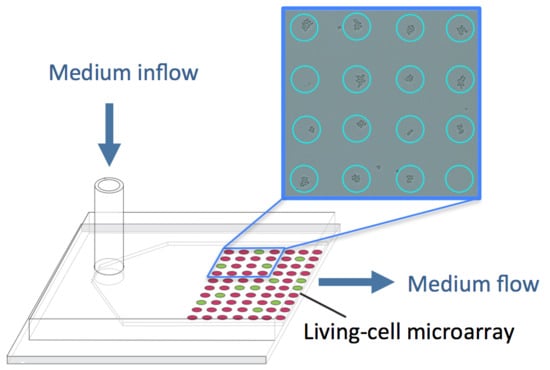
Microfluidic Bioreactors for Cellular Microarrays
Abstract
:1. Introduction
2. Construction Methods of Cellular Array Microfluidic Chips
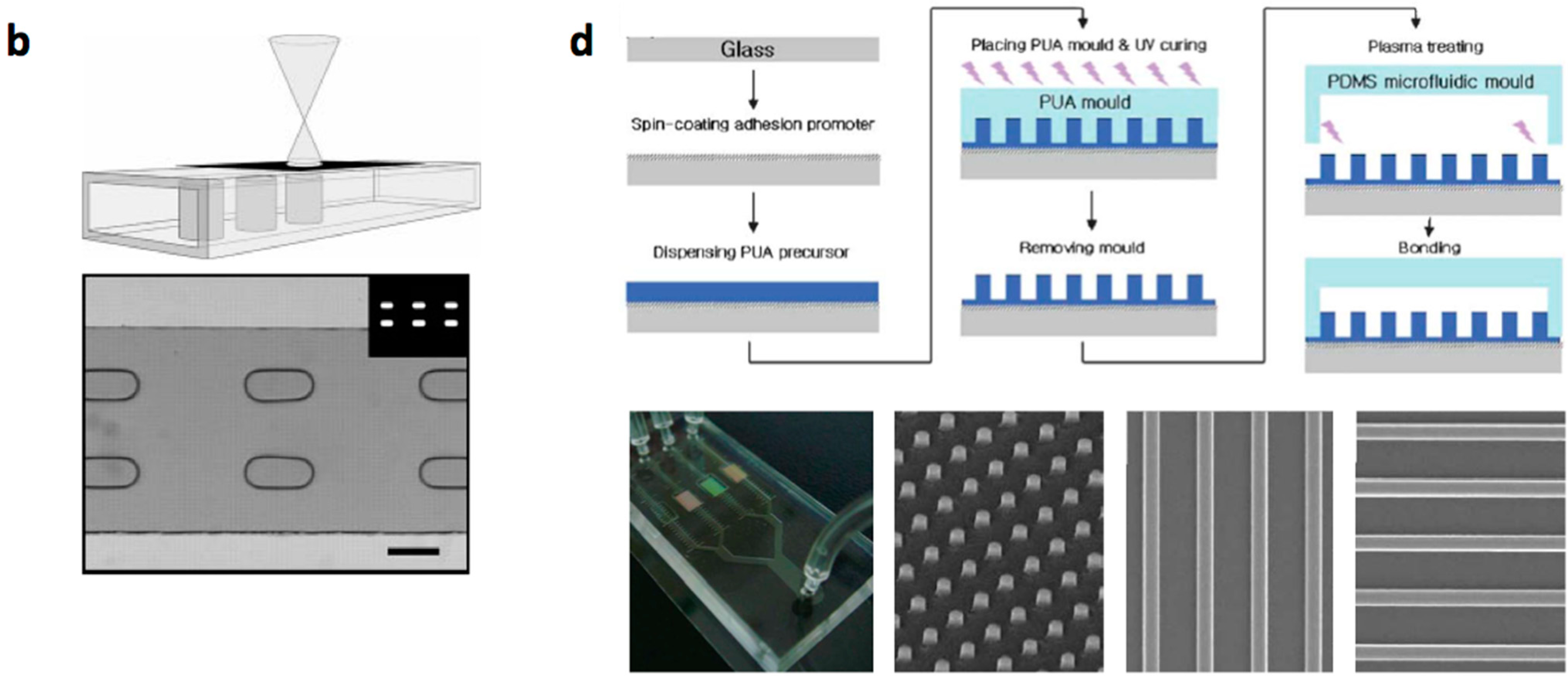
- Conventional MEMS (microelectromechanical systems) processing (photo lithography, electroplating, deposition, and etching) are used to create structures on silicium or glass substrates and soft lithography or etching is used to fabricate channels with PDMS or glass. The microstructered layer is bonded with the microchannel layer to form the microfluidic device [12,13,14,15,16].
- Multilayered PDMS based microfluidic systems that are fabricated with one or multiple uses of soft lithographic process: (a) The PDMS layer with the microstructures and the channel are separately fabricated and bonded with alignment [17,18,19,20]; (b) The channel layer containing the structures is fabricated on a multilayered master mold and is bonded with a flat PDMS or glass substrate [21,22,23].
- Micro/nanostructures are formed on a polymer layer or Si substrate by nanoimprinting, soft lithography or reactive etching following a suitable lithographic process. Next, a microchannel layer of PDMS or glass is bonded to form a microfluidic channel [31].

| Materials | Fabrication Methods | Array Spots | Cell Type | Refs | ||
|---|---|---|---|---|---|---|
| Channel | Micro-Structures | Channel | Micro-Structures | |||
| PDMS, glass | Si, glass, PDMS 1 | Replica moulding, etching | Photo lithography, deposition, etching | 1 | HL60 | [12] |
| PDMS | PDMS | Replica moulding | Replica moulding | 1 16 100 440 8 100/mm2 3600 | Jurkat, U937 C2C12 myoblasts Hela U937, FL 60 HepG2 Hela S. cerevisiae | [18] [19] [22] [23] [32] [33] [20] |
| PDMS, glass | Hydrogel PEG | Replica moulding, etching | Photo-polymerization | >300 | Mouse embryonic stem cells, NIH-3T3 fibroblasts | [24,26,27] |
| PDMS | PDMS, hydrogel, PMMA | Replica molding | Soft lithography | 25,000 | Hybridoma | [29] |
| PDMS, glass Glass, PDMS | Si, hydrogel, PMMA PUA 2 | Replica molding, etching Replica molding | Nanoimprinting, soft lithography, reactive ion etching Capillary molding | 4 | MCF10A, MCF7 | [31] |
3. Cell Arraying Methods
3.1. Cell Patterning on Adhesive Micropatterns

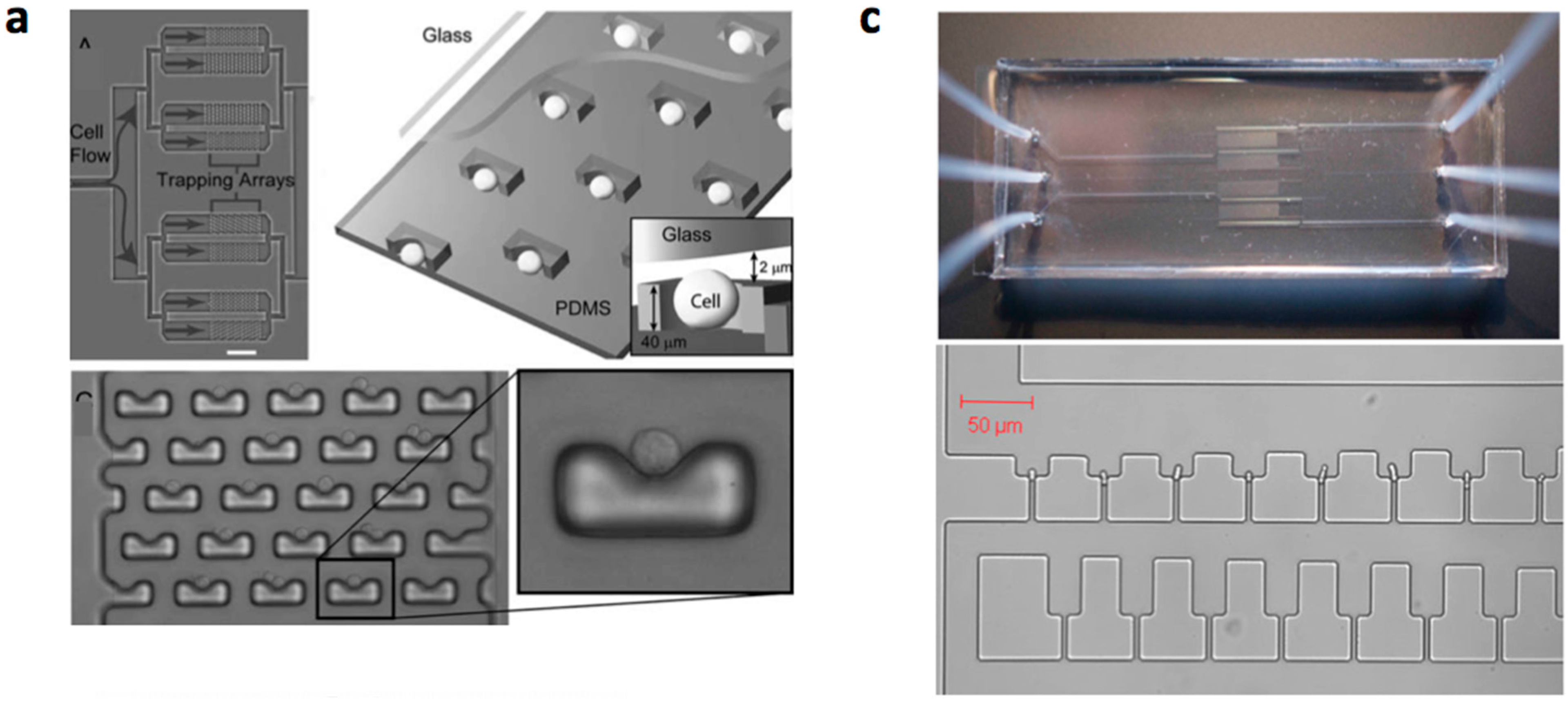
3.2. Mechanical Cell Patterning

| Material | Diameter/Width | Cell Type | Characteristics | Refs |
|---|---|---|---|---|
| PDMS | 20–40 µm | NIH3T3 1, RBL-1 2 | Single-cell screening | [111] |
| 25 µm | Hepatocyte | Single cell oxidative stress screening | [131] | |
| Photoresist 1002F | 50 µm | HeLa | Viability assessment | [132] |
| 50 µm | H1299, MEF 3 | RhoA GTPase biosensor | [133] | |
| Poly(ethylene glycol) | 40–150 µm | Murine embryonic stem cells | The formation of homogeneous embryoid bodies | [134] |
| Polystyrene | 10 µm | B-cells | Detecting activated cells | [135] |
| Optical imaging fibers | 7 µm | NIH 3T3 mouse fibroblast cells | Observation of cell fluorescence | [136] |
| Etched glass | 20 µm | Non-adherent cell lines | Individual cell based assays | [137] |
| SU-8 on coverslip | 90 µm2 squares | Adult neural stem cell | Culturing and dynamic monitoring of stem cell proliferation | [138] |
| PDMS | 25–50 µm | BCE 4 | PDMS wells coated with fibronectin | [139] |
| 160 µm | HSC 5 | HSC proliferation control at the single-cell level | [119] | |
| 70 µm | HeLa | Single cell analysis | [140] | |
| Poly(ethylene glycol) diacrylate | 229–442 µm | Murine ES cells 6 | Wiping technique to localize cells in microwells | [141] |
3.3. Robotic Cell Printing
3.4. Selection of Cell Arraying Method
| Method | Advantages | Limitations |
|---|---|---|
| Adhesive micropatterns |
|
|
Mechanical cell patterning
|
|
|
Robotic printing
|
|
|
4. Operation and Applications of Microfluidic Cell Microarrays
4.1. Microfluidic Operation
4.2. Single-Cell Measurement/Monitoring in Microfluidic Devices
4.3. Microfluidic Cell Array Culture Applications
| Scope | Measurement | Cell Type | Cell-Array Type/Methodology | Cell-Array Size | Fluidic Conditions | Refs |
|---|---|---|---|---|---|---|
| Adherent Cell Types | ||||||
| Drug screening: high-content analysis of cell signaling | On chip off-line immune-cytochemistry. | Mouse fibroblasts (NIH-3T3) | Mechanical cell patterning (PDMS microchambers) | 32 (4 × 8) 4 groups of 8 parallel channel chambers; 8 chambers for each parallel channel | 4 common inlets for 32 parallel channel chambers and 1 additional inlet for a group of 8 channels | [118] |
| Drug screening: screening of chemotherapeutic compounds | Cell imaging with fluorescence microscopy | Human breast cancer (MCF-7) and HepG2 cell lines | Mechanical cell patterning (PDMS microchambers) | 24 (4 × 6) 4 channels rows containing 6 microchambers columns | 1 inlet (from gradient generator) for each column of 4 microchambers | [253] |
| Cytotoxicity evaluation | Real-time microscopy, albumin concentration determination in the effluent (ELISA assay) | Rat hepatocytes in coculture with fibroblasts (3T3-J2) | Cell patterning on adhesive micropatterns (collagen) in a culture chamber (glass bottom, PDMS walls) | 64 (8 × 8) 8 rows and 8 columns of culture chambers | 1 medium inlet for each row of, culture chambers and 1 gas perfusion inlet for each column of chambers | [173] |
| Cytotoxicity evaluation | Real-time cell viability screening by fluorescence microscopy | Human lung carcinoma (A549) cell line | Mechanical cell patterning in microchamber (PDMS with glass bottom) | 25 (5 × 5) 5 parallel channels containing 5 chambers in series | 2 inlets into a gradient concentrator that feeds each parallel channel | [269] |
| Cytotoxicity evaluation | Fluorescent measurement (microscopy) of cytosolic calcium | Mouse leukemic monocyte macrophage (RAW) cell line | Mechanical cell trap barrier (glass) | 1 chamber | 1 inlet | [270] |
| Cytotoxicity evaluation | Fluorescent measurement (microscopy) of cell viability (ethidium homodimer-1) | Human HeLa cancer cell line | Mechanical cell patterning (PDMS microwells) | 3000 (500 × 6) 500 microwells in each parallel channel | 1 chemical inlet followed by a gradient generator, which feeds a different concentration in each parallel channel | [271] |
| Cell biology research: analysis of cell proliferation and migration | Cell number and position (microscopy) | Human breast cancer cells (MDA-MB-231) | Mechanical cell patterning in microchambers (PDSE1 and AZ4562 photoresist) | 1600 (40 × 40) Each chamber is linked together by channels | 1 inlet for all chambers | [272] |
| Cell biology research: long-term cellular monitoring for high-throughput cell-based assays | Fluorescence localization of calcein AM (microscopy) | Human HeLa cancer cell line | Mechanical cell trap barrier (PDMS) | 100 (10 × 10) 10 culture chambers row and per column | 1 inlet per column (from a gradient generator) and 1 inlet for all rows | [22] |
| Cell biology research: continuous monitoring of gene expression | Fluorescence microscopy of eGFP-tagged reporter gene | Human HeLa cancer cell line | Mechanical cell patterning in microchambers (PDMS) | 40 (8 × 5) 5 microchambers for each of the 8 parallel channels | 1 inlet per parallel channel (from a gradient generator) | [273] |
| Cell biology research: adherent cell culture over a logarithmic range of flow rates | Cell number and morphology (microscopy) | Fibroblasts 3T3 cell line | Mechanical cell patterning in microchambers (PDMS with glass bottom) | 16 (4 × 4) 4 groups of 4 chambers in parallel (including the logarithmic diluter) | 1 input per group of 4 chambers | [274] |
| Cell biology research: fully automated cell culture screening system | Differentiation and proliferation; cell number/morphology, cell nucleus staining, alkaline phosphatase activity (microscopy) | Human mesenchymal stem cells | Mechanical cell patterning in microchambers (PDMS) | 96 (2 × 48) 2 rows of 48 parallel chambers | 16 inlets per row | [116] |
| Cell biology research: real-time gene expression monitoring | Fluorescence microscopy of eGFP-tagged genes | Hepatocytes (H35) cell line | Mechanical cell patterning in microchambers (PDMS) | 256 (8 × 8 × 4) 8 rows and 8 columns; each matrix point is composed of a 2 × 2 subarray | 8 inlets for each row and 8 inlets for each column | [117] |
| Non-Adherent Cell Types | ||||||
| Drug screening: antifungal evaluation | Effect of antifungal on viability (fluorescence microscopy) | Saccharomyces cerevisiae | Mechanical cell patterning in microwells (glass) | 44,000 (2 × 22,000) Two arrays of 22,000 microwells | Digital microfluidic platform: microwell seeding by shuttling a cell-containing droplet; droplet of antifungal on the array. | [252] |
| Drug screening: real-time screening of anticancer drugs | Online fluorescence imaging | Human histiocytic leukaemia (U937) and promyelocytic leukaemia (HL60) cell lines | Mechanical cell trap barrier (PDMS) | 440 All traps in a triangular chambers | 1 inlet and 6 outlets (base of the triangle). | [23] |
| Cell biology research: High-throughput analysis of single hematopoietic stem cell proliferation | Proliferation; cell number/morphology; live-cell immunostaining; microscopy | Hemapoietic stem cells (HSC) (clonal population) Preleukemic mouse (ND13) | Mechanical cell patterning in microchambers (PDMS) | 6144 (64 × 96) 64 parallel channels (rows); each channels flows over 96 wells (columns) | 1 inlet for each channel; up to 6 different medium conditions can be loaded | [119] |
| Microbiology research: long-term monitoring of bacteria undergoing programmed population control in a microchemostat | Cell number (microscopy) | Escherichia coli | Mechanical cell patterning in microchambers (PDMS) | 6 | Multiple inlets for each culture chamber. Culture chambers are independently operated (including an on chip peristaltic pump). | [112] |
| Microbiology research: microfluidic chemostat growth to high cell densities | Cell number (microscopy) | E. coli, S. cerevisiae | Mechanical cell patterning in microchambers (PDMS) | 320 (16 × 20) 16 parallel channels containing 20 chambers | 1 inlet branching into the array of 16 parallel channels | [114] |
| Microbiology research: high-throughput time-course analysis of single cell responses | Cell number, fluorescent reporter proteins (microscopy) | S. cerevisiae | Mechanical cell patterning in microwells (polyurethane acrylate) | 3906 wells per mm2 (microwell diameter of 8 µm) | 1 inlet to channel that flows over the wells | [275] |
| Microbiology research: microscopic observation of cell behavior at high resolution | Cell number/morphology and single molecule fluorescence imaging (microscopy) | Schizosaccharo-myces pombe | Mechanical cell trap barrier (PDMS) | 7728 (4 × 1932) 4 trapping regions, each with 1932 mechanical cell traps | 3 inlets feeding into 4 trapping regions | [120] |
| Microbiology research: spatio-temporal analysis of growing bacterial microcolonies in perfusion reactor | Cell number and morphology (microscopy) | E. coli Corynebacterium glutamicum | Mechanical cell trap barrier (PDMS) | 30 (6 × 5) 6 parallel channel containing each 5 cell trap barriers | 2 inlets into a gradient generator, which feeds each parallel channel | [265,276] |
| Scope | Measurement | Cell Type | Populationsize | Cell-Array Type/Methodology | Cell-Array Size | Fluidic Conditions | Refs |
|---|---|---|---|---|---|---|---|
| Adherent Cell Types | |||||||
| Cell biology research: cell culture device (multiple cycles of growth and trypsinization) | Microscopic observation of cell morphology and viability | Murine embryonic fibroblast (BALB/3T3) | 1 to 8 | Mechanical cell patterning (8 mechanical cell trap barriers in PDMS/chamber) | 64 (8 × 8) 8 parallel independent rows; 8 cultivation chambers in series per row | 1 inlet for 1 parallel channel | [277] |
| Non-Adherent Cell Types | |||||||
| Microbiology research: studying signaling network dynamics | Time-lapse cell imaging; heme expression by genetically encoded GFP reporter and protein localization (GFP-tagged protein) | S. cerevisiae | 8 strains | Mechanical cell patterning (PDMS microchambers) | 128 (8 × 16) | 8 chemical inlets for 16 parallel rows | [195] |
| Microbiology research: spatio-temporal analysis of the proteome | Time lapse imaging of GFP-tagged strains | S. cerevisiae | 1152 strains | Direct cell printing and mechanical trapping in microchemostat chambers (PDMS) | 1152 (3 × 384) 3 independent sections of 384 chambers | 1 inlet per chamber section | [202] |
| Microbiology research: monitor biofilm formation under near-native conditions | Quantitative cell analysis by bio-impedance measurement and respiration activity measured by electrochemical microelectrodes | Candida albicans | 2 | Mechanical cell patterning (PDMS microchambers) | 2 | 1 inlet per chamber | [278] |
| Microbiology research: detect cellular dynamics in response to drugs and chemicals | Cell number and fluorescent imaging of protein tagged proteins (microscopy) | S. cerevisiae | 2 | Direct cell printing on agarose-coated glass | 10,000 (100 × 100) Spot sizes of around 200 µm | 1 inlet, channel over all spots | [56] |
5. Conclusions and Outlook
Acknowledgments
Conflicts of Interest
References
- Castel, D.; Pitaval, A.; Debily, M.A.; Gidrol, X. Cell microarrays in drug discovery. Drug Discov. Today 2006, 13–14, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Davis, M.M. Molecular and functional analysis using live cell microarrays. Curr. Opin. Chem. Biol. 2006, 10, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Yarmush, M.L.; King, K.R. Living-cell microarrays. Annu. Rev. Biomed. Eng. 2009, 2009, 11, 235–257. [Google Scholar] [CrossRef] [PubMed]
- Velve-Casquillas, G.; le Berre, M.; Piel, M.; Tran, P.T. Microfluidic tools for cell biological research. Nano Today 2010, 5, 28–47. [Google Scholar] [CrossRef] [PubMed]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Wu, X.; Young, A.T.; Haynes, C.L. Microfluidics-based in vivo mimetic systems for the study of cellular biology. Acc. Chem. Res. 2014, 47, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Mehling, M.; Tay, S. Microfluidic cell culture. Curr. Opin. Biotechnol. 2014, 25, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.G.; Diogo, M.M.; Clark, D.S.; Dordick, J.S.; Cabral, J.M. High-throughput cellular microarray platforms: Applications in drug discovery, toxicology and stem cell research. Trends Biotechnol. 2009, 27, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Gidrol, X.; Fouqué, B.; Ghenim, L.; Haguet, V.; Picollet-D’hahan, N.; Schaack, B. 2D and 3D cell microarrays in pharmacology. Curr. Opin. Pharmacol. 2009, 9, 664–648. [Google Scholar] [CrossRef] [PubMed]
- Sia, S.K.; Whitesides, G.M. Microfluidic devices fabricated in poly(dimethylsiloxane) for biological studies. Electrophoresis 2003, 24, 3563–3576. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Lee, S.H.; Suh, K.Y. Cell research with physically modified microfluidic channels: A review. Lab Chip 2008, 8, 1015–1023. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, C.W.; Yang, J. Cell docking and on-chip monitoring of cellular reactions with a controlled concentration gradient on a microfluidic device. Anal. Chem. 2002, 74, 3991–4001. [Google Scholar] [CrossRef] [PubMed]
- Li, C.W.; Cheung, C.N.; Yang, J.; Tzang, C.H.; Yang, M. PDMS-based microfluidic device with multi-height structures fabricated by single-step photo lithography using printed circuit board as masters. Analyst 2003, 128, 1137–1142. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhang, Q.; Feng, H.; Ang, S.; Chau, F.S.; Liu, W.T. Filter-based microfluidic device as a platform for immunofluorescent assay of microbial cells. Lab Chip 2004, 4, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Valero, A.; Merino, F.; Wolbers, F.; Luttge, R.; Vermes, I.; Andersson, H.; van den Berg, A. Apoptotic cell death dynamics of HL60 cells studied using a microfluidic cell trap device. Lab Chip 2005, 5, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Di Carlo, D.; Aghdam, N.; Lee, L.P. Single-cell enzyme concentrations, kinetics, and inhibition analysis using high-density hydrodynamic cell isolation arrays. Anal. Chem. 2006, 78, 4925–4930. [Google Scholar] [CrossRef] [PubMed]
- Fu, A.Y.; Chou, H.P.; Spence, C.; Arnold, F.H.; Quake, S.R. An integrated microfabricated cell sorter. Anal. Chem. 2002, 74, 2451–2457. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, A.R.; Throndset, W.R.; Whelan, R.J.; Leach, A.M.; Zare, R.N.; Liao, Y.H.; Farrell, K.; Manger, I.D.; Daridon, A. Microfluidic device for single-cell analysis. Anal. Chem. 2003, 75, 3581–3586. [Google Scholar] [CrossRef] [PubMed]
- Tourovskaia, A.; Figueroa-Masot, X.; Folch, A. Differentiation-on-a-chip: A microfluidic platform for long-term cell culture studies. Lab Chip 2005, 5, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Ryley, J.; Pereira-Smith, O.M. Microfluidics device for single cell gene expression analysis in Saccharomyces cerevisiae. Yeast 2006, 23, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Unger, M.A.; Chou, H.P.; Thorsen, T.; Scherer, A.; Quake, S.R. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 2000, 288, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Hung, P.J.; Lee, P.J.; Sabounchi, P.; Lin, R.; Lee, L.P. Continuous perfusion microfluidic cell culture array for high-throughput cell-based assays. Biotechnol. Bioeng. 2005, 89, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wlodkowic, D.; Faley, S.; Zagnoni, M.; Wikswo, J.P.; Cooper, J.M. Microfluidic single-cell array cytometry for the analysis of tumor apoptosis. Anal. Chem. 2009, 81, 5517–5523. [Google Scholar] [CrossRef] [PubMed]
- Beebe, D.J.; Moore, J.S.; Bauer, J.M.; Yu, Q.; Liu, R.H.; Devadoss, C.; Jo, B.H. Functional hydrogel structures for autonomous flow control inside microfluidic channels. Nature 2000, 404, 588–590. [Google Scholar] [CrossRef] [PubMed]
- Moorthy, J.; Beebe, D.J. In situ fabricated porous filters for microsystems. Lab Chip 2003, 3, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Peppas, N.A.; Hilt, J.Z.; Khademhosseini, A.; Langer, R. Hydrogels in biology and medicine: From molecular principles to bionanotechnology. Adv. Mater. 2006, 18, 1345–1360. [Google Scholar] [CrossRef]
- Khademhosseini, A.; Yeh, J.; Jon, S.; Eng, G.; Suh, K.Y.; Burdick, J.A.; Langer, R. Molded polyethylene glycol microstructures for capturing cells within microfluidic channels. Lab Chip 2004, 4, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Khademhosseini, A.; Yeh, J.; Eng, G.; Karp, J.; Kaji, H.; Borenstein, J.; Farokhzad, O.C.; Langer, R. Cell docking inside microwells within reversibly sealed microfluidic channels for fabricating multiphenotype cell arrays. Lab Chip 2005, 5, 1380–1386. [Google Scholar] [CrossRef] [PubMed]
- Love, J.C.; Ronan, J.L.; Grotenberg, G.M.; van der Veen, A.G.; Ploegh, H.L. A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nat. Biotechnol. 2006, 24, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Park, M.C.; Hur, J.Y.; Kwon, K.W.; Park, S.H.; Suh, K.Y. Pumpless, selective docking of yeast cells inside a microfluidic channel induced by receding meniscus. Lab Chip 2006, 6, 988–994. [Google Scholar] [CrossRef] [PubMed]
- Kwon, K.W.; Choi, S.S.; Lee, S.H.; Kim, B.; Lee, S.N.; Park, M.C.; Kim, P.; Hwang, S.Y.; Suh, K.Y. Label-free, microfluidic separation and enrichment of human breast cancer cells by adhesion difference. Lab Chip 2007, 7, 1461–1468. [Google Scholar] [CrossRef] [PubMed]
- Sant, S.; Hancock, M.J.; Donnelly, J.P.; Iyer, D.; Khademhosseini, A. Biomimetic gradient hydrogels for tissue engineering. Can. J. Chem. Eng. 2010, 88, 899–911. [Google Scholar] [CrossRef] [PubMed]
- Rimann, M.; Graf-Hausner, U. Synthetic 3D multicellular systems for drug development. Curr. Opin. Biotechnol. 2012, 23, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Breslin, S.; O’Driscoll, L. Three-dimensional cell culture: The missing link in drug discovery. Drug Discov. Today 2013, 18, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Andersen, T.; Auk-Emblem, P.; Dornish, M. 3D cell culture in alginate hydrogels. Microarrays 2015, 4, 133–161. [Google Scholar] [CrossRef]
- Smidsrød, O.; Skjåk-Braek, G. Alginate as immobilization matrix for cells. Trends Biotechnol. 1990, 8, 71–78. [Google Scholar] [CrossRef]
- Willaert, R.; Baron, G. Gel entrapment and micro-encapsulation: Methods, applications and engineering principles. Rev. Chem. Eng. 1996, 12, 5–205. [Google Scholar] [CrossRef]
- Kong, H.J.; Smith, M.K.; Mooney, D.J. Designing alginate hydrogels to maintain viability of immobilized cells. Biomaterials 2003, 24, 4023–4029. [Google Scholar] [CrossRef]
- Nedovic, V.; Willaert, R. Fundamentals of Cell Immobilisation Biotechnology; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2004. [Google Scholar]
- Nedovic, V.; Willaert, R. Applications of Cell Immobilisation Biotechnology; Springer: Dordrecht, The Netherlands, 2005. [Google Scholar]
- Zhang, S.; Gelain, F.; Zhao, X. Designer self-assembling peptide nanofiber scaffolds for 3D tissue cell cultures. Semin. Cancer Biol. 2005, 15, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Altmann, B.; Welle, A.; Giselbrecht, S.; Truckenmüller, R.; Gottwald, E. The famous versus the inconvenient-or the dawn and the rise of 3D-culture systems. World J. Stem Cells 2009, 1, 43–48. [Google Scholar] [CrossRef] [PubMed]
- DeVolder, R.; Kong, H.J. Hydrogels for in vivo-like three-dimensional cellular studies. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012, 4, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Willaert, R. Cell immobilisation and its applications in biotechnology: Current trends and future prospects. In Fermentation Microbiology and Biotechnology, 3rd ed.; El-Mansi, E.M.T., Bryce, C.F.A., Eds.; CRC Press: Boca Raton, FL, USA, 2012; pp. 313–367. [Google Scholar]
- Luo, Z.; Yue, Y.; Zhang, Y.; Yuan, X.; Gong, J.; Wang, L.; He, B.; Liu, Z.; Sun, Y.; Liu, J.; et al. Designer D-form self-assembling peptide nanofiber scaffolds for 3-dimensional cell cultures. Biomaterials 2013, 34, 4902–4913. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Tan, H. Alginate-based biomaterials for regenerative medicine applications. Materials 2013, 6, 1285–1309. [Google Scholar] [CrossRef]
- Sánchez, P.; Hernández, R.M.; Pedraz, J.L.; Orive, G. Encapsulation of cells in alginate gels. Methods Mol. Biol. 2013, 1051, 313–325. [Google Scholar] [PubMed]
- Ahmed, E.M. Hydrogel: Preparation, characterization, and applications: A review. J. Adv. Res. 2015, 6, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Montanez-Sauri, S.I.; Beebe, D.J.; Sung, K.E. Microscale screening systems for 3D cellular microenvironments: Platforms, advances, and challenges. Cell. Mol. Life Sci. 2015, 72, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, H.; Hirano, A.; Nagasaki, Y.; Okano, T.; Horiike, Y.; Kataoka, K. Two-dimensional multiarray formation of hepatocyte spheroids on a microfabricated PEG-brush surface. Chembiochem 2004, 5, 850–855. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.J.; Su, J.; Berglund, J.D.; Russ, B.V.; Meredith, J.C.; Galis, Z.S. The use of temperature-composition combinatorial libraries to study the effects of biodegradable polymer blend surfaces on vascular cells. Biomaterials 2005, 26, 4557–4567. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.G., Jr.; Eidelman, N.; Kennedy, S.B.; Sehgal, A.; Khatri, C.A.; Washburn, N.R. Combinatorial screening of cell proliferation on poly(l-lactic acid)/poly(d,l-lactic acid) blends. Biomaterials. 2005, 26, 6906–6915. [Google Scholar] [CrossRef] [PubMed]
- Khetani, S.R.; Bhatia, S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008, 26, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Sodunke, T.R.; Turner, K.K.; Caldwell, S.A.; McBride, K.W.; Reginato, M.J.; Noh, H.M. Micropatterns of Matrigel for three-dimensional epithelial cultures. Biomaterials 2007, 28, 4006–4016. [Google Scholar] [CrossRef] [PubMed]
- Flaim, C.J.; Teng, D.; Chien, S.; Bhatia, S.N. Combinatorial signaling microenvironments for studying stem cell fate. Stem Cells Dev. 2008, 17, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, M.; Pla-Roca, M.; Safavieh, R.; Nazarova, E.; Safavieh, M.; Li, H.; Vogel, J.; Juncker, D. Microfluidic perfusion system for culturing and imaging yeast cell microarrays and rapidly exchanging media. Lab Chip 2010, 10, 2449–2457. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.B.; Wu, M.H.; Wang, S.S.; Lee, G.B. Microfluidic cell culture chip with multiplexed medium delivery and efficient cell/scaffold loading mechanisms for high-throughput perfusion 3-dimensional cell culture-based assays. Biomed. Microdevices 2011, 13, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Dos Reis, G.; Fenili, F.; Gianfelice, A.; Bongiorno, G.; Marchesi, D.; Scopelliti, P.E.; Borgonovo, A.; Podestà, A.; Indrieri, M.; Ranucci, E.; et al. Direct microfabrication of topographical and chemical cues for the guided growth of neural cell networks on polyamidoamine hydrogels. Macromol. Biosci. 2010, 10, 842–852. [Google Scholar] [CrossRef] [PubMed]
- Skardal, A.; Atala, A. Biomaterials for integration with 3-D bioprinting. Ann. Biomed. Eng. 2015, 43, 730–746. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.Y.; Kumar, R.A.; Sukumaran, S.M.; Hogg, M.G.; Clark, D.S.; Dordick, J.S. Three-dimensional cellular microarray for high-throughput toxicology assays. Proc. Natl. Acad. Sci. USA 2008, 105, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.G.; Kwon, S.J.; Bale, S.S.; Lee, M.Y.; Diogo, M.M.; Clark, D.S.; Cabral, J.M.; Dordick, J.S. Three-dimensional cell culture microarray for high-throughput studies of stem cell fate. Biotechnol. Bioeng. 2010, 106, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Meli, L.; Jordan, E.T.; Clark, D.S.; Linhardt, R.J.; Dordick, J.S. Influence of a three-dimensional, microarray environment on human cell culture in drug screening systems. Biomaterials 2012, 33, 9087–9096. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.J. Nanoimprint Lithography: Methods and Material Requirements. Adv. Mater. 2007, 19, 495–513. [Google Scholar] [CrossRef]
- Yap, F.L.; Zhang, Y. Protein and cell micropatterning and its integration with micro/nanoparticles assembly. Biosens. Bioelectron. 2007, 22, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Anselme, K.; Davidson, P.; Popa, A.M.; Giazzon, M.; Liley, M.; Ploux, L. The interaction of cells and bacteria with surfaces structured at the nanometre scale. Acta Biomater. 2010, 6, 3824–3846. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Xia, Y.; Whitesides, G.M. Soft lithography for micro- and nanoscale patterning. Nat. Protoc. 2010, 5, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Ekerdt, B.L.; Segalman, R.A.; Schaffer, D.V. Spatial organization of cell-adhesive ligands for advanced cell culture. Biotechnol. J. 2013, 8, 1411–1423. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.V.; Patil, R.; Thombre, D.K.; Gade, W.N. Micro-nanopatterning as tool to study the role of physicochemical properties on cell-surface interactions. J. Biomed. Mater. Res. A 2013, 101, 3019–3032. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, S.A.; Chen, C.S. Microcontact printing: A tool to pattern. Soft Matter. 2007, 3, 168–177. [Google Scholar] [CrossRef]
- Mrksich, M. Using self-assembled monolayers to model the extracellular matrix. Acta Biomater. 2009, 5, 832–841. [Google Scholar] [CrossRef] [PubMed]
- Théry, M.; Piel, M. Adhesive micropatterns for cells: A microcontact printing protocol. Cold Spring Harb. Protoc. 2009, 7. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, J.; Takarada, T.; Yamaguchi, K.; Maeda, M. Recent advances in cell micropatterning techniques for bioanalytical and biomedical sciences. Anal. Sci. 2008, 24, 67–72. [Google Scholar] [CrossRef] [PubMed]
- D’Arcangelo, E.; McGuigan, A.P. Micropatterning strategies to engineer controlled cell and tissue architecture in vitro. Biotechniques 2015, 58, 13–23. [Google Scholar] [PubMed]
- McFarland, C.D.; Thomas, C.H.; DeFilippis, C.; Steele, J.G.; Healy, K.E. Protein adsorption and cell attachment to patterned surfaces. J. Biomed. Mater. Res. 2000, 49, 200–210. [Google Scholar] [CrossRef]
- Falconnet, D.; Csucs, G.; Grandin, H.M.; Textor, M. Surface engineering approaches to micropattern surfaces for cell-based assays. Biomaterials 2006, 27, 3044–3063. [Google Scholar] [CrossRef] [PubMed]
- Hook, A.L.; Thissen, H.; Voelcker, N.H. Surface manipulation of biomolecules for cell microarray applications. Trends Biotechnol. 2006, 24, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, H. Nanofabrication of nonfouling surfaces for micropatterning of cell and microtissue. Molecules 2010, 15, 5525–5546. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Robert, L.; Ouyang, Q.; Taddei, F.; Chen, Y.; Lindner, A.B.; Baigl, D. Microcontact printing of living bacteria arrays with cellular resolution. Nano Lett. 2007, 7, 2068–2072. [Google Scholar] [CrossRef] [PubMed]
- Dertinger, S.K.; Jiang, X.; Li, Z.; Murthy, V.N.; Whitesides, G.M. Gradients of substrate-bound laminin orient axonal specification of neurons. Proc. Natl. Acad. Sci. USA 2002, 99, 12542–12547. [Google Scholar] [CrossRef] [PubMed]
- Cosson, S.; Lutolf, M.P. Microfluidic patterning of protein gradients on biomimetic hydrogel substrates. Methods Cell Biol. 2014, 121, 91–102. [Google Scholar] [PubMed]
- Théry, M. Micropatterning as a tool to decipher cell morphogenesis and functions. J. Cell Sci. 2010, 123, 4201–4213. [Google Scholar] [CrossRef] [PubMed]
- Balakirev, M.Y.; Porte, S.; Vernaz-Gris, M.; Berger, M.; Arié, J.P.; Fouqué, B.; Chatelain, F. Photochemical patterning of biological molecules inside a glass capillary. Anal. Chem. 2005, 77, 5474–5479. [Google Scholar] [CrossRef] [PubMed]
- Bélisle, J.M.; Correia, J.P.; Wiseman, P.W.; Kennedy, T.E.; Costantino, S. Patterning protein concentration using laser-assisted adsorption by photobleaching, LAPAP. Lab Chip 2008, 8, 2164–2167. [Google Scholar] [CrossRef] [PubMed]
- Bélisle, J.M.; Kunik, D.; Costantino, S. Rapid multicomponent optical protein patterning. Lab Chip 2009, 9, 3580–3585. [Google Scholar] [CrossRef] [PubMed]
- Bélisle, J.M.; Mazzaferri, J.; Costantino, S. Laser-assisted adsorption by photobleaching. Methods Cell Biol. 2014, 119, 125–140. [Google Scholar] [PubMed]
- Dillmore, W.S.; Yousaf, M.N.; Mrksich, M. A photochemical method for patterning the immobilization of ligands and cells to self-assembled monolayers. Langmuir 2004, 20, 7223–7231. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Choi, J.C.; Jung, H.R.; Katz, J.S.; Kim, M.G.; Doh, J. Addressable micropatterning of multiple proteins and cells by microscope projection photo lithography based on a protein friendly photoresist. Langmuir 2010, 26, 12112–12118. [Google Scholar] [CrossRef] [PubMed]
- Fink, J.; Théry, M.; Azioune, A.; Dupont, R.; Chatelain, F.; Bornens, M.; Piel, M. Comparative study and improvement of current cell micro-patterning techniques. Lab Chip 2007, 7, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Azioune, A.; Storch, M.; Bornens, M.; Théry, M.; Piel, M. Simple and rapid process for single cell micro-patterning. Lab Chip 2009, 9, 1640–1642. [Google Scholar] [CrossRef] [PubMed]
- Itoga, K.; Kobayashi, J.; Yamato, M.; Okano, T. Micropatterning with a liquid crystal display (LCD) projector. Methods Cell Biol. 2014, 119, 141–158. [Google Scholar] [PubMed]
- Doyle, A.D. Generation of micropatterned substrates using micro photopatterning. Curr. Protoc. Cell Biol. 2009. [Google Scholar] [CrossRef]
- Miller, J.S.; Béthencourt, M.I.; Hahn, M.; Lee, T.R.; West, J.L. Laser-scanning lithography (LSL) for the soft lithographic patterning of cell-adhesive self-assembled monolayers. Biotechnol. Bioeng. 2006, 93, 1060–1068. [Google Scholar] [CrossRef] [PubMed]
- Slater, J.H.; West, J.L. Fabrication of multifaceted, micropatterned surfaces and image-guided patterning using laser scanning lithography. Methods Cell Biol. 2014, 119, 193–217. [Google Scholar] [PubMed]
- Piner, R.D.; Zhu, J.; Xu, F.; Hong, S.; Mirkin, C.A. “Dip-Pen” nanolithography. Science 1999, 283, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Reinhoudt, D.N.; Otto, C.; Subramaniam, V.; Velders, A.H. Strategies for patterning biomolecules with dip-pen nanolithography. Small 2011, 7, 989–1002. [Google Scholar] [CrossRef] [PubMed]
- Hirtz, M.; Greiner, A.M.; Landmann, T.; Bastmeyer, M.; Fuchs, H. Click-chemistry based multi-component microarrays by quill-like pens. Adv. Mater. Interfaces 2014, 1, 1300129. [Google Scholar] [CrossRef]
- Bog, U.; Laue, T.; Grossmann, T.; Beck, T.; Wienhold, T.; Richter, B.; Hirtz, M.; Fuchs, H.; Kalt, H.; Mappes, T. On-chip microlasers for biomolecular detection via highly localized deposition of a multifunctional phospholipid ink. Lab Chip 2013, 13, 2701–2707. [Google Scholar] [CrossRef] [PubMed]
- Rath, P.; Hirtz, M.; Lewes-Malandrakis, G.; Brink, D.; Nebel, C.; Pernice, W.H.P. Diamond nanophotonic circuits functionalized by dip-pen nanolithography. Adv. Opt. Mater. 2015, 3, 328–335. [Google Scholar] [CrossRef]
- Huo, F.; Zheng, Z.; Zheng, G.; Giam, L.R.; Zhang, H.; Mirkin, C.A. Polymer pen lithography. Science 2008, 321, 1658–1660. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, F.; Hirtz, M.; Greiner, A.M.; Weschenfelder, M.; Waterkotte, B.; Bastmeyer, M.; Fuchs, H. Interdigitated multicolored bioink micropatterns by multiplexed polymer pen lithography. Small 2013, 9, 3266–3275. [Google Scholar] [CrossRef] [PubMed]
- Truskett, V.N.; Watts, M.P. Trends in imprint lithography for biological applications. Trends Biotechnol. 2006, 24, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Shi, J.; Zhang, F.; Lei, L.; Li, X.; Wang, L.; Liub, L.; Chen, Y. High resolution and hybrid patterning for single cell attachment. Microelectron. Eng. 2010, 87, 726–729. [Google Scholar] [CrossRef]
- Pesen, D.; Erlandsson, A.; Ulfendahl, M.; Haviland, D.B. Image reversal for direct electron beam patterning of protein coated surfaces. Lab Chip 2007, 7, 1603–1606. [Google Scholar] [CrossRef] [PubMed]
- Rundqvist, J.; Mendoza, B.; Werbin, J.L.; Heinz, W.F.; Lemmon, C.; Romer, L.H.; Haviland, D.B.; Hoh, J.H. High fidelity functional patterns of an extracellular matrix protein by electron beam-based inactivation. J. Am. Chem. Soc. 2007, 129, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Krsko, P.; McCann, T.E.; Thach, T.T.; Laabs, T.L.; Geller, H.M.; Libera, M.R. Length-scale mediated adhesion and directed growth of neural cells by surface-patterned poly(ethylene glycol) hydrogels. Biomaterials 2009, 30, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Medvedev, N.; Palankar, R.; Krauel, K.; Greinacher, A.; Delcea, M. Micropatterned array to assess the interaction of single platelets with platelet factor 4-heparin-IgG complexes. Thromb. Haemost. 2014, 111, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.M.; Jang, S.G.; Choi, D.G.; Kim, S.; Yu, H.K. Nanomachining by colloidal lithography. Small 2006, 2, 458–475. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.Y.; Donahue, H.J. Cell sensing and response to micro- and nanostructured surfaces produced by chemical and topographic patterning. Tissue Eng. 2007, 8, 1879–1891. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.A. Colloidal lithography and current fabrication techniques producing in-plane nanotopography for biological applications. J. R. Soc. Interface 2007, 4, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Dalby, M.J.; McCloy, D.; Robertson, M.; Agheli, H.; Sutherland, D.; Affrossman, S.; Oreffo, R.O. Osteoprogenitor response to semi-ordered and random nanotopographies. Biomaterials 2006, 27, 2980–1987. [Google Scholar] [CrossRef] [PubMed]
- Rettig, J.R.; Folch, A. Large-scale single-cell trapping and imaging using microwell arrays. Anal. Chem. 2005, 77, 5628–5634. [Google Scholar] [CrossRef] [PubMed]
- Balagaddé, F.K.; You, L.; Hansen, C.L.; Arnold, F.H.; Quake, S.R. Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science 2005, 309, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Balagaddé, F.K.; Song, H.; Ozaki, J.; Collins, C.H.; Barnet, M.; Arnold, F.H.; Quake, S.R.; You, L. A synthetic Escherichia coli predator-prey ecosystem. Mol. Syst. Biol. 2008, 4, 187. [Google Scholar] [CrossRef] [PubMed]
- Groisman, A.; Lobo, C.; Cho, H.; Campbell, J.K.; Dufour, Y.S.; Stevens, A.M.; Levchenko, A. A microfluidic chemostat for experiments with bacterial and yeast cells. Nat. Methods 2005, 2, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Falconnet, D.; Niemistö, A.; Taylor, R.J.; Ricicova, M.; Galitski, T.; Shmulevich, I.; Hansen, C.L. High-throughput tracking of single yeast cells in a microfluidic imaging matrix. Lab Chip 2011, 11, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Sjöberg, R.; Leyrat, A.A.; Pirone, D.M.; Chen, C.S.; Quake, S.R. Versatile, fully automated, microfluidic cell culture system. Anal. Chem. 2007, 79, 8557–8563. [Google Scholar] [CrossRef] [PubMed]
- King, K.R.; Wang, S.; Irimia, D.; Jayaraman, A.; Toner, M.; Yarmush, M.L. A high-throughput microfluidic real-time gene expression living cell array. Lab Chip 2007, 7, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Cheong, R.; Wang, C.J.; Levchenko, A. Using a microfluidic device for high-content analysis of cell signaling. Sci. Signal. 2009, 2, l2. [Google Scholar] [CrossRef] [PubMed]
- Lecault, V.; Vaninsberghe, M.; Sekulovic, S.; Knapp, D.J.; Wohrer, S.; Bowden, W.; Viel, F.; McLaughlin, T.; Jarandehei, A.; Miller, M.; et al. High-throughput analysis of single hematopoietic stem cell proliferation in microfluidic cell culture arrays. Nat. Methods 2011, 8, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Long, Z.; Nugent, E.; Javer, A.; Cicuta, P.; Sclavi, B.; Cosentino Lagomarsino, M.; Dorfman, K.D. Microfluidic chemostat for measuring single cell dynamics in bacteria. Lab Chip 2013, 13, 947–954. [Google Scholar] [CrossRef] [PubMed]
- Bell, L.; Seshia, A.; Lando, D.; Laue, E.; Palayret, M.; Lee, S.F.; Klenerman, D. A microfluidic device for the hydrodynamic immobilisation of living fission yeast cells for super-resolution imaging. Sens. Actuators B Chem. 2014, 192, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Robert, L.; Pelletier, J.; Dang, W.L.; Taddei, F.; Wright, A.; Jun, S. Robust growth of Escherichia coli. Curr. Biol. 2010, 20, 1099–1103. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Morgan, M.; Sachs, A.N.; Samorezov, J.; Teller, R.; Shen, Y.; Pienta, K.J.; Takayama, S. Single cell trapping in larger microwells capable of supporting cell spreading and proliferation. Microfluid. Nanofluidics 2010, 8, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Cookson, S.; Ostroff, N.; Pang, W.L.; Volfson, D.; Hasty, J. Monitoring dynamics of single-cell gene expression over multiple cell cycles. Mol. Syst. Biol. 2005, 1, 0024. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.R.; Pang, W.L.; Ostroff, N.A.; Baumgartner, B.L.; Nayak, S.; Tsimring, L.S.; Hasty, J. Metabolic gene regulation in a dynamically changing environment. Nature 2008, 454, 1119–1122. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Evander, M.; Hammarström, B.; Laurell, T. Review of cell and particle trapping in microfluidic systems. Anal. Chim. Acta 2009, 649, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Friedman, N.; Xie, X.S. Stochastic protein expression in individual cells at the single molecule level. Nature 2006, 440, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Di Carlo, D.; Wu, L.Y.; Lee, L.P. Dynamic single cell culture array. Lab Chip 2006, 6, 1445–1449. [Google Scholar] [CrossRef] [PubMed]
- Skelley, A.M.; Kirak, O.; Suh, H.; Jaenisch, R.; Voldman, J. Microfluidic control of cell pairing and fusion. Nat. Methods 2009, 6, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Roach, K.L.; King, K.R.; Uygun, B.E.; Kohane, I.S.; Yarmush, M.L.; Toner, M. High throughput single cell bioinformatics. Biotechnol. Prog. 2009, 25, 1772–1779. [Google Scholar] [CrossRef] [PubMed]
- Pai, J.H.; Wang, Y.; Salazar, G.T.; Sims, C.E.; Bachman, M.; Li, G.P.; Allbritton, N.L. Photoresist with low fluorescence for bioanalytical applications. Anal. Chem. 2007, 79, 8774–8780. [Google Scholar] [CrossRef] [PubMed]
- Shadpour, H.; Zawistowski, J.S.; Herman, A.; Hahn, K.; Allbritton, N.L. Patterning pallet arrays for cell selection based on high-resolution measurements of fluorescent biosensors. Anal. Chim. Acta 2011, 696, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Karp, J.M.; Yeh, J.; Eng, G.; Fukuda, J.; Blumling, J.; Suh, K.Y.; Cheng, J.; Mahdavi, A.; Borenstein, J.; Langer, R.; et al. Controlling size, shape and homogeneity of embryoid bodies using poly(ethylene glycol) microwells. Lab Chip 2007, 7, 786–794. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, S.; Kishi, H.; Tokimitsu, Y.; Kondo, S.; Honda, R.; Rao, S.R.; Omori, M.; Tamiya, E.; Muraguchi, A. Single-cell microarray for analyzing cellular response. Anal. Chem. 2005, 77, 8050–8056. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.C.; Walt, D.R. Application of high-density optical microwell arrays in a live-cell biosensing system. Anal. Biochem. 2000, 278, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, M.; Deutsch, A.; Shirihai, O.; Hurevich, I.; Afrimzon, E.; Shafran, Y.; Zurgil, N. A novel miniature cell retainer for correlative high-content analysis of individual untethered non-adherent cells. Lab Chip 2006, 6, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Chin, V.I.; Taupin, P.; Sanga, S.; Scheel, J.; Gage, F.H.; Bhatia, S.N. Microfabricated platform for studying stem cell fates. Biotechnol. Bioeng. 2004, 88, 399–415. [Google Scholar] [CrossRef] [PubMed]
- Ostuni, E.; Chen, C.S.; Ingber, D.E.; Whitesides, G.M. Selective deposition of proteins and cells in arrays of microwells. Langmuir 2001, 17, 2828–2834. [Google Scholar] [CrossRef]
- Wang, Y.; Salazar, G.T.; Pai, J.H.; Shadpour, H.; Sims, C.E.; Allbritton, N.L. Micropallet arrays with poly(ethylene glycol) walls. Lab Chip 2008, 8, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Hancock, M.J.; Brigham, M.D.; Khademhosseini, A. Cell confinement in patterned nanoliter droplets in a microwell array by wiping. J. Biomed. Mater. Res. A 2010, 93, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Barbulovic-Nad, I.; Lucente, M.; Sun, Y.; Zhang, M.; Wheeler, A.R.; Bussmann, M. Bio-microarray fabrication techniques--a review. Crit. Rev. Biotechnol. 2006, 26, 237–259. [Google Scholar] [CrossRef] [PubMed]
- Guillemot, F.; Souquet, A.; Catros, S.; Guillotin, B.; Lopez, J.; Faucon, M.; Pippenger, B.; Bareille, R.; Rémy, M.; Bellance, S.; et al. High-throughput laser printing of cells and biomaterials for tissue engineering. Acta Biomater. 2010, 6, 2494–2500. [Google Scholar] [CrossRef] [PubMed]
- Schaack, B.; Reboud, J.; Combe, S.; Fouqué, B.; Berger, F.; Boccard, S.; Filhol-Cochet, O.; Chatelain, F.A. “DropChip” cell array for DNA and siRNA transfection combined with drug screening. NanoBiotech 2005, 1, 183–189. [Google Scholar] [CrossRef]
- Xu, T.; Jin, J.; Gregory, C.; Hickman, J.J.; Boland, T. Inkjet printing of viable mammalian cells. Biomaterials 2005, 26, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Ringeisen, B.R.; Othon, C.M.; Barron, J.A.; Young, D.; Spargo, B.J. Jet-based methods to print living cells. Biotechnol. J. 2006, 1, 930–948. [Google Scholar] [CrossRef] [PubMed]
- Roth, E.A.; Xu, T.; Das, M.; Gregory, C.; Hickman, J.J.; Boland, T. Inkjet printing for high-throughput cell patterning. Biomaterials 2004, 25, 3707–3715. [Google Scholar] [CrossRef] [PubMed]
- Guillemot, F.; Mironov, V.; Nakamura, M. Bioprinting is coming of age: Report from the International Conference on Bioprinting and Biofabrication in Bordeaux (3B’09). Biofabrication 2010, 2, 010201. [Google Scholar] [CrossRef] [PubMed]
- Ferris, C.J.; Gilmore, K.G.; Wallace, G.G. In het Panhuis, M. Biofabrication: An overview of the approaches used for printing of living cells. Appl. Microbiol. Biotechnol. 2013, 97, 4243–4258. [Google Scholar] [CrossRef] [PubMed]
- Narayanaswamy, R.; Niu, W.; Scouras, A.D.; Hart, G.T.; Davies, J.; Ellington, A.D.; Iyer, V.R.; Marcotte, E.M. Systematic profiling of cellular phenotypes with spotted cell microarrays reveals mating-pheromone response genes. Genome Biol. 2006, 7, R6. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Niu, W.; Yao, J.; Mohr, S.; Marcotte, E.M.; Lambowitz, A.M. Group II intron protein localization and insertion sites are affected by polyphosphate. PLoS Biol. 2008, 6, e150. [Google Scholar] [CrossRef] [PubMed]
- Hart, T.; Zhao, A.; Garg, A.; Bolusani, S.; Marcotte, E.M. Human cell chips: Adapting DNA microarray spotting technology to cell-based imaging assays. PLoS ONE 2009, 4, e7088. [Google Scholar] [CrossRef] [PubMed]
- Odde, D.J.; Renn, M.J. Laser-guided direct writing for applications in biotechnology. Trends Biotechnol. 1999, 17, 385–389. [Google Scholar] [CrossRef]
- Nahmias, Y.; Odde, D.J. Micropatterning of living cells by laser-guided direct writing: Application to fabrication of hepatic-endothelial sinusoid-like structures. Nat. Protoc. 2006, 1, 2288–2296. [Google Scholar] [CrossRef] [PubMed]
- Tasoglu, S.; Demirci, U. Bioprinting for stem cell research. Trends Biotechnol. 2013, 31, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.K.; Ringeisen, B.R.; Callahan, J.; Brooks, M.; Bubb, D.M.; Wu, H.D.; Piqué, A.; Spargo, B.; McGill, R.A.; Chrisey, D.B. The deposition, structure, pattern deposition, and activity of biomaterial thin-films by matrix-assisted pulsed-laser evaporation (MAPLE) and MAPLE direct write. Thin Solid Films 2001, 398–399, 607–614. [Google Scholar] [CrossRef]
- Barron, J.A.; Rosen, R.; Jones-Meehan, J.; Spargo, B.J.; Belkin, S.; Ringeisen, B.R. Biological laser printing of genetically modified Escherichia coli for biosensor applications. Biosens. Bioelectron. 2004, 15, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Barron, J.A.; Wu, P.; Ladouceur, H.D.; Ringeisen, B.R. Biological laser printing: A novel technique for creating heterogeneous 3-dimensional cell patterns. Biomed. Microdevices 2004, 6, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Guillotin, B.; Souquet, A.; Catros, S.; Duocastella, M.; Pippenger, B.; Bellance, S.; Bareille, R.; Rémy, M.; Bordenave, L.; Amédée, J.; et al. Laser assisted bioprinting of engineered tissue with high cell density and microscale organization. Biomaterials 2010, 31, 7250–7256. [Google Scholar] [CrossRef] [PubMed]
- Schiele, N.R.; Chrisey, D.B.; Corr, D.T. Gelatin-based laser direct-write technique for the precise spatial patterning of cells. Tissue Eng. Part C Methods 2011, 17, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Schiele, N.R.; Corr, D.T.; Huang, Y.; Raof, N.A.; Xie, Y.; Chrisey, D.B. Laser-based direct-write techniques for cell printing. Biofabrication 2010, 2, 032001. [Google Scholar] [CrossRef] [PubMed]
- Demirci, U.; Montesano, G. Single cell epitaxy by acoustic picolitre droplets. Lab Chip 2007, 7, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.Y.; Diamond, S.L. Advancing microarray assembly with acoustic dispensing technology. Anal. Chem. 2009, 81, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Frampton, J.P.; Raghavan, S.; Sabahi-Kaviani, R.; Luker, G.; Deng, C.X.; Takayama, S. Rapid generation of multiplexed cell cocultures using acoustic droplet ejection followed by aqueous two-phase exclusion patterning. Tissue Eng. Part C Methods 2012, 18, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Torr, G.R. The acoustic radiation force. Am. J. Phys. 1984, 52, 402–408. [Google Scholar] [CrossRef]
- Elrod, S.A.; Hadimioglu, B.; Khuri-Yakub, B.T.; Rawson, E.G.; Richley, E.; Quate, C.F.; Mansour, N.N.; Lundgren, T.S. Nozzleless droplet formation with focused acoustic beams. J. Appl. Phys. 1989, 65, 3441–3447. [Google Scholar] [CrossRef]
- Shi, J.; Ahmed, D.; Mao, X.; Lin, S.C.; Lawit, A.; Huang, T.J. Acoustic tweezers: Patterning cells and microparticles using standing surface acoustic waves (SSAW). Lab Chip 2009, 9, 2890–2895. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Lin, S.C.; Kiraly, B.; Yue, H.; Li, S.; Chiang, I.K.; Shi, J.; Benkovic, S.J.; Huang, T.J. On-chip manipulation of single microparticles, cells, and organisms using surface acoustic waves. Proc. Natl. Acad. Sci. USA 2012, 109, 11105–11109. [Google Scholar] [CrossRef] [PubMed]
- Demirci, U.; Montesano, G. Cell encapsulating droplet vitrification. Lab Chip 2007, 7, 1428–1433. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.S.; Adler, D.; Xu, F.; Kayaalp, E.; Nureddin, A.; Anchan, R.M.; Maas, R.L.; Demirci, U. Vitrification and levitation of a liquid droplet on liquid nitrogen. Proc. Natl. Acad. Sci. USA 2010, 107, 4596–4600. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.; Kim, Y.G.; Dong, L.; Lombardi, M.; Haeggstrom, E.; Jensen, R.V.; Hsiao, L.L.; Demirci, U. Drop-on-demand single cell isolation and total RNA analysis. PLoS ONE 2011, 6, e17455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, J.W.; Khetani, S.R.; Bhatia, S.N. In vitro zonation and toxicity in a hepatocyte bioreactor. Toxicol. Sci. 2005, 84, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Kane, B.J.; Zinner, M.J.; Yarmush, M.L.; Toner, M. Liver-specific functional studies in a microfluidic array of primary mammalian hepatocytes. Anal. Chem. 2006, 78, 4291–4298. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.N.; Jiang, X.; Ryan, D.; Whitesides, G.M. Compatibility of mammalian cells on surfaces of poly(dimethylsiloxane). Langmuir 2004, 20, 11684–11691. [Google Scholar] [CrossRef] [PubMed]
- Stangegaard, M.; Petronis, S.; Jørgensen, A.M.; Christensen, C.B.; Dufva, M. A biocompatible micro cell culture chamber (microCCC) for the culturing and on-line monitoring of eukaryote cells. Lab Chip 2006, 6, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- Brischwein, M.; Motrescu, E.R.; Cabala, E.; Otto, A.M.; Grothe, H.; Wolf, B. Functional cellular assays with multiparametric silicon sensor chips. Lab Chip 2003, 3, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Peterman, M.C.; Mehenti, N.Z.; Bilbao, K.V.; Lee, C.J.; Leng, T.; Noolandi, J.; Bent, S.F.; Blumenkranz, M.S.; Fishman, H.A. The Artificial Synapse Chip: A flexible retinal interface based on directed retinal cell growth and neurotransmitter stimulation. Artif. Organs 2003, 27, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.G.; Levenberg, S.; Langer, R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat. Biotechnol. 2004, 22, 863–866. [Google Scholar] [CrossRef] [PubMed]
- Bettinger, C.J.; Weinberg, E.J.; Kulig, K.M.; Vacanti, J.P.; Wang, Y.; Borenstein, J.T.; Langer, R. Three-dimensional microfluidic tissue-engineering scaffolds using a flexible biodegradable polymer. Adv. Mater. 2005, 18, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, R.; Solomon, M.; Satyamoorthy, K.; Martin, L.E.; Lingen, M.W. Tissue microarray-a high-throughput molecular analysis in head and neck cancer. J. Oral Pathol. Med. 2008, 37, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Zhu, X.; Futai, N.; Cho, B.S.; Takayama, S. Computerized microfluidic cell culture using elastomeric channels and Braille displays. Proc. Natl. Acad. Sci. USA 2004, 101, 15861–1586. [Google Scholar] [CrossRef] [PubMed]
- Burks, G.A.; Velegol, S.B.; Paramonova, E.; Lindenmuth, B.E.; Feick, J.D.; Logan, B.E. Macroscopic and nanoscale measurements of the adhesion of bacteria with varying outer layer surface composition. Langmuir 2003, 19, 2366–2371. [Google Scholar] [CrossRef]
- Katsikogianni, M.G.; Missirlis, Y.F. Bacterial adhesion onto materials with specific surface chemistries under flow conditions. J. Mater. Sci. Mater. Med. 2010, 21, 963–968. [Google Scholar] [CrossRef] [PubMed]
- Kasas, S.; Ruggeri, F.S.; Benadiba, C.; Maillard, C.; Stupar, P.; Tournu, H.; Dietler, G.; Longo, G. Detecting nanoscale vibrations as signature of life. Proc. Natl. Acad. Sci. USA 2015, 112, 378–381. [Google Scholar] [CrossRef] [PubMed]
- Ming, F.; Eisenthal, R.; Whish, W.J.; Hubble, J. The kinetics of affinity-mediated cell-surface attachment. Enzym. Microb. Technol. 2000, 26, 216–221. [Google Scholar] [CrossRef]
- Uhlendorf, J.; Miermont, A.; Delaveau, T.; Charvin, G.; Fages, F.; Bottani, S.; Batt, G.; Hersen, P. Long-term model predictive control of gene expression at the population and single-cell levels. Proc. Natl. Acad. Sci. USA 2012, 109, 14271–14276. [Google Scholar] [CrossRef] [PubMed]
- Yamazoe, H.; Tanabe, T. Cell micropatterning on an albumin-based substrate using an inkjet printing technique. J. Biomed. Mater. Res. A 2009, 91, 1202–1209. [Google Scholar] [CrossRef] [PubMed]
- Douglas, E.S.; Chandra, R.A.; Bertozzi, C.R.; Mathies, R.A.; Francis, M.B. Self-assembled cellular microarrays patterned using DNA barcodes. Lab Chip 2007, 7, 1442–1448. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.S.; Mrksich, M.; Huang, S.; Whitesides, G.M.; Ingber, D.E. Geometric control of cell life and death. Science 1997, 276, 1425–1428. [Google Scholar] [CrossRef] [PubMed]
- Degot, S.; Auzan, M.; Chapuis, V.; Béghin, A.; Chadeyras, A.; Nelep, C.; Calvo-Muñoz, M.L.; Young, J.; Chatelain, F.; Fuchs, A. Improved visualization and quantitative analysis of drug effects using micropatterned cells. J. Vis. Exp. 2010, 46, 2514. [Google Scholar] [CrossRef] [PubMed]
- Schauer, K.; Duong, T.; Bleakley, K.; Bardin, S.; Bornens, M.; Goud, B. Probabilistic density maps to study global endomembrane organization. Nat. Methods 2010, 7, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.N.; Yarmush, M.L.; Toner, M. Controlling cell interactions by micropatterning in co-cultures: Hepatocytes and 3T3 fibroblasts. J. Biomed. Mater. Res. 1997, 34, 189–199. [Google Scholar] [CrossRef]
- Wang, X.; Song, W.; Kawazoe, N.; Chen, G. The osteogenic differentiation of mesenchymal stem cells by controlled cell-cell interaction on micropatterned surfaces. J. Biomed. Mater. Res. A 2013, 101, 3388–3395. [Google Scholar] [CrossRef] [PubMed]
- Bhadriraju, K.; Chen, C.S. Engineering cellular microenvironments to improve cell-based drug testing. Drug Discov. Today 2002, 7, 612–620. [Google Scholar] [CrossRef]
- De Gans, B.J.; Schubert, U.S. Inkjet printing of polymer micro-arrays and libraries: Instrumentation, requirements, and perspectives. Macromol. Rapid Commun. 2003, 24, 659–666. [Google Scholar] [CrossRef]
- Kovarik, M.L.; Gach, P.C.; Ornoff, D.M.; Wang, Y.; Balowski, J.; Farrag, L.; Allbritton, N.L. Micro total analysis systems for cell biology and biochemical assays. Anal. Chem. 2012, 84, 516–540. [Google Scholar] [CrossRef] [PubMed]
- Quake, S.R.; Scherer, A. From micro- to nanofabrication with soft materials. Science 2000, 290, 1536–1540. [Google Scholar] [CrossRef] [PubMed]
- Thorsen, T.; Maerkl, S.J.; Quake, S.R. Microfluidic large-scale integration. Science 2002, 298, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Melin, J.; Quake, S.R. Microfluidic large-scale integration: The evolution of design rules for biological automation. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 213–231. [Google Scholar] [CrossRef] [PubMed]
- Groisman, A.; Enzelberger, M.; Quake, S.R. Microfluidic memory and control devices. Science 2003, 300, 955–958. [Google Scholar] [CrossRef] [PubMed]
- Toepke, M.W.; Abhyankar, V.V.; Beebe, D.J. Microfluidic logic gates and timers. Lab Chip 2007, 7, 1449–1453. [Google Scholar] [CrossRef] [PubMed]
- Dénervaud, N.; Becker, J.; Delgado-Gonzalo, R.; Damay, P.; Rajkumar, A.S.; Unser, M.; Shore, D.; Naef, F.; Maerkl, S.J. A chemostat array enables the spatio-temporal analysis of the yeast proteome. Proc. Natl. Acad. Sci. USA 2013, 110, 15842–15847. [Google Scholar] [CrossRef] [PubMed]
- Leibacher, I.; Schoendube, J.; Dual, J.; Zengerle, R.; Koltay, P. Enhanced single-cell printing by acoustophoretic cell focusing. Biomicrofluidics 2015, 9, 024109. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.J.; di Carlo, D. Single cell analysis for quantitative systems biology. In Single Cell Analysis: Technologies and Applications; Anselmetti, D., Ed.; Wiley-VCH Verlag GmbH & Co.: Weinheim, Germany, 2009; pp. 135–160. [Google Scholar]
- Rao, C.V.; Wolf, D.M.; Arkin, A.P. Control, exploitation and tolerance of intracellular noise. Nature 2002, 420, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Lidstrom, M.E.; Meldrum, D.R. Life-on-a-chip. Nat. Rev. Microbiol. 2003, 1, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Sigal, A.; Milo, R.; Cohen, A.; Geva-Zatorsky, N.; Klein, Y.; Liron, Y.; Rosenfeld, N.; Danon, T.; Perzov, N.; Alon, U. Variability and memory of protein levels in human cells. Nature 2006, 444, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.R.; Ghaemmaghami, S.; Ihmels, J.; Breslow, D.K.; Noble, M.; DeRisi, J.L.; Weissman, J.S. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 2006, 441, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Xiao, J.; Ren, X.; Lao, K.; Xie, X.S. Probing gene expression in live cells, one protein molecule at a time. Science 2006, 311, 1600–1603. [Google Scholar] [CrossRef] [PubMed]
- Elowitz, M.B.; Levine, A.J.; Siggia, E.D.; Swain, P.S. Stochastic gene expression in a single cell. Science 2002, 297, 1183–1186. [Google Scholar] [CrossRef] [PubMed]
- Levsky, J.M.; Singer, R.H. Gene expression and the myth of the average cell. Trends Cell Biol. 2003, 13, 4–6. [Google Scholar] [CrossRef]
- Lahav, G.; Rosenfeld, N.; Sigal, A.; Geva-Zatorsky, N.; Levine, A.J.; Elowitz, M.B.; Alon, U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 2004, 36, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Lidstrom, M.E.; Konopka, M.C. The role of physiological heterogeneity in microbial population behavior. Nat. Chem. Biol. 2010, 6, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Bodovitz, S. Single cell analysis: The new frontier in “omics”. Trends Biotechnol. 2010, 28, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Raser, J.M.; O’Shea, E.K. Noise in gene expression: Origins, consequences, and control. Science 2005, 309, 2010–2013. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, N.; Young, J.W.; Alon, U.; Swain, P.S.; Elowitz, M.B. Gene regulation at the single-cell level. Science 2005, 307, 1962–195. [Google Scholar] [CrossRef] [PubMed]
- Mettetal, J.T.; Muzzey, D.; Pedraza, J.M.; Ozbudak, E.M.; van Oudenaarden, A. Predicting stochastic gene expression dynamics in single cells. Proc. Natl. Acad. Sci. USA 2006, 103, 7304–7309. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, U.S. Signaling in small subcellular volumes. I. Stochastic and diffusion effects on individual pathways. Biophys. J. 2004, 87, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Kholodenko, B.N.; Hancock, J.F.; Kolch, W. Signalling ballet in space and time. Nat. Rev. Mol. Cell Biol. 2010, 11, 414–426. [Google Scholar] [CrossRef] [PubMed]
- Le Gac, S.; van den Berg, A. Single cells as experimentation units in lab-on-a-chip devices. Trends Biotechnol. 2010, 28, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Z.Z.; Li, Q. Microfluidic techniques for dynamic single-cell analysis. Microchim. Acta 2010, 168, 177–195. [Google Scholar] [CrossRef]
- Walling, M.A.; Shepard, J.R. Cellular heterogeneity and live cell arrays. Chem. Soc. Rev. 2011, 40, 4049–4076. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Marshall, D. Microfluidics for single cell analysis. Curr. Opin. Biotechnol. 2012, 23, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Dusny, C.; Schmid, A. Microfluidic single-cell analysis links boundary environments and individual microbial phenotypes. Environ. Microbiol. 2015, 17, 1839–1856. [Google Scholar] [CrossRef] [PubMed]
- Junkin, M.; Tay, S. Microfluidic single-cell analysis for systems immunology. Lab Chip 2014, 14, 1246–1260. [Google Scholar] [CrossRef] [PubMed]
- Pedraza, J.M.; van Oudenaarden, A. Noise propagation in gene networks. Science 2005, 307, 1965–1969. [Google Scholar] [CrossRef] [PubMed]
- Charvin, G.; Cross, F.R.; Siggia, E.D. A microfluidic device for temporally controlled gene expression and long-term fluorescent imaging in unperturbed dividing yeast cells. PLoS ONE 2008, 3, e1468. [Google Scholar] [CrossRef] [PubMed]
- Charvin, G.; Cross, F.R.; Siggia, E.D. Forced periodic expression of G1 cyclins phase-locks the budding yeast cell cycle. Proc. Natl. Acad. Sci. USA 2009, 106, 6632–6637. [Google Scholar] [CrossRef] [PubMed]
- Rowat, A.C.; Bird, J.C.; Agresti, J.J.; Rando, O.J.; Weitz, D.A. Tracking lineages of single cells in lines using a microfluidic device. Proc. Natl. Acad. Sci. USA 2009, 106, 18149–18154. [Google Scholar] [CrossRef] [PubMed]
- Howson, R.; Huh, W.K.; Ghaemmaghami, S.; Falvo, J.V.; Bower, K.; Belle, A.; Dephoure, N.; Wykoff, D.D.; Weissman, J.S.; O’Shea, E.K. Construction, verification and experimental use of two epitope-tagged collections of budding yeast strains. Comp. Funct. Genomics 2005, 6, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.A.; Geva-Zatorsky, N.; Eden, E.; Frenkel-Morgenstern, M.; Issaeva, I.; Sigal, A.; Milo, R.; Cohen-Saidon, C.; Liron, Y.; Kam, Z.; et al. Dynamic proteomics of individual cancer cells in response to a drug. Science 2008, 322, 1511–1516. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.L.; Haskins, J.R.; Giuliano, K. (Eds.) High Content Screening: A Powerful Approach to Systems Cell Biology and Drug Discovery; Humana Press: Totowa, NJ, USA, 2007.
- Haney, S.A. (Ed.) High Content Screening: Science, Techniques, and Applications; John Wiley&Sons: Hoboken, NJ, USA, 2008.
- Lipp, P.; Kaestner, L. Image-based high-content screening–a view from basic sciences. In High-Throughput Screening in Drug Discovery; Hüser, J., Ed.; Wiley VCH: Weinheim, Germany, 2006; pp. 129–149. [Google Scholar]
- Huh, W.K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Conrad, C.; Erfle, H.; Warnat, P.; Daigle, N.; Lörch, T.; Ellenberg, J.; Pepperkok, R.; Eils, R. Automatic identification of subcellular phenotypes on human cell arrays. Genome Res. 2004, 14, 1130–1136. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.L.; Ohtani, M.; Sawai, H.; Sano, F.; Saka, A.; Watanabe, D.; Yukawa, M.; Ohya, Y.; Morishita, S. SCMD: Saccharomyces cerevisiae Morphological Database. Nucleic Acids Res. 2004, 32, D319–D322. [Google Scholar] [CrossRef] [PubMed]
- Vaisberg, E.A.; Lenzi, D.; Hansen, R.L.; Keon, B.H.; Finer, J.T. An infrastructure for high-throughput microscopy: Instrumentation, informatics, and integration. Methods Enzymol. 2006, 414, 484–512. [Google Scholar] [PubMed]
- Loo, L.H.; Wu, L.F.; Altschuler, S.J. Image-based multivariate profiling of drug responses from single cells. Nat. Methods 2007, 4, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, A.E. Image-based chemical screening. Nat. Chem. Biol. 2007, 3, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Locke, J.C.; Elowitz, M.B. Using movies to analyse gene circuit dynamics in single cells. Nat. Rev. Microbiol. 2009, 7, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, Y.; Choi, P.J.; Li, G.W.; Chen, H.; Babu, M.; Hearn, J.; Emili, A.; Xie, X.S. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 2010, 329, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Derveaux, S.; Stubbe, B.G.; Braeckmans, K.; Roelant, C.; Sato, K.; Demeester, J.; de Smedt, S.C. Synergism between particle-based multiplexing and microfluidics technologies may bring diagnostics closer to the patient. Anal. Bioanal. Chem. 2008, 391, 2453–2467. [Google Scholar] [CrossRef] [PubMed]
- Situma, C.; Hashimoto, M.; Soper, S.A. Merging microfluidics with microarray-based bioassays. Biomol. Eng. 2006, 23, 213–231. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, D.; Dittrich, P.S. Advances in microfluidics for drug discovery. Expert Opin. Drug Discov. 2010, 5, 1081–1094. [Google Scholar] [CrossRef] [PubMed]
- Wlodkowic, D.; Cooper, J.M. Microfluidic cell arrays in tumor analysis: New prospects for integrated cytomics. Expert Rev. Mol. Diagn. 2010, 10, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Marimuthu, M.; Kim, S. Microfluidic cell coculture methods for understanding cell biology, analyzing bio/pharmaceuticals, and developing tissue constructs. Anal. Biochem. 2011, 413, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Neuži, P.; Giselbrecht, S.; Länge, K.; Huang, T.J.; Manz, A. Revisiting lab-on-a-chip technology for drug discovery. Nat. Rev. Drug Discov. 2012, 11, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.T.; Yu, L.; Li, P.; Dong, H.; Wang, Y.; Liu, Y.; Li, C.M. On-chip investigation of cell-drug interactions. Adv. Drug Deliv. Rev. 2013, 65, 1556–1574. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wu, J.; Wang, S.; Durmus, N.G.; Gurkan, U.A.; Demirci, U. Microengineering methods for cell-based microarrays and high-throughput drug-screening applications. Biofabrication 2011, 3, 034101. [Google Scholar] [CrossRef] [PubMed]
- Angres, B. Cell microarrays. Expert Rev. Mol. Diagn. 2005, 5, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.T.; Vriens, K.; Cornaglia, M.; Gijs, M.; Kokalj, T.; Thevissen, K.; Geeraerd, A.; Cammue, B.P.; Puers, R.; Lammertyn, J. Digital microfluidics for time-resolved cytotoxicity studies on single non-adherent yeast cells. Lab Chip 2015, 15, 1852–1860. [Google Scholar] [CrossRef] [PubMed]
- Pasirayi, G.; Scott, S.M.; Islam, M.; O’Hare, L.; Bateson, S.; Ali, Z. Low cost microfluidic cell culture array using normally closed valves for cytotoxicity assay. Talanta 2014, 129, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Tourovskaia, A.; Folch, A. Biology on a chip: Microfabrication for studying the behavior of cultured cells. Crit. Rev. Biomed. Eng. 2003, 31, 423–488. [Google Scholar] [CrossRef] [PubMed]
- Meyvantsson, I.; Beebe, D.J. Cell culture models in microfluidic systems. Annu. Rev. Anal. Chem. (Palo Alto Calif.) 2008, 1, 423–449. [Google Scholar] [CrossRef] [PubMed]
- Salieb-Beugelaar, G.B.; Simone, G.; Arora, A.; Philippi, A.; Manz, A. Latest developments in microfluidic cell biology and analysis systems. Anal. Chem. 2010, 82, 4848–4864. [Google Scholar] [CrossRef] [PubMed]
- Berthuy, O.I.; Blum, L.J.; Marquette, C.A. Cells on chip for multiplex screening. Biosens. Bioelectron. 2015. [Google Scholar] [CrossRef] [PubMed]
- Cambier, T.; Honegger, T.; Vanneaux, V.; Berthier, J.; Peyrade, D.; Blanchoin, L.; Larghero, J.; Théry, M. Design of a 2D no-flow chamber to monitor hematopoietic stem cells. Lab Chip 2015, 15, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Lattermann, C.; Büchs, J. Microscale and miniscale fermentation and screening. Curr. Opin. Biotechnol. 2014, 23, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Marques, M.P.; Fernandes, P. Microfluidic devices: Useful tools for bioprocess intensification. Molecules 2011, 16, 8368–8401. [Google Scholar] [CrossRef] [PubMed]
- Love, K.R.; Panagiotou, V.; Jiang, B.; Stadheim, T.A.; Love, J.C. Integrated single-cell analysis shows Pichia pastoris secretes protein stochastically. Biotechnol. Bioeng. 2010, 106, 319–325. [Google Scholar] [PubMed]
- Dai, J.; Yoon, S.H.; Sim, H.Y.; Yang, Y.S.; Oh, T.K.; Kim, J.F.; Hong, J.W. Charting microbial phenotypes in multiplex nanoliter batch bioreactors. Anal. Chem. 2013, 85, 5892–5899. [Google Scholar] [CrossRef] [PubMed]
- Grünberger, A.; Wiechert, W.; Kohlheyer, D. Single-cell microfluidics: Opportunity for bioprocess development. Curr. Opin. Biotechnol. 2014, 29, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Mustafi, N.; Grünberger, A.; Mahr, R.; Helfrich, S.; Nöh, K.; Blombach, B.; Kohlheyer, D.; Frunzke, J. Application of a genetically encoded biosensor for live cell imaging of l-valine production in pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum strains. PLoS ONE 2014, 9, e85731. [Google Scholar] [CrossRef] [PubMed]
- Unthan, S.; Grünberger, A.; van Ooyen, J.; Gätgens, J.; Heinrich, J.; Paczia, N.; Wiechert, W.; Kohlheyer, D.; Noack, S. Beyond growth rate 0.6: What drives Corynebacterium glutamicum to higher growth rates in defined medium. Biotechnol. Bioeng. 2014, 111, 359–371. [Google Scholar] [CrossRef] [PubMed]
- El-Ali, J.; Sorger, P.K.; Jensen, K.F. Cells on chips. Nature 2006, 442, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Szita, N.; Boccazzi, P.; Zhang, Z.; Boyle, P.; Sinskey, A.J.; Jensen, K.F. Development of a multiplexed microbioreactor system for high-throughput bioprocessing. Lab Chip 2005, 5, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Boccazzi, P.; Zanzotto, A.; Szita, N.; Bhattacharya, S.; Jensen, K.F.; Sinskey, A.J. Gene expression analysis of Escherichia coli grown in miniaturized bioreactor platforms for high-throughput analysis of growth and genomic data. Appl. Microbiol. Biotechnol. 2005, 68, 518–532. [Google Scholar] [CrossRef] [PubMed]
- Ziolkowska, K.; Jedrych, E.; Kwapiszewski, R.; Lopacinska, J.M.; Skolimowski, M.; Chudy, M. PDMS/glass microfluidic cell culture system for cytotoxicity tests and cells passage. Sens. Actuators B: Chem. 2010, 145, 533–542. [Google Scholar] [CrossRef]
- Li, X.; Xue, X.; Li, P.C. Real-time detection of the early event of cytotoxicity of herbal ingredients on single leukemia cells studied in a microfluidic biochip. Integr. Biol. (Camb.) 2009, 1, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, M.; Hayashi, T.; Mori, T.; Yoshino, T.; Nakasono, S.; Matsunaga, T. Microfluidic device with chemical gradient for single-cell cytotoxicity assays. Anal. Chem. 2011, 83, 3648–3654. [Google Scholar] [CrossRef] [PubMed]
- Chanasakulniyom, M.; Glidle, A.; Cooper, J.M. Cell proliferation and migration inside single cell arrays. Lab Chip 2015, 15, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; King, K.R.; Wieder, K.J.; Toner, M.; Yarmush, M.L.; Jayaraman, A. Dynamic gene expression profiling using a microfabricated living cell array. Anal. Chem. 2004, 76, 4098–4103. [Google Scholar] [CrossRef] [PubMed]
- Kim, L.; Vahey, M.D.; Lee, H.Y.; Voldman, J. Microfluidic arrays for logarithmically perfused embryonic stem cell culture. Lab Chip 2006, 6, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Park, M.C.; Hur, J.Y.; Cho, H.S.; Park, S.H.; Suh, K.Y. High-throughput single-cell quantification using simple microwell-based cell docking and programmable time-course live-cell imaging. Lab Chip 2011, 11, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Grünberger, A.; Paczia, N.; Probst, C.; Schendzielorz, G.; Eggeling, L.; Noack, S.; Wiechert, W.; Kohlheyer, D. A disposable picolitre bioreactor for cultivation and investigation of industrially relevant bacteria on the single cell level. Lab Chip 2012, 12, 2060–2068. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Kim, M.C.; Thorsen, T.; Wang, Z. A self-contained microfluidic cell culture system. Biomed. Microdevices 2009, 11, 1233–1237. [Google Scholar] [CrossRef] [PubMed]
- Gottschamel, J.; Richter, L.; Mak, A.; Jungreuthmayer, C.; Birnbaumer, G.; Milnera, M.; Brückl, H.; Ertl, P. Development of a disposable microfluidic biochip for multiparameter cell population measurements. Anal. Chem. 2009, 81, 8503–8512. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Willaert, R.G.; Goossens, K. Microfluidic Bioreactors for Cellular Microarrays. Fermentation 2015, 1, 38-78. https://doi.org/10.3390/fermentation1010038
Willaert RG, Goossens K. Microfluidic Bioreactors for Cellular Microarrays. Fermentation. 2015; 1(1):38-78. https://doi.org/10.3390/fermentation1010038
Chicago/Turabian StyleWillaert, Ronnie G., and Katty Goossens. 2015. "Microfluidic Bioreactors for Cellular Microarrays" Fermentation 1, no. 1: 38-78. https://doi.org/10.3390/fermentation1010038