, A. Adjustment of protein crystal porosity for biotemplating: Chemical and protein engineering tools. In Porous Media and Its Applications in Science, Engineering and Industry; Vafai, K., Ed.; American Institute of Physics: College Park, MD, USA, 2010; pp. 198–203. [Google Scholar]
- Cohen-Hadar, N.; Lagziel-Simis, S.; Wine, Y.; Frolow, F.; Freeman, A. Re-structuring protein crystals porosity for biotemplating by chemical modification of lysine residues. Biotechnol. Bioeng. 2011, 108, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Kondow, T.; Mafune, F. Self-assembly of gold nanoparticles in protein crystal. Chem. Phys. Lett. 2011, 504, 175–179. [Google Scholar] [CrossRef]
- Liang, M.; Wang, L.; Su, R.; Qi, W.; Wang, M.; Yu, Y.; He, Z. Synthesis of silver nanoparticles within crosslinked lysozyme crystals as recyclable catalysts for 4-nitrophenol reduction. Catal. Sci. Technol. 2013, 3, 1910–1914. [Google Scholar] [CrossRef]
- Liu, M.; Wang, L.; Huang, R.; Yu, Y.; Su, R.; Qi, W.; He, Z. Superior catalytic performance of gold nanoparticles within small cross-linked lysozyme crystals. Langmuir 2016, 32, 10895–10904. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Mathew, B.; Mao, C. Biotemplated synthesis of hollow double-layered core/shell titania/silica nanotubes under ambient conditions. Small 2012, 8, 3691–3697. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Nimmo, S.L.; Cao, B.; Mao, C. Oxide formation on biological nanostructures via a structure-directing agent: Towards an understanding of precise structural transcription. Chem. Sci. 2012, 3, 2639–2645. [Google Scholar] [CrossRef] [PubMed]
- Gopinathan, P.; Ashok, A.M.; Selvakumar, R. Bacterial flagella as biotemplate for the synthesis of silver nanoparticle impregnated bionanomaterial. Appl. Surf. Sci. 2013, 276, 717–722. [Google Scholar] [CrossRef]



© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Freeman, A. Protein-Mediated Biotemplating on the Nanoscale. Biomimetics 2017, 2, 14. https://doi.org/10.3390/biomimetics2030014
Freeman A. Protein-Mediated Biotemplating on the Nanoscale. Biomimetics. 2017; 2(3):14. https://doi.org/10.3390/biomimetics2030014
Chicago/Turabian StyleFreeman, Amihay. 2017. "Protein-Mediated Biotemplating on the Nanoscale" Biomimetics 2, no. 3: 14. https://doi.org/10.3390/biomimetics2030014




