Modeling the Growth of Lactococcus lactis NCIM 2114 under Differently Aerated and Agitated Conditions in Broth Medium
Abstract
:1. Introduction
2. Experimental Section
2.1. Growth of L. lactis NCIM 2114
2.2. Bacterial Strains, Media, and Culture Conditions
2.3. Experimental Runs
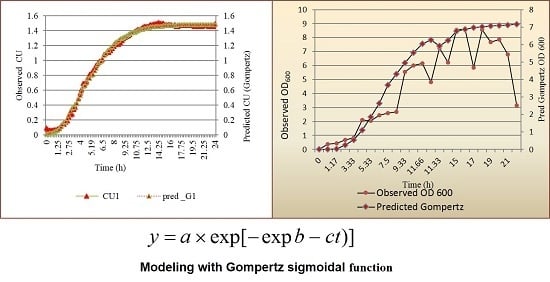
2.4. Fitting of Data and Modeling Growth of L. lactis as a Function of Time
2.5. F Test
2.6. Growth Characteristics
- The specific growth rate maxima μ, the slope of the line when the organism grows exponentially and is drawn on the inflection point of the tangent drawn;
- The lag time, λ, as t, on X-axis intercept of this tangent passed through inflection point on exponential curve;
- The asymptote (A = “a” computed) as the maximal value of growth reached, t approaching infinity. The decline of growth was not studied.
- Using Equation (6) a = A
- Using Equation (7) μm = ac/e {Specific growth rate maxima, h−1}
- Using Equations (7) and (8) λ = (b − 1)/c {lag time, h}
3. Results and Discussion
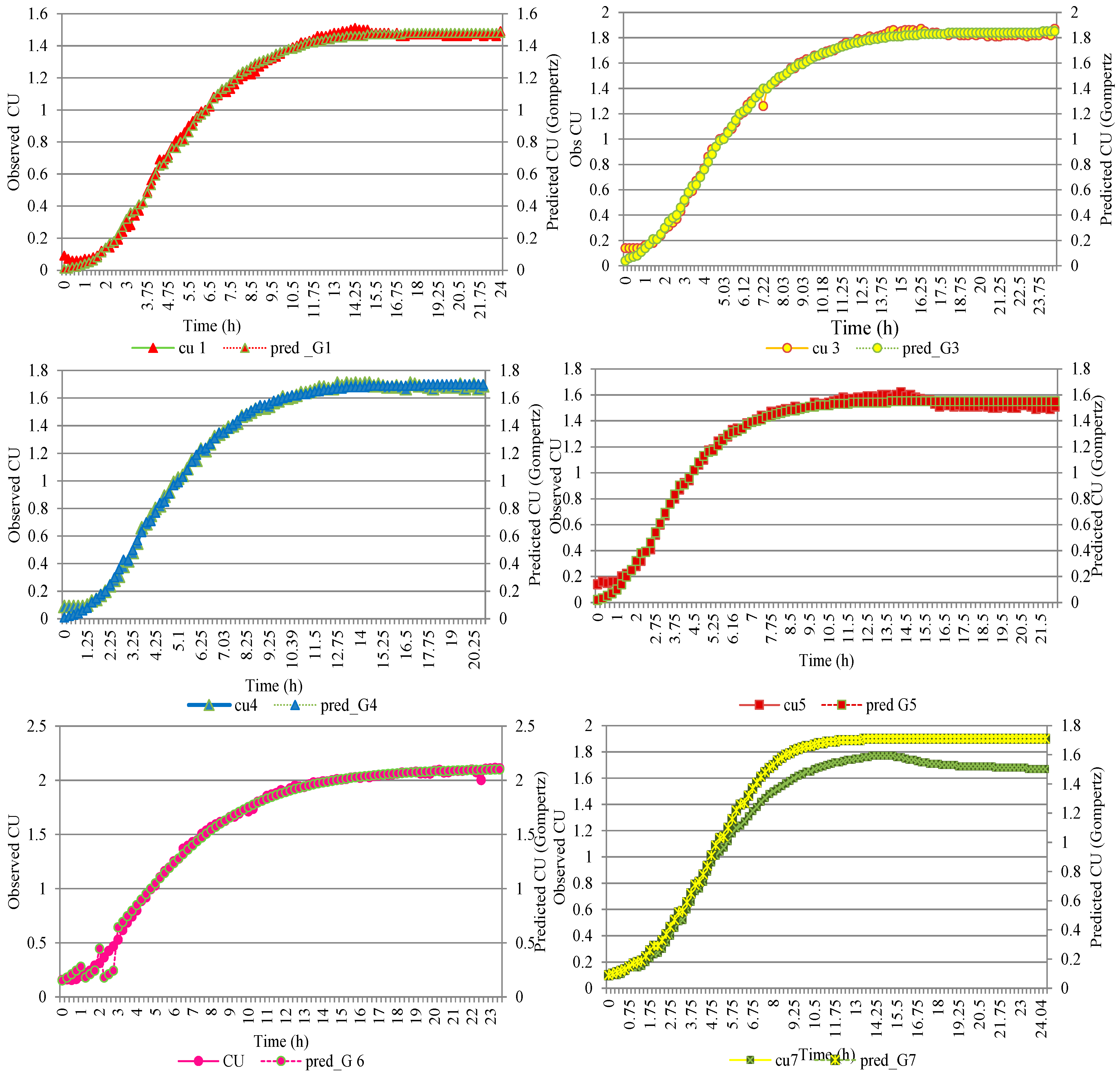
3.1. Modeling Growth of L. lactis
| S. No | Factors Used in Different Batch Runs | Sigmoidal functions | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Logistic | Gompertz | Richards | ||||||||||
| Agitation (rpm) | Aeration (vvm) | |||||||||||
| a | b | c | a | b | c | a | b | c | d | |||
| 1 | 25 | 0.0 | 1.46 | 3.04 | 0.60 | 1.48 | 1.68 | 0.42 | 1.48 | 0.03 | 0.42 | 4.09 |
| 2 | 25 | 0.0 | 1.76 | 2.92 | 0.64 | 2.04 | 1.34 | 0.34 | 2.54 | −0.55 | 0.18 | 3.30 |
| 3 | 25 | 0.2 | 1.68 | 3.00 | 0.58 | 1.70 | 1.62 | 0.40 | 1.75 | −1.54 | 0.21 | 0.70 |
| 4 | 25 | 0.4 | 1.68 | 3.02 | 0.65 | 1.70 | 1.66 | 0.45 | 1.70 | 0.14 | 0.47 | 3.88 |
| 5 | 50 | 0.0 | 1.54 | 2.78 | 0.76 | 1.55 | 1.53 | 0.53 | 4.82 | −3.41 | 0.06 | 0.13 |
| 6 | 50 | 0.0 | 2.05 | 2.27 | 0.43 | 2.09 | 1.16 | 0.30 | 2.09 | −0.07 | 0.29 | 3.75 |
| 7 | 50 | 0.2 | 1.71 | 2.84 | 0.62 | 1.73 | 1.51 | 0.43 | 1.77 | −2.09 | 0.22 | −0.36 |
| 8 | 50 | 0.0 | 1.74 | 1.97 | 0.53 | 1.76 | 0.97 | 0.37 | 1.82 | −1.17 | 0.22 | 0.04 |
| 9 | 50 | 0.4 | 2.03 | 2.04 | 0.44 | 2.05 | 1.02 | 0.31 | 2.05 | 0.33 | 3.53 | 0.14 |
| 10 | 100 | 0.0 | 1.97 | 2.61 | 0.49 | 2.00 | 1.34 | 0.33 | 1.99 | 0.14 | 0.35 | 4.32 |
| 11 | 100 | 0.2 | 1.82 | 2.28 | 0.56 | 1.85 | 1.14 | 0.38 | 1.84 | 0.32 | 0.44 | 3.42 |
| 12 | 100 | 0.4 | 1.82 | 2.67 | 0.54 | 1.84 | 1.40 | 0.37 | 1.88 | −1.10 | 0.24 | 1.64 |
| S. No | Factors | R2 | |||
|---|---|---|---|---|---|
| Agitation (rpm) | Aeration (vvm) | ||||
| Logistic | Gompertz | Richards | |||
| 1 | 25 | 0.0 | 0.993 | 0.993 | 0.997 |
| 2 | 25 | 0.0 | 0.995 | 0.991 | 0.986 |
| 3 | 25 | 0.2 | 0.992 | 0.998 | 0.992 |
| 4 | 25 | 0.4 | 0.996 | 0.998 | 0.998 |
| 5 | 50 | 0.0 | 0.995 | 0.992 | ND |
| 6 | 50 | 0.0 | 0.994 | 0.999 | 0.999 |
| 7 | 50 | 0.2 | 0.997 | 0.995 | 0.967 |
| 8 | 50 | 0.0 | 0.994 | 0.998 | 0.989 |
| 9 | 50 | 0.4 | 0.883 | 0.885 | 0.885 |
| 10 | 100 | 0.0 | 0.996 | 0.998 | 0.998 |
| 11 | 100 | 0.2 | 0.997 | 0.998 | 0.999 |
| 12 | 100 | 0.4 | 0.996 | 0.999 | 0.992 |
3.2. F Test
| S. No | Run with Factors | fd Logistic | fd Gompertz | F Value | p Value # | |
|---|---|---|---|---|---|---|
| Agitation (rpm) | Aeration (vvm) | |||||
| 1 | 25 | 0.0 | 4.91 | 0 | 3.98 | 0.05 |
| 2 | 25 | 0.0 | −2.07 | −0.55 | 7.00 | 0.05 |
| 3 | 25 | 0.2 | 2.18 | −2.32 | 3.98 | 0.05 |
| 4 | 25 | 0.4 | 6.12 | 0.25 | 6.64 | 0.22 |
| 5 | 50 | 0.0 | 5.07 | 10.04 | 6.97 | 0.05 |
| 6 | 50 | 0.0 | 12.44 | 0.15 | 3.96 | 0.05 |
| 7 | 50 | 0.2 | −2.89 | −2.14 | 3.98 | 0.05 |
| 8 | 50 | 0.0 | −0.74 | −2.93 | 7.00 | 0.05 |
| 9 | 50 | 0.4 | 0.07 | 0.003 | 3.89 | 0.05 |
| 10 | 100 | 0.0 | 6.27 | 0.30 | 3.97 | 0.05 |
| 11 | 100 | 0.2 | 5.94 | 2.06 | 3.96 | 0.05 |
| 12 | 100 | 0.4 | −0.22 | −2.53 | 3.98 | 0.05 |
| % Acceptance of given model: among the 12 batch runs | 67% | 92% | ||||
3.3. Derivation of the Growth Characteristics of the Organism
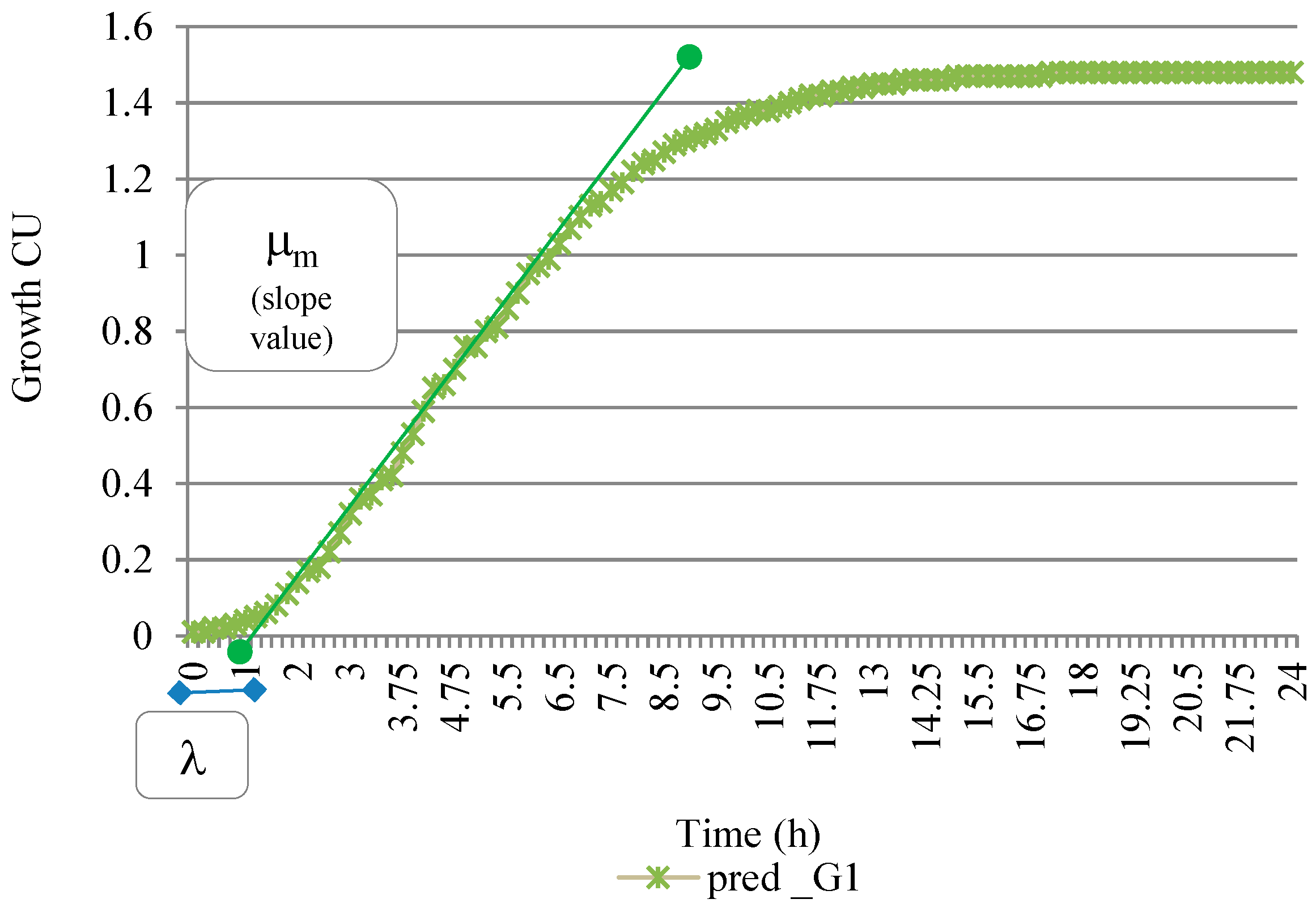
| S. No | Growth Factors Used | Specific Growth Rate Maxima µmax (h−1) | Inf pt: t (h) | Lag Time λ (h) | Time between Lag Time and Inflection Point T (h) | |
|---|---|---|---|---|---|---|
| Agitation (rpm) | Aeration (vvm) | |||||
| 1 | 25 | 0.0 | 0.23 | 4.04 | 1.63 | 2.41 |
| 2 | 25 | 0.0 | 0.25 | 3.99 | 1.02 | 2.98 |
| 3 | 25 | 0.2 | 0.25 | 4.11 | 1.58 | 2.53 |
| 4 | 25 | 0.4 | 0.28 | 3.71 | 1.48 | 2.24 |
| 5 | 50 | 0.0 | 0.30 | 2.87 | 0.99 | 1.87 |
| 6 | 50 | 0.0 | 0.23 | 3.89 | 0.53 | 3.36 |
| 7 | 50 | 0.2 | 0.27 | 3.52 | 1.19 | 2.33 |
| 8 | 50 | 0.0 | 0.24 | 2.61 | −0.08 | 2.68 |
| 9 | 50 | 0.4 | 0.23 | 3.28 | 0.06 | 3.23 |
| 10 | 100 | 0.0 | 0.24 | 4.09 | 1.03 | 3.06 |
| 11 | 100 | 0.2 | 0.26 | 2.99 | 0.37 | 2.62 |
| 12 | 100 | 0.4 | 0.25 | 3.80 | 1.09 | 2.71 |
| One Tailed Correlations | Specific Growth Rate Maxima: µmax(h−1) | Inflection Point: t (h) | Lag Time: λ (h) | Time between Lag Time and Inflection Point: T (h) |
|---|---|---|---|---|
| Maximum specific growth rate µmax | 1 | |||
| Infpt: t | −0.39 | 1 | ||
| Lag time: λ(h) | 0.23 | 0.69 | 1 | |
| Time between lag time and inflection point: T(h) | −0.77 | 0.30 | −0.49 | 1 |

4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dalgaard, P.; Koutsoumanis, K. Comparison of maximum specific growth rates and lag time estimated from absorbance and viable count data by different mathematical models. J. Microbiol. Methods 2001, 43, 183–196. [Google Scholar] [CrossRef]
- Annadurai, G.; Rajesh Babu, S.; Srinivasamurthy, V.R. Development of mathematical models (Logistic, Gompertz and Richards models) describing the growth pattern of Pseudomonas putida (NICM 2174). Bioproc. Eng. 2000, 23, 607–612. [Google Scholar] [CrossRef]
- Zwiettering, M.H.; Jongenburger, I.; Rombouts, F.M.; van’T Riet, K. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 1990, 56, 1875–1881. [Google Scholar]
- Kristel, B.; Els, D.; Karen, V.; Geeraerd, A.H.; Standaert, A.R.; Devlieghere, F.; Debevere, J.; van Impe, J.F. Concepts and Tools for Predictive Modeling of Microbial Dynamics. J. Food Prot. 2004, 67, 2041–2052. [Google Scholar]
- Frederick, B.; Fleming, H.P. Modeling of the Competitive Growth of Listeria monocytogenes and Lactococcus lactis in Vegetable Broth. Appl. Environ. Microbiol. 1998, 64, 3159–3165. [Google Scholar]
- Matthias, K.; Guido, H.; Hella, L.-F.; Jost, L.; Maik, K. Grofit: Fitting Biological Growth Curves with R. J. Stat. Softw. 2010, 33, 1–21. [Google Scholar]
- Birch, C.A. new generalized logistic sigmoid growth equation compared with the Richards growth equation. Ann. Bot. 1999, 83, 713–723. [Google Scholar] [CrossRef]
- De Vuyst, L.; Vandamme, E.J. Influence of the carbon source on nisin production in Lactococcus lactis subsp. lactis batch fermentations. J. Gen. Microbiol. 1992, 138, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Van Impe, J.F.; Nicolaï, B.M.; Martens, T.; de Baerdemaeker, J.; Vandewalle, J. Dynamic mathematical model to predict microbial growth and inactivation during food processing. Appl. Environ. Microbiol. 1992, 58, 2901–2909. [Google Scholar]
- STR, Sartorius. BIOSTAT Fermenter Systems Handbook—Laboratory Scale Fermenters, Pilot and Production Plant Fermenters: BIOSTAT- A-plus; Sartorius BBI Systems GmbH: Melsungen, Germany, 2005; pp. 1–14. [Google Scholar]
- American Public Health Association. Standard Methods for the Examination of Water and Wastewater, 19th ed.; Eaton, A.D., Clesceri, L.S., Greenberg, A.E., Eds.; Washington, DC, USA, 1995; pp. 4–99. [Google Scholar]
- Senese, F. How can I Predict Oxygen Solubility in Water? Available online: http://antoine.frostburg.edu/chem/senese/101/solutions/faq/predictingDO.shtml (accessed on 2 May 2014).
- Schleifer, K.H.; Kraus, J.; Dvorak, C.; Kilpper-Bälz, R.; Collins, M.D.; Fischer, W. Transfer of Streptococcus lactis and related streptococci to the genus Lactococcus gen. nov. System. Appl. Microbiol. 1985, 6, 183–195. [Google Scholar] [CrossRef]
- De Man, J.C.; Rogosa, M.; Sharpe, M.E. A medium for the cultivation of lactobacilli. J. Appl. Microbiol. 1960, 23, 130–135. [Google Scholar] [CrossRef]
- Behnken, D.W.; Draper, N.R. Residuals and their variance patterns. Technometrics 1972, 14, 101–111. [Google Scholar] [CrossRef]
- Timm, N.H. Appendix Tables -Upper percentage points of the F distribution. In Applied Multivariate Analysis; Pub Springer: Secaucus, NJ, USA, 2002. [Google Scholar]
- Gompertz, B. On the nature of the function expressive of the law of human mortality, and on a new mode determining the value of life contingencies. Philos. Trans. R. Soc. Lond. 1825, 115, 513–585. [Google Scholar] [CrossRef]
- Marquardt, D.W. An algorithm for least-squares estimation of nonlinear parameters. J. Soc. Ind. Appl. Math. 1963, 11, 431–441. [Google Scholar] [CrossRef]
- Soper, D. p Value Calculator. Available online: http://www.danielsoper.com/statcalc3/calc.aspx?id=7 (accessed on 23 August 2014).
- Mikkel, N.; Jensen, S.; Bang, N.; John, W. Glucose metabolism in Lactococcus lactis MG1363 under different aeration conditions: Requirement of acetate to sustain growth under microaerobic conditions. Appl. Environ. Microbiol. 2003, 69, 3462–3468. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, S.; Singh, K.N.; Mandjiny, S.; Holmes, L. Modeling the Growth of Lactococcus lactis NCIM 2114 under Differently Aerated and Agitated Conditions in Broth Medium. Fermentation 2015, 1, 86-97. https://doi.org/10.3390/fermentation1010086
Singh S, Singh KN, Mandjiny S, Holmes L. Modeling the Growth of Lactococcus lactis NCIM 2114 under Differently Aerated and Agitated Conditions in Broth Medium. Fermentation. 2015; 1(1):86-97. https://doi.org/10.3390/fermentation1010086
Chicago/Turabian StyleSingh, Sunita, Kamalesh N. Singh, Siva Mandjiny, and Leonard Holmes. 2015. "Modeling the Growth of Lactococcus lactis NCIM 2114 under Differently Aerated and Agitated Conditions in Broth Medium" Fermentation 1, no. 1: 86-97. https://doi.org/10.3390/fermentation1010086