Application of NMR Screening Methods with 19F Detection to Fluorinated Compounds Bound to Proteins
Abstract
:1. Introduction
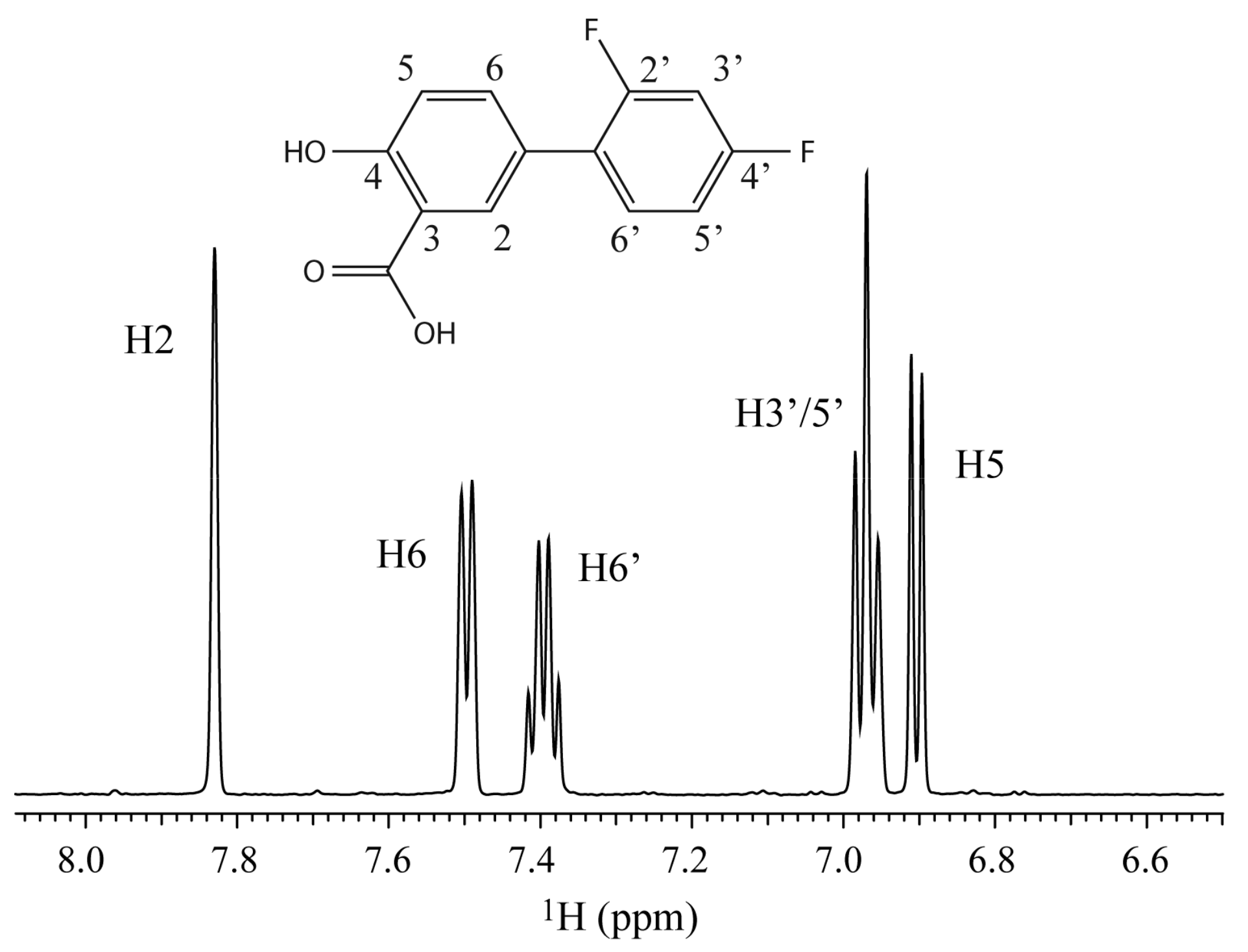
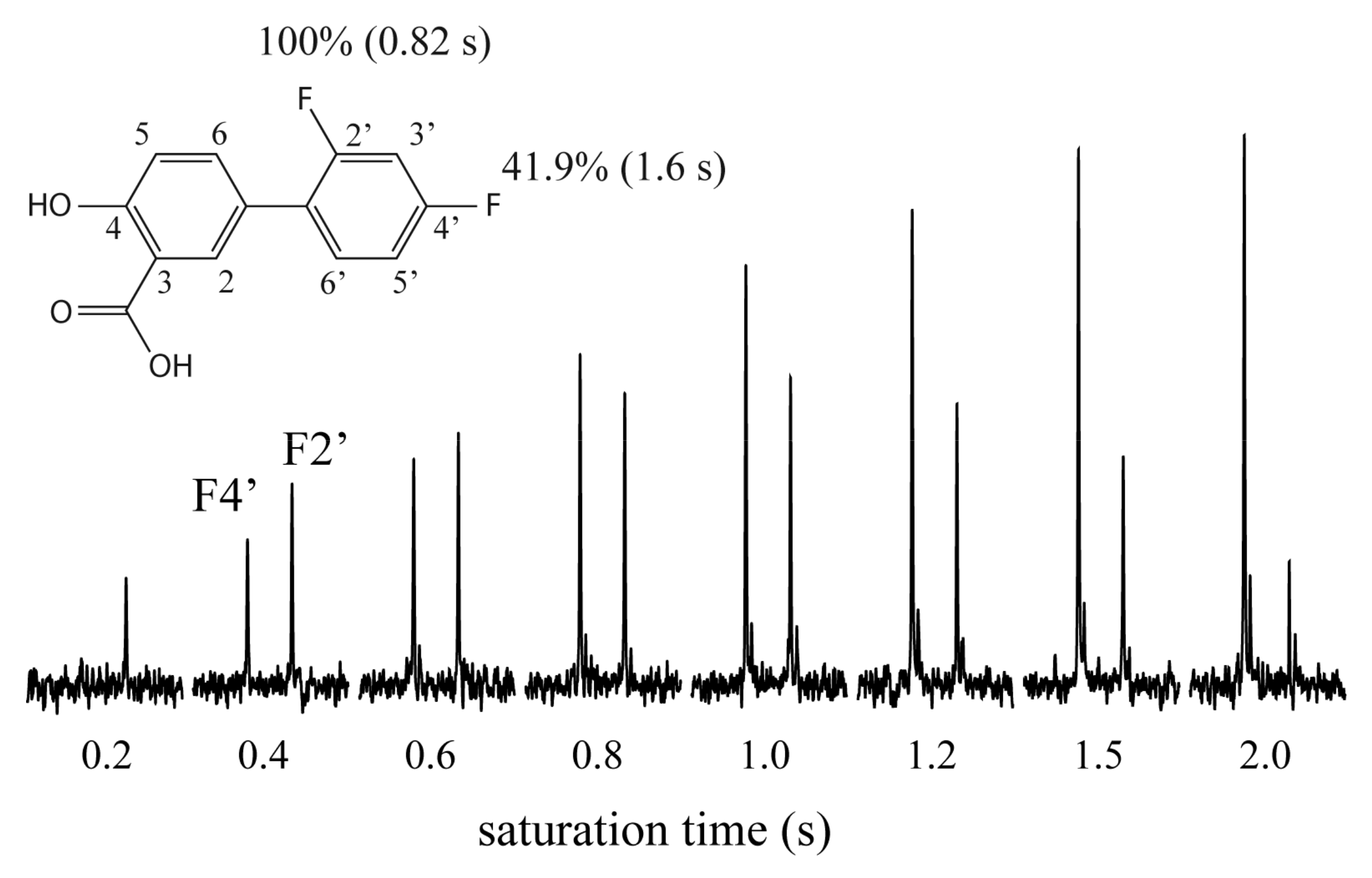
2. Results and Discussion
3. Materials and Methods
3.1. Instrumentation and Chemicals
3.2. NMR Spectroscopy
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Chen, A.; Shapiro, M.J. NOE Pumping: A novel NMR technique for identification of compounds with binding affinity to macromolecules. J. Am. Chem. Soc. 1998, 120, 10258–10259. [Google Scholar] [CrossRef]
- Maye, M.; Meyer, B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 2001, 123, 6108–6117. [Google Scholar]
- Dalvit, C.; Pevarello, P.; Tatò, M.; Veronesi, M.; Vulpetti, A.; Sundström, M. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J. Biomol. NMR 2000, 18, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C.; Fogliatto, G.P.; Stewart, A.; Veronesi, M.; Stockman, B.J. WaterLOGSY as a method for primary NMR screening: Practical aspects and range of applicability. J. Biomol. NMR 2001, 21, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Shapiro, M.J. NOE Pumping. 2. A high-throughput method to determine compounds with binding affinity to macromolecules by NMR. J. Am. Chem. Soc. 2000, 122, 414–415. [Google Scholar] [CrossRef]
- Dalvit, C.; Fagerness, P.E.; Hadden, S.T.A.; Sarver, R.W.; Stockman, B.J. Fluorine-NMR experiments for high-throughput screening: Theoretical aspects, practical considerations, and range of applicability. J. Am. Chem. Soc. 2003, 125, 7696–7703. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C.; Flocco, M.; Stockman, B.J.; Veronesi, M. Competition binding experiments for rapidly ranking lead molecules for their binding affinity to human serum albumin. Comb. Chem. High Throughput Screen. 2002, 5, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, C.; Kurita, J.; Furihata, K.; Tashiro, M. Achievement of 1H-19F heteronuclear experiments using the conventional spectrometer with a shared single high band amplifier. Magn. Reson. Chem. 2015, 53, 327–329. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.; James, T.L. NMR-based characterization of phenothiazines as a RNA binding scaffold. J. Am. Chem. Soc. 2004, 126, 4453–4460. [Google Scholar] [CrossRef] [PubMed]
- Mizukoshi, Y.; Abe, A.; Takizawa, T.; Hanzawa, H.; Fukunishi, Y.; Shimada, I.; Takahashi, H. An accurate pharmacophore mapping method by NMR spectroscopy. Angew. Chem. Int. Ed. 2012, 51, 1362–1365. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furihata, K.; Usui, M.; Tashiro, M. Application of NMR Screening Methods with 19F Detection to Fluorinated Compounds Bound to Proteins. Magnetochemistry 2018, 4, 3. https://doi.org/10.3390/magnetochemistry4010003
Furihata K, Usui M, Tashiro M. Application of NMR Screening Methods with 19F Detection to Fluorinated Compounds Bound to Proteins. Magnetochemistry. 2018; 4(1):3. https://doi.org/10.3390/magnetochemistry4010003
Chicago/Turabian StyleFurihata, Kazuo, Moe Usui, and Mitsuru Tashiro. 2018. "Application of NMR Screening Methods with 19F Detection to Fluorinated Compounds Bound to Proteins" Magnetochemistry 4, no. 1: 3. https://doi.org/10.3390/magnetochemistry4010003



