MicroRNA Expression in Bovine Cumulus Cells in Relation to Oocyte Quality
Abstract
:1. Introduction
2. Results
2.1. Comparison of Individual versus Group Culture
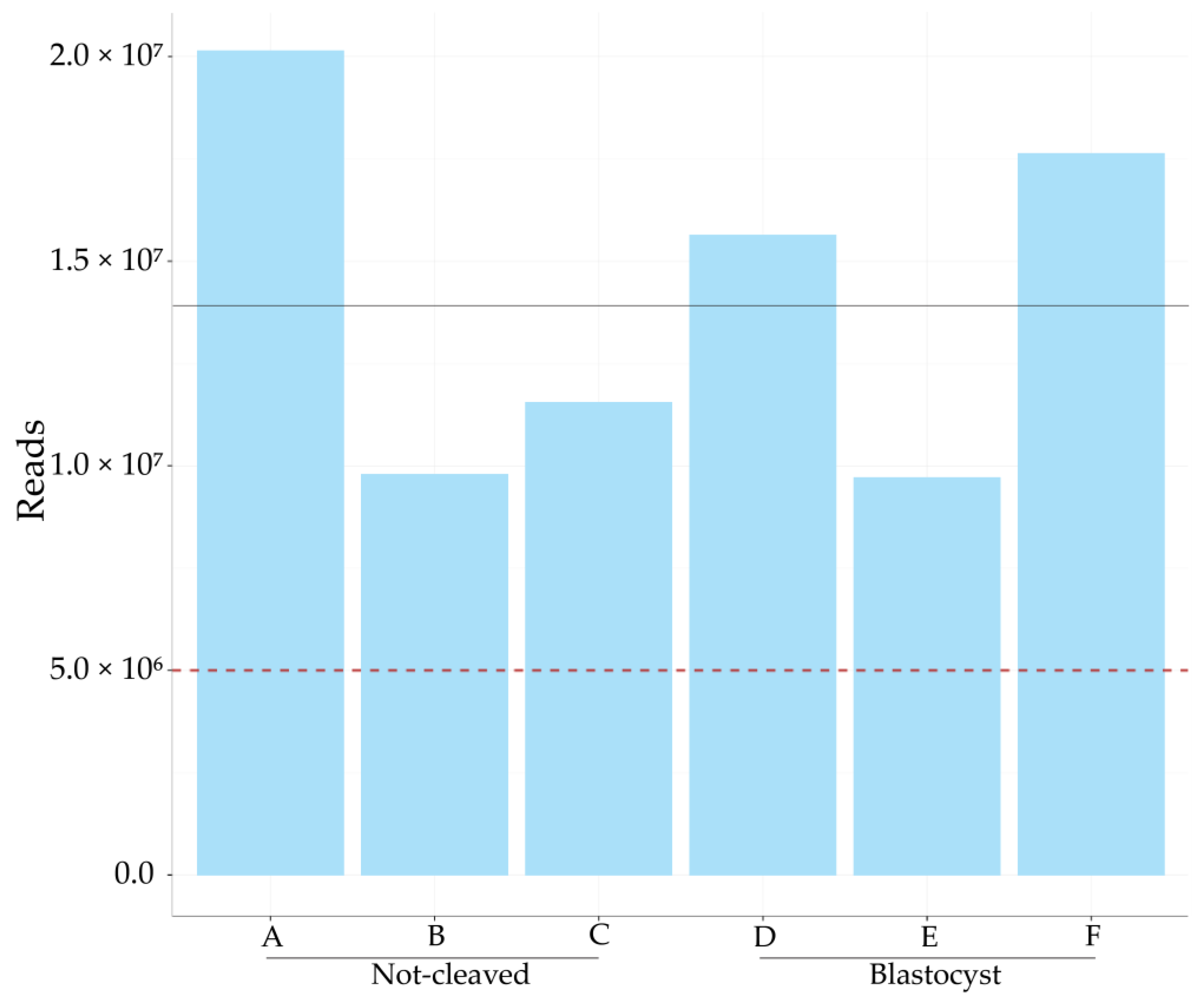
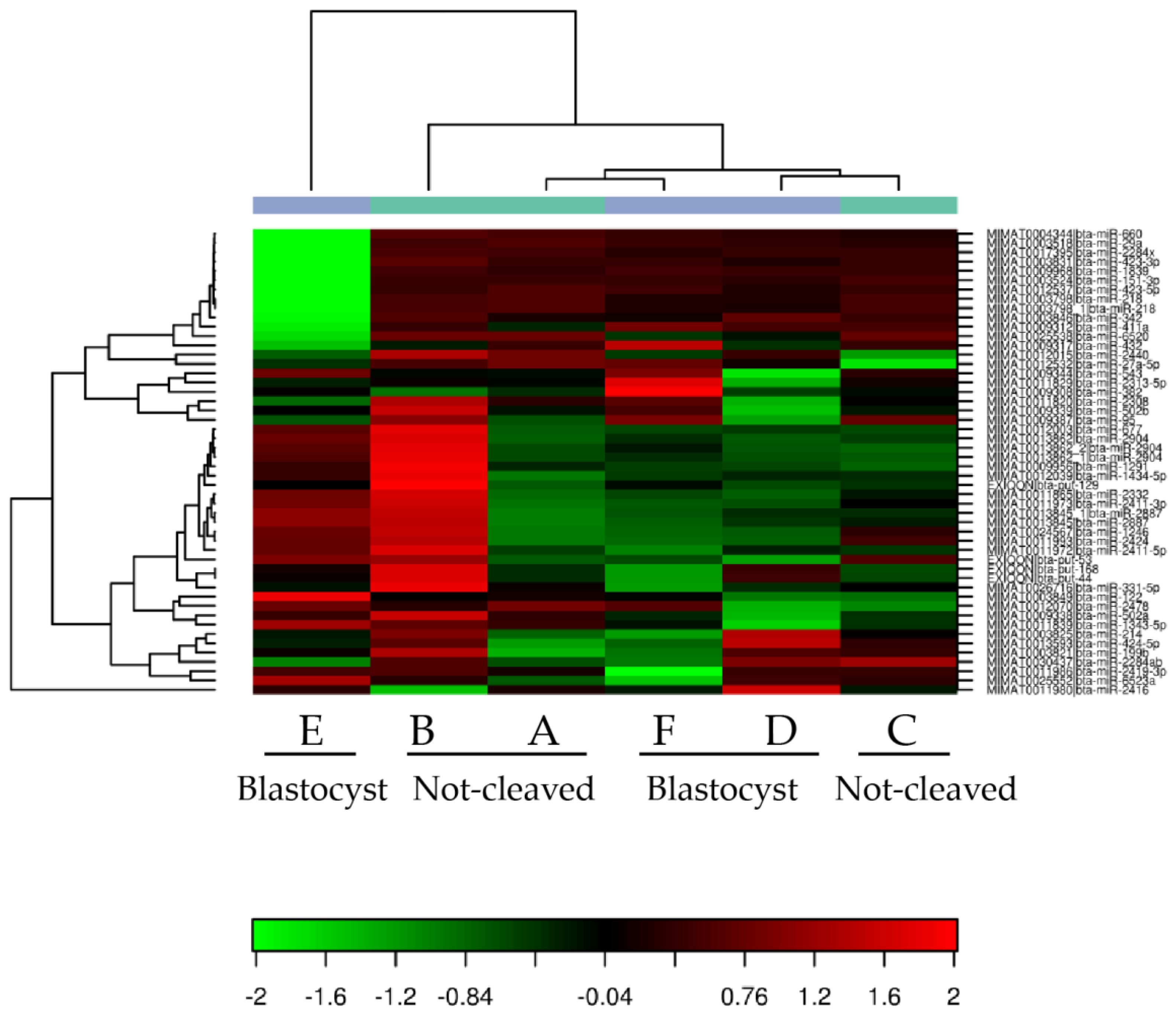
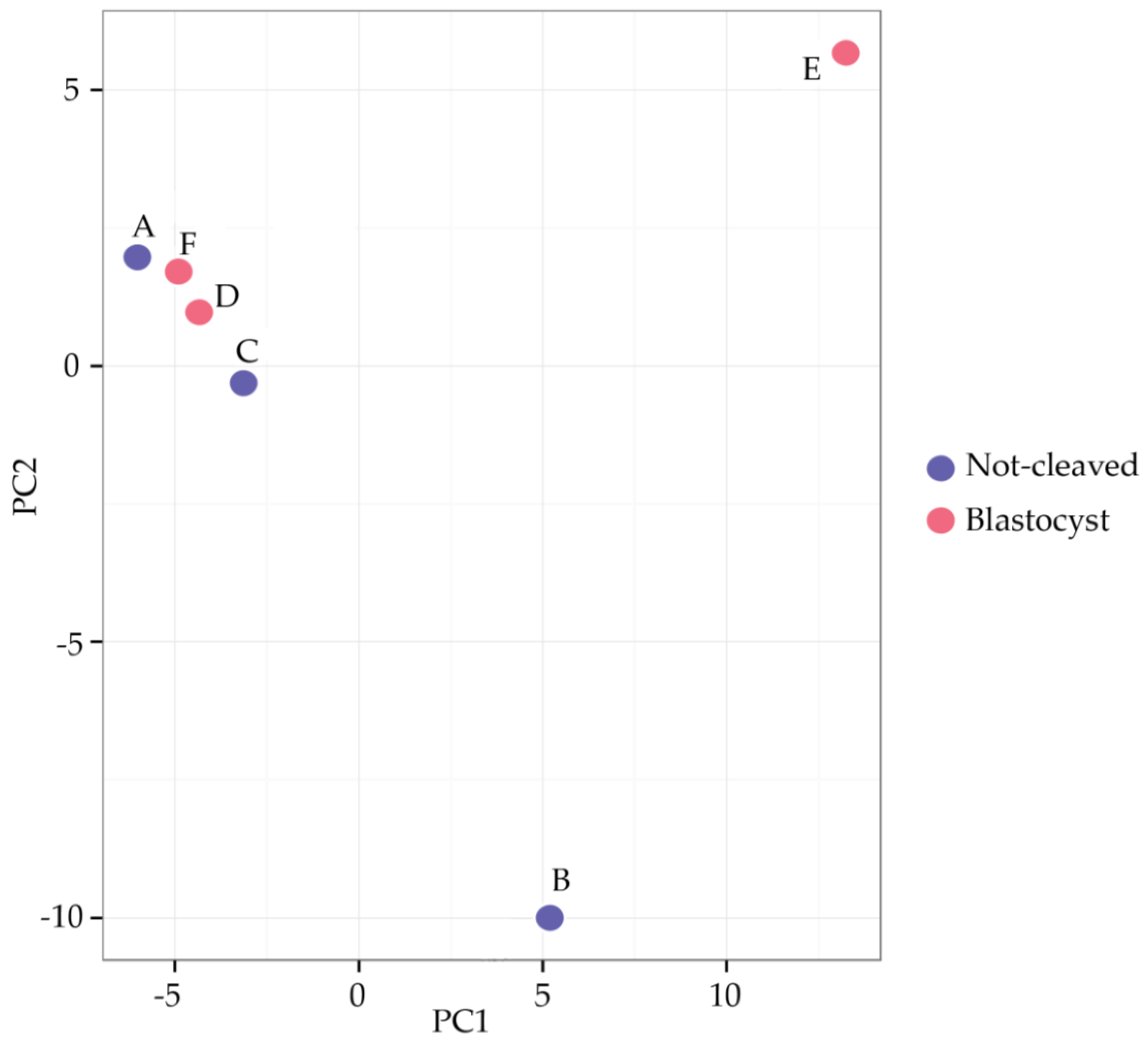
2.2. Sequencing of miRNAs in Cumulus Cells
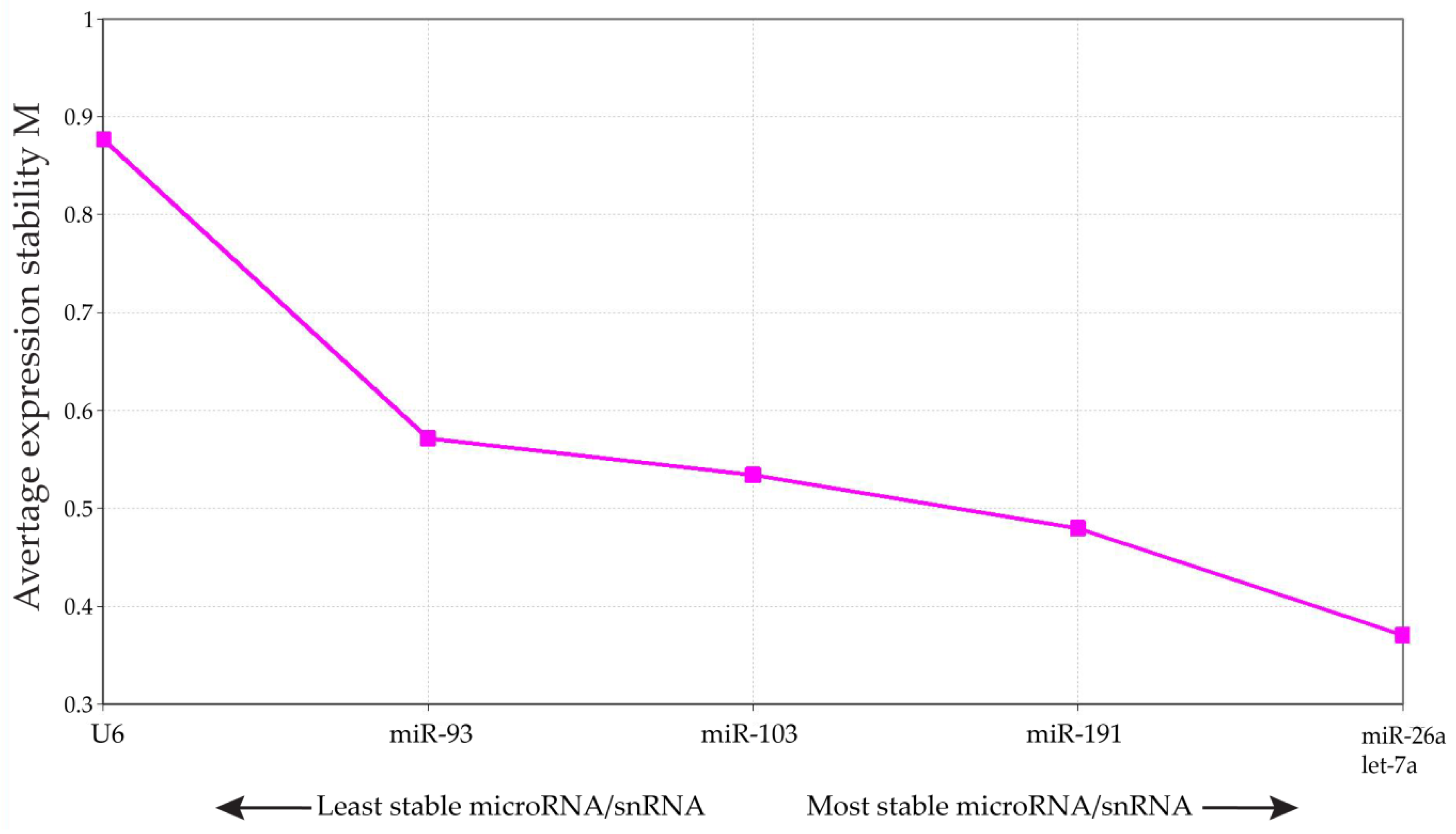
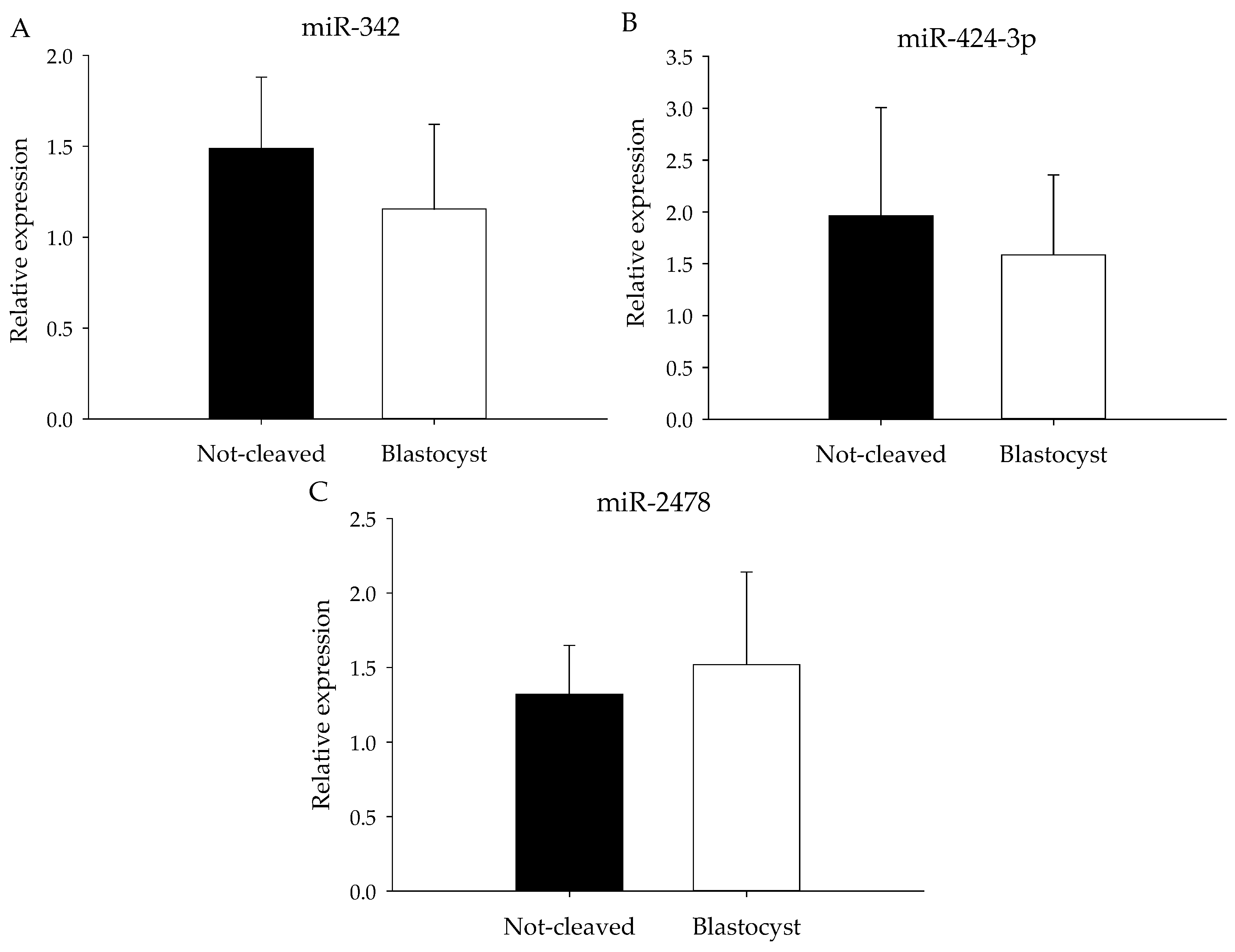
2.3. Quantitative Real-Time Reverse Transcription-PCR (qRT-PCR) from Pooled Cumulus Complex Samples
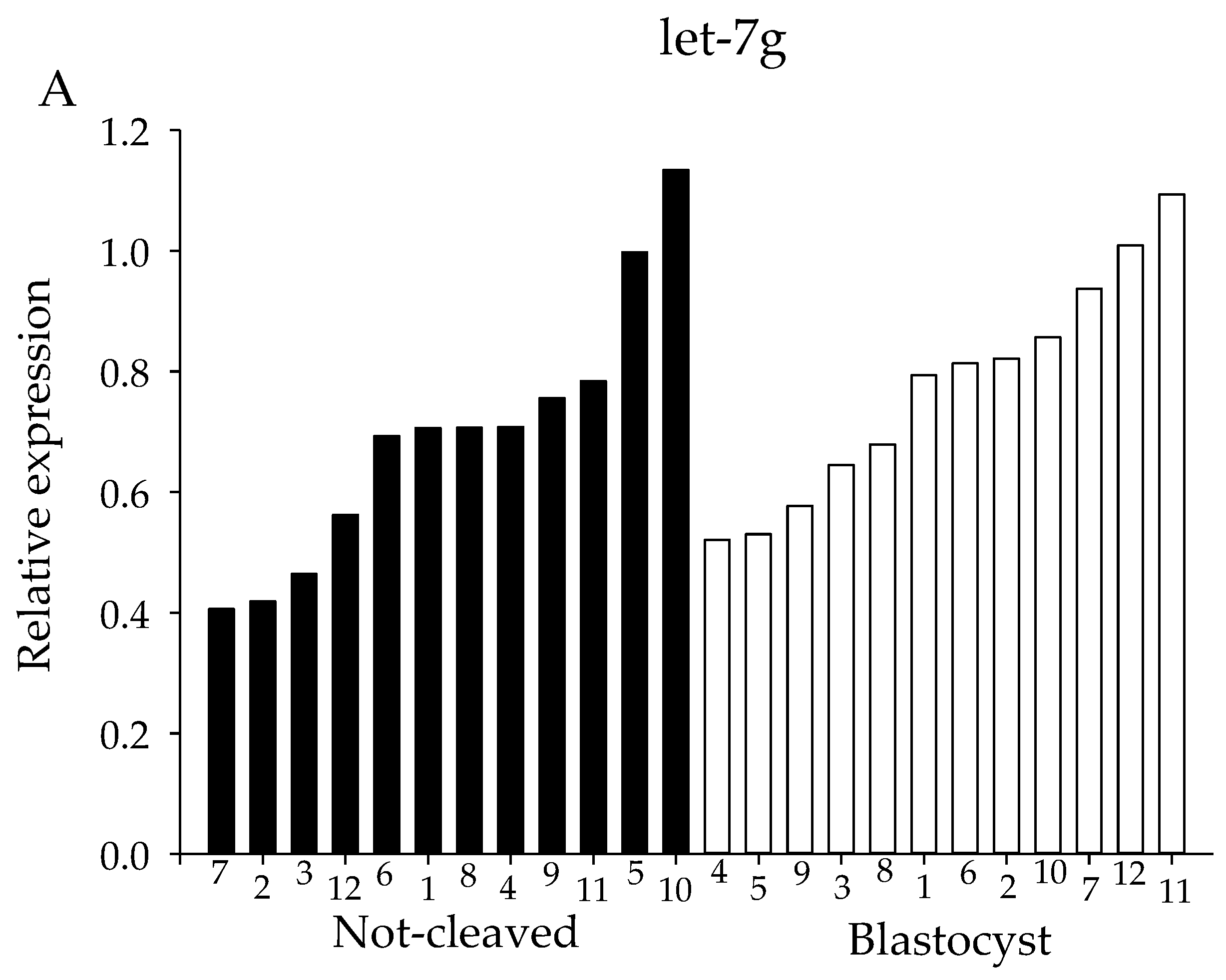
2.4. qRT-PCR from Individual Cumulus Complex Samples
3. Discussion
4. Materials and Methods
4.1. Collection of Cumulus−Oocyte−Complexes, In Vitro Maturation and Fertilization
4.2. RNA Isolation
4.3. Library Preparation and Next Generation Sequencing
4.4. First-Strand Synthesis and qRT-PCR
4.5. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
References
- Findlay, J.K.; Hutt, K.J.; Hickey, M.; Anderson, R.A. How is the number of primordial follicles in the ovarian reserve established? Biol. Reprod. 2015, 93, 111. [Google Scholar] [CrossRef]
- Cohen, P.E.; Holloway, J.K. Predicting gene networks in human oocyte meiosis. Biol. Reprod. 2010, 82, 469–472. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; McKenzie, L.J.; Matzuk, M.M. Revisiting oocyte-somatic cell interactions: In search of novel intrafollicular predictors and regulators of oocyte developmental competence. Mol. Hum. Reprod. 2008, 14, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Coticchio, G.; Dal Canto, M.; Mignini Renzini, M.; Guglielmo, M.C.; Brambillasca, F.; Turchi, D.; Novara, P.V.; Fadini, R. Oocyte maturation: Gamete-somatic cells interactions, meiotic resumption, cytoskeletal dynamics and cytoplasmic reorganization. Hum. Reprod. Update 2015, 21, 427–454. [Google Scholar] [CrossRef]
- Tanghe, S.; Van Soom, A.; Nauwynck, H.; Coryn, M.; de Kruif, A. Minireview: Functions of the cumulus oophorus during oocyte maturation, ovulation, and fertilization. Mol. Reprod. Dev. 2002, 61, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Buccione, R.; Schroeder, A.C.; Eppig, J.J. Interactions between somatic cells and germ cells throughout mammalian oogenesis. Biol. Reprod. 1990, 43, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Albertini, D.F. The road to maturation: Somatic cell interaction and self-organization of the mammalian oocyte. Nat. Rev. Mol. Cell Biol. 2013, 14, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Hussein, T.S.; Thompson, J.G.; Gilchrist, R.B. Oocyte-secreted factors enhance oocyte developmental competence. Dev. Biol. 2006, 296, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, R.B.; Lane, M.; Thompson, J.G. Oocyte-secreted factors: Regulators of cumulus cell function and oocyte quality. Hum. Reprod. Update 2008, 14, 159–177. [Google Scholar] [CrossRef] [PubMed]
- Albertini, D.; Combelles, C.; Benecchi, E.; Carabatsos, M. Cellular basis for paracrine regulation of ovarian follicle development. Reproduction 2001, 121, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Macaulay, A.D.; Gilbert, I.; Caballero, J.; Barreto, R.; Fournier, E.; Tossou, P.; Sirard, M.A.; Clarke, H.J.; Khamdjian, E.W.; Richard, F.J.; et al. The gametic synapse: RNA transfer to the bovine oocyte. Biol. Reprod. 2014, 91, 90. [Google Scholar] [CrossRef] [PubMed]
- Macaulay, A.D.; Gilbert, I.; Scantland, S.; Fournier, E.; Ashkar, F.; Bastien, A.; Saadi, H.A.; Gagné, D.; Sirard, M.A.; Khandjian, É.W.; et al. Cumulus cell transcripts transit to the bovine oocyte in preparation for maturation. Biol. Reprod. 2016, 94, 16. [Google Scholar] [CrossRef] [PubMed]
- Eppig, J.J.; Wigglesworth, K.; Pendola, F.L. The mammalian oocyte orchestrates the rate of ovarian follicular development. Proc. Natl. Acad. Sci. USA 2002, 99, 2890–2894. [Google Scholar] [CrossRef] [PubMed]
- Assou, S.; Al-edani, T.; Haouzi, D.; Philippe, N.; Lecellier, C.H.; Piquemal, D.; Commes, T.; Aït-Ahmed, O.; Dechaud, H.; Hamamah, S. MicroRNAs: New candidates for the regulation of the human cumulus–oocyte complex. Hum. Reprod. 2013, 28, 3038–3049. [Google Scholar] [CrossRef] [PubMed]
- Li, G.P.; Bunch, T.D.; White, K.L.; Rickords, L.; Liu, Y.; Sessions, B.R. Denuding and centrifugation of maturing bovine oocytes alters oocyte spindle integrity and the ability of cytoplasm to support parthenogenetic and nuclear transfer embryo development. Mol. Reprod. Dev. 2006, 73, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Balaban, B.; Urman, B.; Sertac, A.; Alatas, C.; Aksoy, S.; Mercan, R. Oocyte morphology does not affect fertilization rate, embryo quality and implantation rate after intracytoplasmic sperm injection. Hum. Reprod. 1998, 13, 3431–3433. [Google Scholar] [CrossRef] [PubMed]
- Coticchio, G.; Sereni, E.; Serrao, L.; Mazzone, S.; Iadarola, I.; Borini, A. What criteria for the definition of oocyte quality? Ann. N. Y. Acad. Sci. 2004, 1034, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Nachtigall, R.D.; Becker, G.; Friese, C.; Butler, A.; MacDougall, K. Parents’ conceptualization of their frozen embryos complicates the disposition decision. Fertil. Steril. 2005, 84, 431–434. [Google Scholar] [CrossRef] [PubMed]
- Driancourt, M. Regulation of ovarian follicular dynamics in farm animals. Implications for manipulation of reproduction. Theriogenology 2001, 55, 1211–1239. [Google Scholar] [CrossRef]
- Baerwald, A.R.; Adams, G.P.; Pierson, R.A. Ovarian antral folliculogenesis during the human menstrual cycle: A review. Hum. Reprod. Update 2012, 18, 73–91. [Google Scholar] [CrossRef] [PubMed]
- Rizos, D.; Gutiérrez-Adán, A.; Pérez-Garnelo, S.; de la Fuente, J.; Boland, M.P.; Lonergan, P. Bovine embryo culture in the presence or absence of serum: Implications for blastocyst development, cryotolerance, and messenger RNA expression. Biol. Reprod. 2003, 68, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Albuz, F.K.; Sasseville, M.; Lane, M.; Armstrong, D.T.; Thompson, J.G.; Gilchrist, R.B. Simulated physiological oocyte maturation (SPOM): A novel in vitro maturation system that substantially improves embryo yield and pregnancy outcomes. Hum. Reprod. 2010, 25, 2999–3011. [Google Scholar] [CrossRef] [PubMed]
- Assou, S.; Haouzi, D.; De Vos, J.; Hamamah, S. Human cumulus cells as biomarkers for embryo and pregnancy outcomes. Mol. Hum. Reprod. 2010, 16, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, K.M.; Feil, D.K.; Dunning, K.R.; Lane, M.; Russel, D.L. Human cumulus cell gene expression as a biomarker of pregnancy outcome after single embryo transfer. Fertil. Steril. 2011, 96, 47–52.e2. [Google Scholar] [CrossRef] [PubMed]
- Matoba, S.; Bender, K.; Fahey, A.G.; Mamo, S.; Brennan, L.; Lonergan, P.; Fair, T. Predictive value of bovine follicular components as markers of oocyte developmental potential. Reprod. Fertil. Dev. 2014, 26, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Bunel, A.; Jorssen, E.P.; Merckx, E.; Leroy, J.L.; Bols, P.E.; Sirard, M.A. Individual bovine in vitro embryo production and cumulus cell transcriptomic analysis to distinguish cumulus-oocyte complexes with high or low developmental potential. Theriogenology 2015, 83, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Wathlet, S.; Adriaenssens, T.; Segers, I.; Verheyen, G.; Van de Velde, H.; Coucke, W.; Ron El, R.; Devroey, P.; Smitz, J. Cumulus cell gene expression predicts better cleavage-stage embryo or blastocyst development and pregnancy for ICSI patients. Hum. Reprod. 2011, 26, 1035–1051. [Google Scholar] [CrossRef] [PubMed]
- O’Gorman, A.; Wallace, M.; Cottell, E.; Gibney, M.J.; McAuliffe, F.M.; Wingfield, M.; Brennan, L. Metabolic profiling of human follicular fluid identifies potential biomarkers of oocyte developmental competence. Reproduction 2013, 146, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Behm-Ansmant, I.; Rehwinkel, J.; Izaurralde, E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Kim, S.Y.; Carmell, M.A.; Murchison, E.P.; Alcorn, H.; Li, M.Z.; Mills, A.A.; Elledge, S.J.; Anderson, K.V.; Hannon, G.J. Dicer is essential for mouse development. Nat. Genet. 2003, 35, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Abd El Naby, W.S.; Hagos, T.H.; Hossain, M.M.; Salilew-Wondim, D.; Gad, A.Y.; Rings, F.; Cinar, M.U.; Tholen, E.; Looft, C.; Schellander, K.; et al. Expression analysis of regulatory microRNAs in bovine cumulus oocyte complex and preimplantation embryos. Zygote 2013, 21, 31–51. [Google Scholar] [CrossRef] [PubMed]
- Tesfaye, D.; Worku, D.; Rings, F.; Phatsara, C.; Thoen, E.; Schellander, K.; Hoelker, M. Identification and expression profiling of microRNAs during bovine oocyte maturation using heterologous approach. Mol. Reprod. Dev. 2009, 76, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Kaneda, M.; O’Carroll, D.; Hajkova, P.; Barton, S.C.; Sun, Y.A.; Lee, C.; Tarakhovsky, A.; Lao, K.; Surani, M.A. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007, 21, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Mondou, E.; Dufort, I.; Gohin, M.; Fournier, E.; Sirard, M. Analysis of microRNAs and their precursors in bovine early embryonic development. Mol. Hum. Reprod. 2012, 18, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.W.; Wang, B.; Ding, C.H.; Li, T.; Gu, F.; Zhou, C. Differentially expressed micoRNAs in human oocytes. J. Assist. Reprod. Genet. 2011, 28, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.H.; Xu, B.; Zhang, Y.W.; Liu, Y.S.; Ma, C.H. Research resources: Comparative microRNA profiles in human corona radiata cells and cumulus oophorus cells detected by next-generation small RNA sequencing. PLoS ONE 2014, 9, e106706. [Google Scholar] [CrossRef] [PubMed]
- Carletti, M.Z.; Fiedler, S.D.; Christenson, L.K. MicroRNA 21 blocks apoptosis in mouse periovulatory granulosa cells. Biol. Reprod. 2010, 83, 286–295. [Google Scholar] [CrossRef]
- Wright, E.C.; Hale, B.J.; Yang, C.X.; Njoka, J.G.; Ross, J.W. MicroRNA-21 and PDCD4 expression during in vitro oocyte maturation in pigs. Reprod. Biol. Endocrinol. 2016, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Sohel, M.M.H.; Hoelker, M.; Noferesti, S.S.; Salilew-Wondim, D.; Tholen, E.; Looft, C.; Rings, F.; Uddin, M.J.; Spencer, T.E.; Schellander, K.; et al. Exosomal and non-exosomal transport of extra-cellular microRNAs in follicular fluid: Implications for bovine oocyte developmental competence. PLoS ONE 2013, 8, e78505. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, S.M.; Matzuk, M.M. Oocyte-somatic cell communication and microRNA function in the ovary. Ann. Endocrinol. 2010, 71, 144–148. [Google Scholar] [CrossRef]
- Bussing, I.; Slack, F.J.; Grosshans, H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008, 14, 400–409. [Google Scholar] [CrossRef]
- Tam, O.H.; Aravin, A.A.; Stein, P.; Girard, A.; Murchison, E.P.; Cheloufi, S.; Hodges, E.; Anger, M.; Sachidanandam, R.; Schultz, R.M.; et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 2008, 453, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Chen, Z.; Liu, X.; Zhou, X. Evaluating the microRNA targeting sites by luciferase reporter gene assay. Methods Mol. Biol. 2013, 936, 117–127. [Google Scholar] [PubMed]
- Van Tol, H.T.A.; van Eerdenburg, F.J.C.M.; Colenbrander, B.; Roelen, B.A.J. Enhancement of Bovine oocyte maturation by leptin is accompanied by an upregulation in mRNA expression of leptin receptor isoforms in cumulus cells. Mol. Reprod. Dev. 2008, 75, 578–587. [Google Scholar] [CrossRef] [PubMed]
- Parrish, J.J.; Susko-Parrish, J.; Winer, M.A.; First, N.L. Capacitation of bovine sperm by heparin. Biol. Reprod. 1988, 38, 1171–1180. [Google Scholar] [CrossRef] [PubMed]
- A Guide to the microRNA and Small RNA Sequencing Service. Available online: https://www.exiqon.com/ls/Documents/Scientific/Service-Customer-Information-NGS.pdf (accessed on 2 February 2017).
- Reducing Whole-Genome Data Storage Footprint. Available online: http://www.illumina.com/documents/products/whitepapers/whitepaper_datacompression.pdf (accessed on 18 October 2016).
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Day 5 | Day 8 | |||||
|---|---|---|---|---|---|---|
| Culture type | Total | Not–cleaved | 2 to 8 | >8 | Blastocyst | |
| Group | n | 481 | 121 | 207 | 153 | 129 |
| % | 25.2 | 43 | 31.8 | 26.8 | ||
| Individual | n | 188 | 35 | 86 | 67 | 53 |
| % | 18.6 | 45.7 | 35.6 | 28.2 | ||
| miRNA | Counts NC | Counts Blast |
|---|---|---|
| bta-miR-21-5p | 2,324,236 | 2,915,237 |
| bta-let-7f | 431,569 | 335,113 |
| bta-let-7i | 302,000 | 256,981 |
| bta-let-7g | 228,393 | 177,529 |
| bta-miR-155 | 225,179 | 158,161 |
| bta-let-7a-5p | 217,033 | 158,238 |
| bta-miR-99a-5p | 129,482 | 112,348 |
| bta-miR-30d | 85,620 | 75,966 |
| bta-miR-26a | 73,820 | 56,991 |
| bta-miR-320a | 59,660 | 53,015 |
| bta-miR-92a | 55,108 | 47,255 |
| bta-miR-10b | 54,473 | 66,439 |
| bta-miR-202 | 38,887 | 32,030 |
| bta-miR-148a | 38,574 | 40,607 |
| bta-let-7b | 36,653 | 29,117 |
| bta-miR-532 | 34,632 | 32,815 |
| bta-miR-99b | 29,386 | 31,287 |
| bta-let-7e | 25,088 | 20,497 |
| bta-miR-125a | 22,159 | 20,436 |
| bta-let-7c | 23,668 | 17,542 |
| miRNA | Not-cleaved | Blastocyst | ||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | F | |
| bta–miR–214 | 35 | 33 | 30 | 119 | 22 | 13 |
| bta–miR–218 | 4,546 | 1,451 | 2,198 | 3,004 | 86 | 1,411 |
| bta–miR–222 | 3,228 | 1,175 | 2,090 | 1,724 | 1,291 | 1,151 |
| bta–miR–342 | 1,267 | 802 | 963 | 2,856 | 20 | 515 |
| bta–miR–424–3p | 354 | 62 | 166 | 159 | 208 | 181 |
| bta–miR–424–5p | 27 | 26 | 34 | 113 | 20 | 16 |
| bta–miR–2284ab | 32 | 20 | 41 | 70 | 12 | 13 |
| bta–miR–2478 | 357 | 92 | 66 | 105 | 154 | 150 |
| bta–let–7i | 428,545 | 199,640 | 277,815 | 396,881 | 225,420 | 148,643 |
| bta–let–7g | 332,160 | 154,890 | 198,131 | 237,141 | 163,940 | 131,506 |
| Name | Chr | START | STOP | SEQUENCE |
| bta-put-45 | chr7 | 12981779 | 12981801 | CCGUGCCUACUGAGCUGAAACAC |
| bta-put-129 | chr21 | 69641635 | 69641655 | UGCAAGCAACACUCUGUGGCA |
| bta-put-53 | chr7 | 53516556 | 53516579 | UAUACUCUGAUUGGUUCAUUAUGA |
| bta-put-79 | chr29 | 1067001 | 1067020 | AUGGUCAUUACCAAGGCUUU |
| bta-put-168 | chr7 | 5206807 | 5206828 | UCAAAGUGAAUUUGGAGGUUCU |
| bta-put-44 | chr7 | 5206802 | 5206823 | UCAAAGUGAAUUUGGAGGUUCU |
| bta-put-82 | chr29 | 41693966 | 41693989 | GAUCCGCGUAAUGUACGGAGGUAG |
| bta-put-156 | chr5 | 103546363 | 103546386 | GGACCUCAGUUCCAAACCUCUGCC |
| bta-put-25 | chr5 | 104238656 | 104238676 | AUGUGGACCCAGGGAGCUGGG |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uhde, K.; Van Tol, H.T.A.; Stout, T.A.E.; Roelen, B.A.J. MicroRNA Expression in Bovine Cumulus Cells in Relation to Oocyte Quality. Non-Coding RNA 2017, 3, 12. https://doi.org/10.3390/ncrna3010012
Uhde K, Van Tol HTA, Stout TAE, Roelen BAJ. MicroRNA Expression in Bovine Cumulus Cells in Relation to Oocyte Quality. Non-Coding RNA. 2017; 3(1):12. https://doi.org/10.3390/ncrna3010012
Chicago/Turabian StyleUhde, Karen, Helena T. A. Van Tol, Tom A. E. Stout, and Bernard A. J. Roelen. 2017. "MicroRNA Expression in Bovine Cumulus Cells in Relation to Oocyte Quality" Non-Coding RNA 3, no. 1: 12. https://doi.org/10.3390/ncrna3010012







