GPU Accelerated Image Processing in CCD-Based Neutron Imaging
Abstract
:1. Introduction
2. Materials and Methods
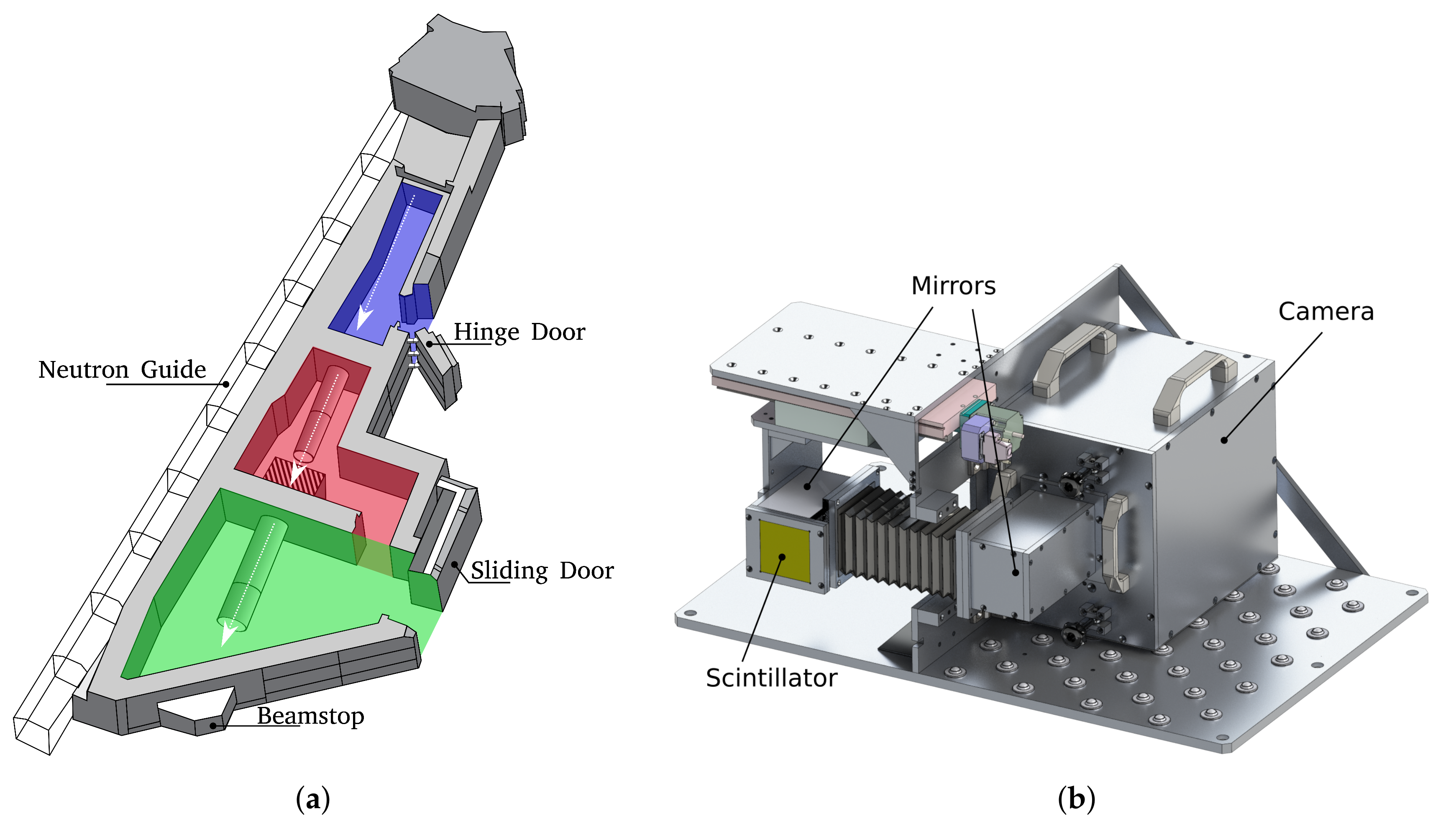
2.1. Setup
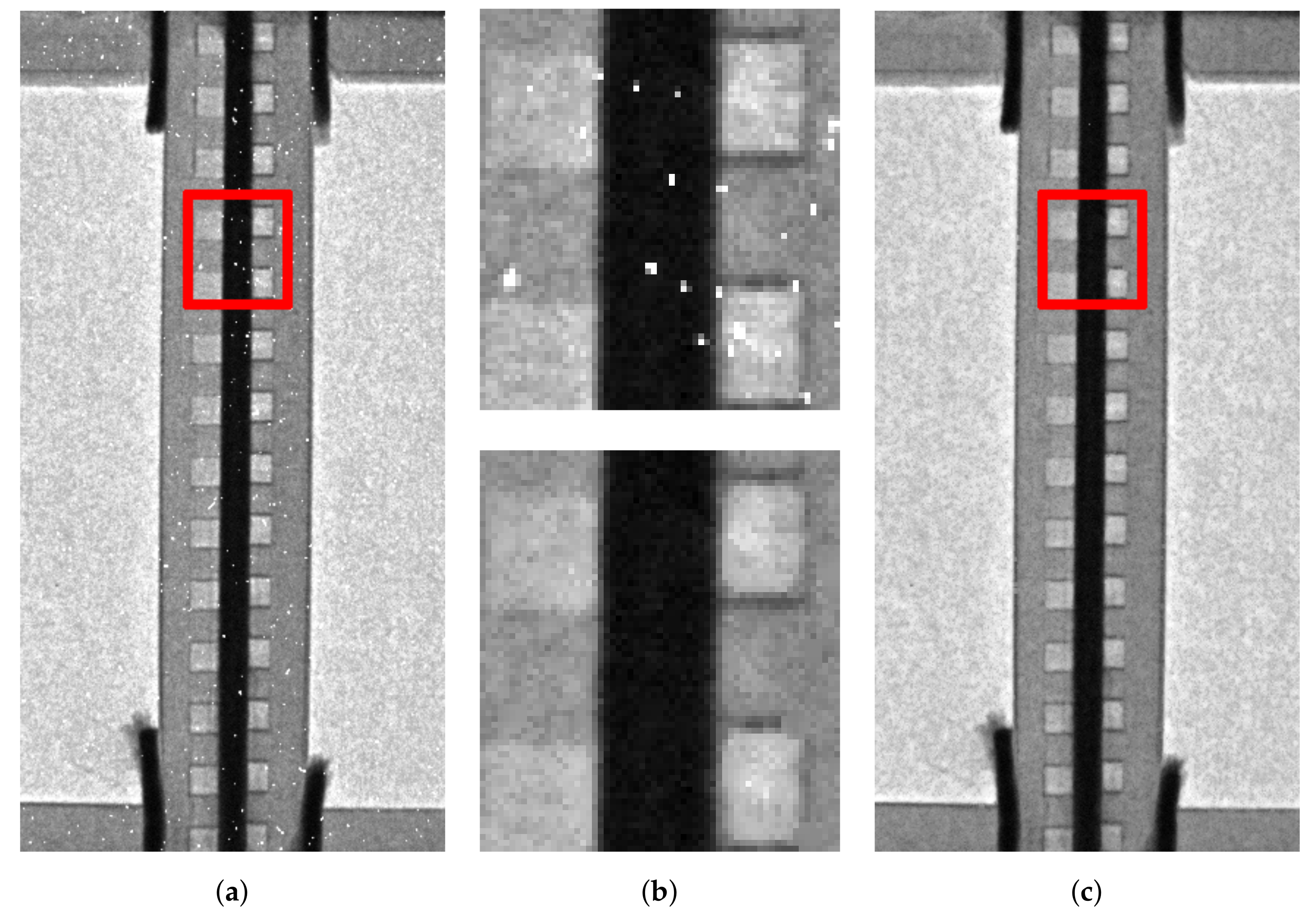
2.2. Noise Removal
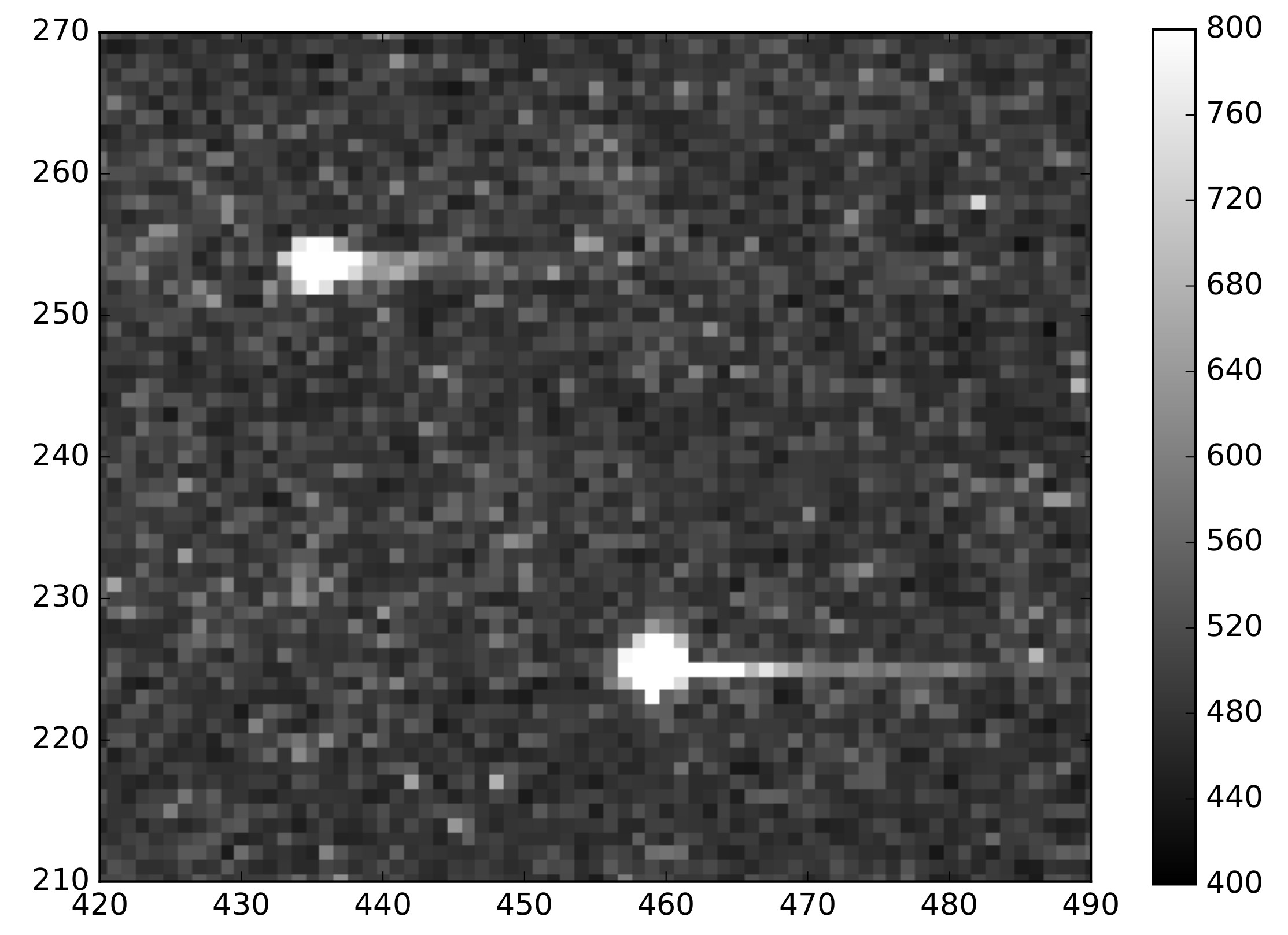
2.3. Sample
2.4. Implementation
3. Results and Discussion
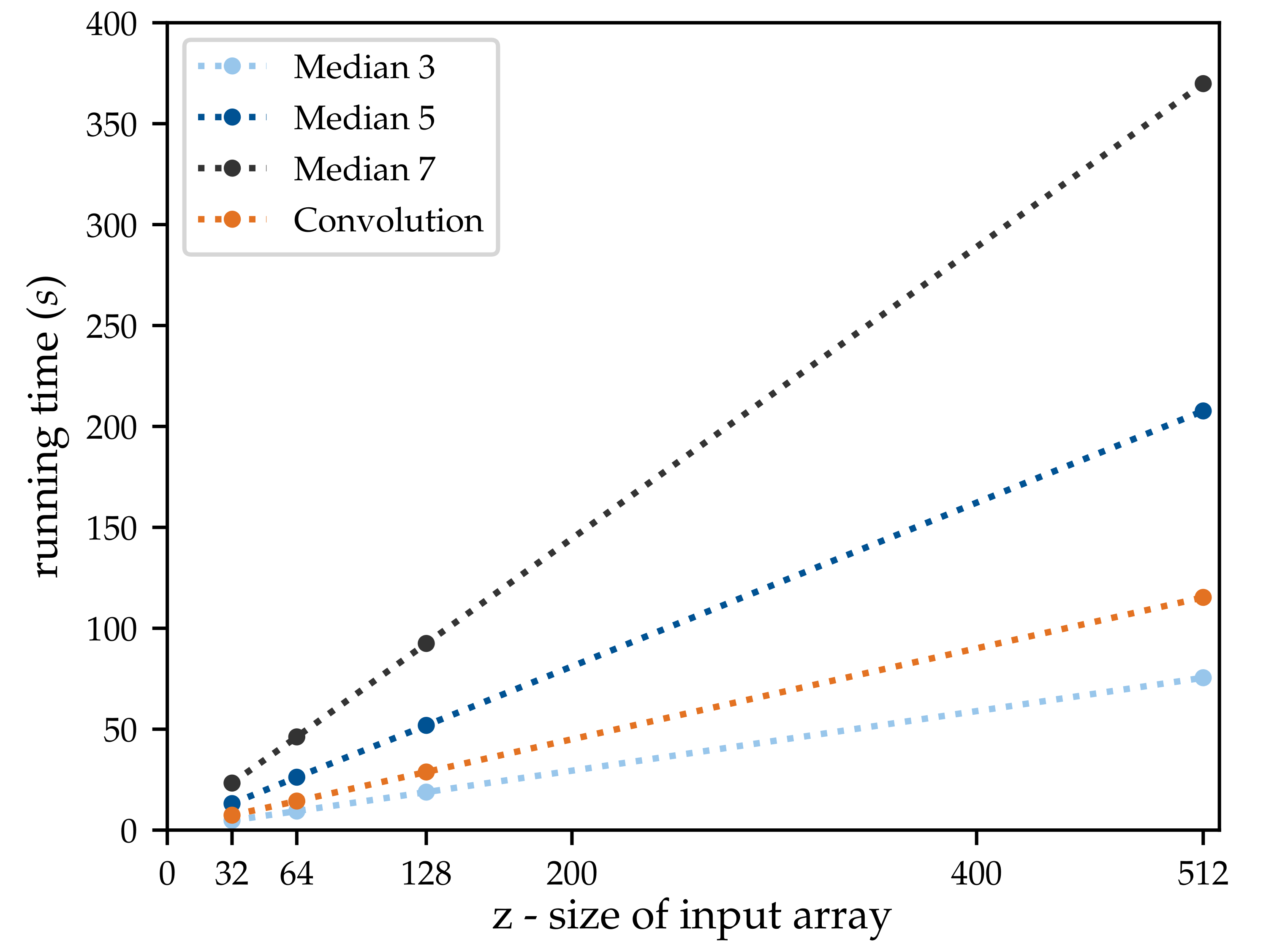
3.1. Profiling
3.2. Optimisation
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| API | Application Programming Interface |
| CPU | Central Processing Unit |
| DSP | Digital Signal Processor |
| FFT | Fast Fourier Transform |
| FPGA | Field Programmable Gate Array |
| GPU | Graphics Processing Unit |
| LoG | Laplacian of Gaussian |
Appendix A. Source Code
Appendix A.1. LoG Convolution
1 __kernel void LoG(__read_only image3d_t d_image, 2 __read_only image3d_t d_coefficients, 3 __write_only image3d_t d_out_image, 4 int size, // The dimension of the convolution kernel 5 int offset_x, // optional if origin is not at (0,0) 6 int offset_y) 7 { 8 // Get the work element: 9 int x_i = get_global_id(0)+offset_x; 10 int y_i = get_global_id(1)+offset_y; 11 int z = get_global_id(2); 12 //Initialise values: 13 float gauss_laplacian = 0.0; 14 float weight = 0.0; 15 int get_x = 0; 16 int get_y = 0; 17 int offset = (size-1)/2; 18 // Convolution: 19 for ( int y = y_i-offset; y <= y_i+offset; y++) 20 { 21 get_x = 0; 22 for ( int x = x_i-offset; x <= x_i+offset; x++) 23 { 24 weight = read_imagef( 25 d_coefficients, 26 sampler, 27 (int4)(get_x, get_y, 0, 0)). s0; 28 gauss_laplacian += (read_imagef( 29 d_image, 30 sampler, 31 (int4)(x, y, z, 0)). s0 * weight); 32 get_x++; 33 } 34 get_y++; 35 } 36 37 // Write the output value to the result array: 38 write_imagef( 39 d_out_image, 40 (int4)(x_i, y_i, z, 0), 41 (float4)(gauss_laplacian)); 42 }
Appendix A.2. Adaptive Median
1 __kernel void a_median(__read_only image3d_t d_image,
2 __read_only image3d_t d_mask,
3 __write_only image3d_t d_out_image,
4 int t_low,
5 int t_middle,
6 int t_high, // Threshold levels
7 int offset_x,
8 int offset_y)
9 {
10 __private int x = get_global_id(0)+offset_x;
11 __private int y = get_global_id(1)+offset_y;
12 __private int z = get_global_id(2);
13 __private int get_x, get_y, i, j;
14 __private int pos = 0;
15 __private float mask_value = read_imagef(
16 d_mask,
17 sampler,
18 (int4)(x, y, z, 0)). s0;
19 __private int size;
20 // Set the median size:
21 if (mask_value < t_low)
22 {
23 size = 1;
24 }
25 else if (t_low<=mask_value && mask_value<t_middle)
26 {
27 size = 3;
28 }
29 else if (t_middle<=mask_value && mask_value<t_high)
30 {
31 size = 5;
32 }
33 else if (mask_value>=t_high)
34 {
35 size = 7;
36 }
37 __private int offset = (size − 1)/2;
38 __private int window_size = size*size;
39 __private float window[81] = {0};
40 __private float temp, median;
41 __private float win_value;
42 // Getting all values from the window
43 for (get_x=-1*offset; get_x<=offset; get_x++)
44 {
45 for (get_y=-1*offset; get_y<=offset; get_y++)
46 {
47 win_value = read_imagef(
48 d_image,
49 sampler,
50 (int4)(x+get_x, y+get_y, z, 0)). s0;
51 window[pos] = win_value;
52 pos++;
53 }
54 }
55
56 // Sorting the array in ascending order
57 for (i = 0; i < window_size-1; i++) {
58 for (j = 0; j < window_size-i-1; j++) {
59 if (window[j] > window[j + 1]) {
60 // swap elements
61 temp = window[j];
62 window[j] = window[j + 1];
63 window[j + 1] = temp;
64 }
65 }
66 }
67 // Calculate and output median
68 median = window[((size*size) − 1)/2];
69 write_imagef(
70 d_out_image,
71 (int4)(x, y, z, 0),
72 (float4)(median));
73
74 }
References
- Sears, V. Neutron scattering lengths and cross sections. Neutron News 1992, 3, 26–37. [Google Scholar] [CrossRef]
- Schreyer, A. Physical Properties of Photons and Neutrons. In Neutrons and Synchrotron Radiation in Engineering Materials Science; Wiley-VCH Verlag GmbH & Co. KGaA: Hoboken, NJ, USA, 2008; pp. 79–89. [Google Scholar]
- Lehmann, E.H.; Ridikas, D. Status of Neutron Imaging—Activities in a Worldwide Context. Phys. Procedia 2015, 69, 10–17. [Google Scholar] [CrossRef]
- Anderson, I.S.; Andreani, C.; Carpenter, J.M.; Festa, G.; Gorini, G.; Loong, C.K.; Senesi, R. Research opportunities with compact accelerator-driven neutron sources. Phys. Rep. 2016, 654, 1–58. [Google Scholar] [CrossRef] [Green Version]
- Gulo, C.A.S.J.; Sementille, A.C.; Tavares, J.M.R.S. Techniques of medical image processing and analysis accelerated by high-performance computing: A systematic literature review. J. Real-Time Image Process. 2017, 1–18. [Google Scholar] [CrossRef]
- Pfleger, S.G.; Plentz, P.D.M.; Rocha, R.C.O.; Pereira, A.D.; Castro, M. Real-time video denoising on multicores and GPUs with Kalman-based and Bilateral filters fusion. J. Real-Time Image Process. 2017, 1–14. [Google Scholar] [CrossRef]
- Calzada, E.; Gruenauer, F.; Mühlbauer, M.; Schillinger, B.; Schulz, M. New design for the ANTARES-II facility for neutron imaging at FRM II. Nuclear Instrum. Methods Phys. Res. Sec. A Accel. Spectrom. Detect. Assoc. Equip. 2009, 605, 50–53. [Google Scholar] [CrossRef]
- Schulz, M.; Schillinger, B. ANTARES: Cold neutron radiography and tomography facility. J. Large-Scale Res. Facil. 2015, 1, A17. [Google Scholar] [CrossRef]
- Li, H.; Schillinger, B.; Calzada, E.; Yinong, L.; Muehlbauer, M. An adaptive algorithm for gamma spots removal in CCD-based neutron radiography and tomography. Nuclear Instrum. Methods Phys. Res. 2006, 564, 405–413. [Google Scholar] [CrossRef]
- Breitwieser, M.; Moroni, R.; Schock, J.; Schulz, M.; Schillinger, B.; Pfeiffer, F.; Zengerle, R.; Thiele, S. Water management in novel direct membrane deposition fuel cells under low humidification. Int. J. Hydrog. Energy 2016, 41. [Google Scholar] [CrossRef]
- Cooley, J.W.; Tukey, J.W. An algorithm for the machine calculation of complex Fourier series. Math. Comput. 1965, 19, 297–301. [Google Scholar] [CrossRef]
- Hegland, M. A self-sorting in-place fast Fourier transform algorithm suitable for vector and parallel processing. Numer. Math. 1994, 547, 507–547. [Google Scholar] [CrossRef]
- Moreland, K.; Angel, E. The FFT on a GPU. In Proceedings of the ACM SIGGRAPH/EUROGRAPHICS Conference On Graphics Hardware, San Diego, CA, USA, 26–27 July 2003; pp. 112–120. [Google Scholar]
- Fialka, O.; Čadík, M. FFT and convolution performance in image filtering on GPU. In Proceedings of the International Conference on Information Visualisation, London, UK, 5–7 July 2006; pp. 609–614. [Google Scholar] [CrossRef]
- Klöckner, A.; Pinto, N.; Lee, Y.; Catanzaro, B.; Ivanov, P.; Fasih, A. PyCUDA and PyOpenCL: A scripting-based approach to GPU run-time code generation. Parallel Comput. 2012, 38, 157–174. [Google Scholar] [CrossRef] [Green Version]
- Khronos Group. OpenCL Specification; Technical Report; Khronos Group: Beaverton, OR, USA, 2015. [Google Scholar]
- Jones, E.; Oliphant, T.; Peterson, P. SciPy: Open Source Scientific Tools for Python, 2001. Available online: http://www.scipy.org/ (accessed on 14 August 2018).

| Isotope | ||
|---|---|---|
| 1H | 82.02 | 0.3326 |
| H | 7.64 | 0.000519 |
| 5B | 5.24 | 767 |
| 13Al | 1.503 | 0.231 |
| 64Gd | 180 | 49,700 |
| Gd | 1044 | 259,000 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schock, J.; Michael, S.; Pfeiffer, F. GPU Accelerated Image Processing in CCD-Based Neutron Imaging. J. Imaging 2018, 4, 104. https://doi.org/10.3390/jimaging4090104
Schock J, Michael S, Pfeiffer F. GPU Accelerated Image Processing in CCD-Based Neutron Imaging. Journal of Imaging. 2018; 4(9):104. https://doi.org/10.3390/jimaging4090104
Chicago/Turabian StyleSchock, Jonathan, Schulz Michael, and Franz Pfeiffer. 2018. "GPU Accelerated Image Processing in CCD-Based Neutron Imaging" Journal of Imaging 4, no. 9: 104. https://doi.org/10.3390/jimaging4090104





