MicroRNA and Breast Cancer: Understanding Pathogenesis, Improving Management
Abstract
:1. Introduction
2. Function and Biosynthesis of MicroRNA
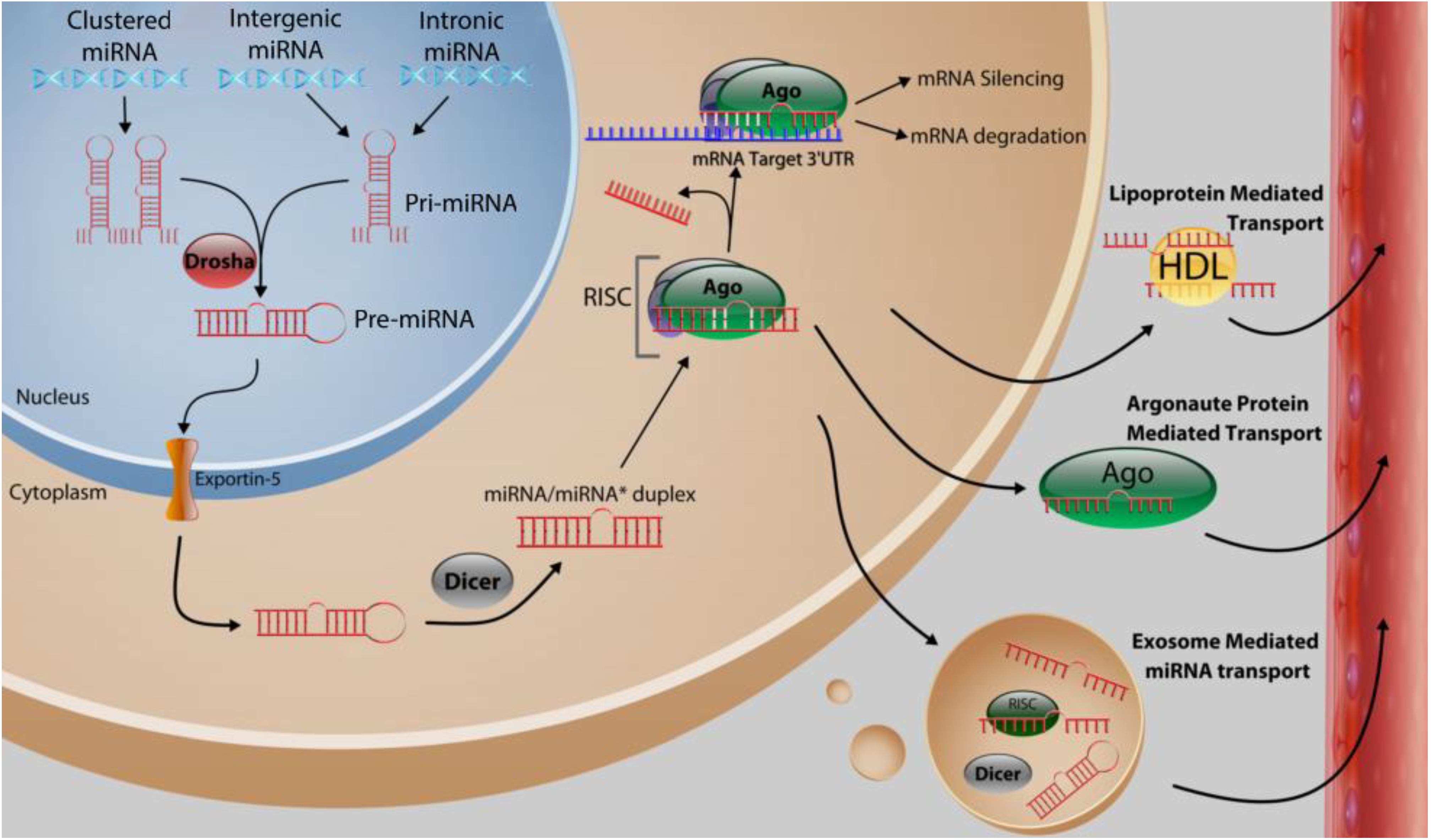
3. MicroRNA Control of Tumor Initiation
3.1. Let-7 MicroRNA Family
3.2. MicroRNA-17-92 Cluster
3.3. MicroRNA-34 Family, miR-379 and miR-497
3.4. MicroRNA-22
3.5. MicroRNA-21
3.6. MicroRNA-23/27/24 Cluster
3.7. MicroRNA-155

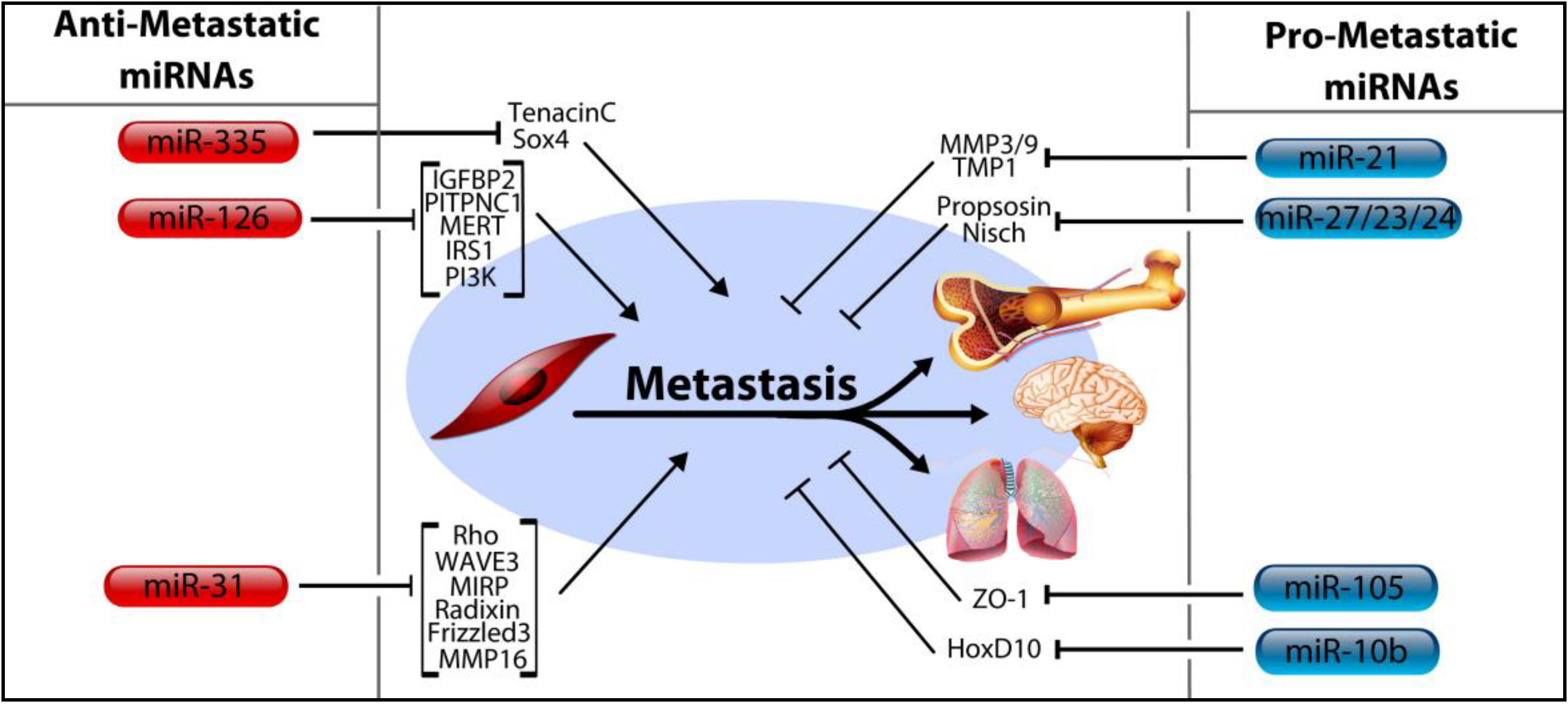
4. MicroRNA Control of Tumor Progression, Metastasis
4.1. MicroRNA-206, miR-126, and miR-335
4.2. MicroRNA-31
4.3. MicroRNA-10b, miRNA-105, and the miRNA-191/425 Cluster
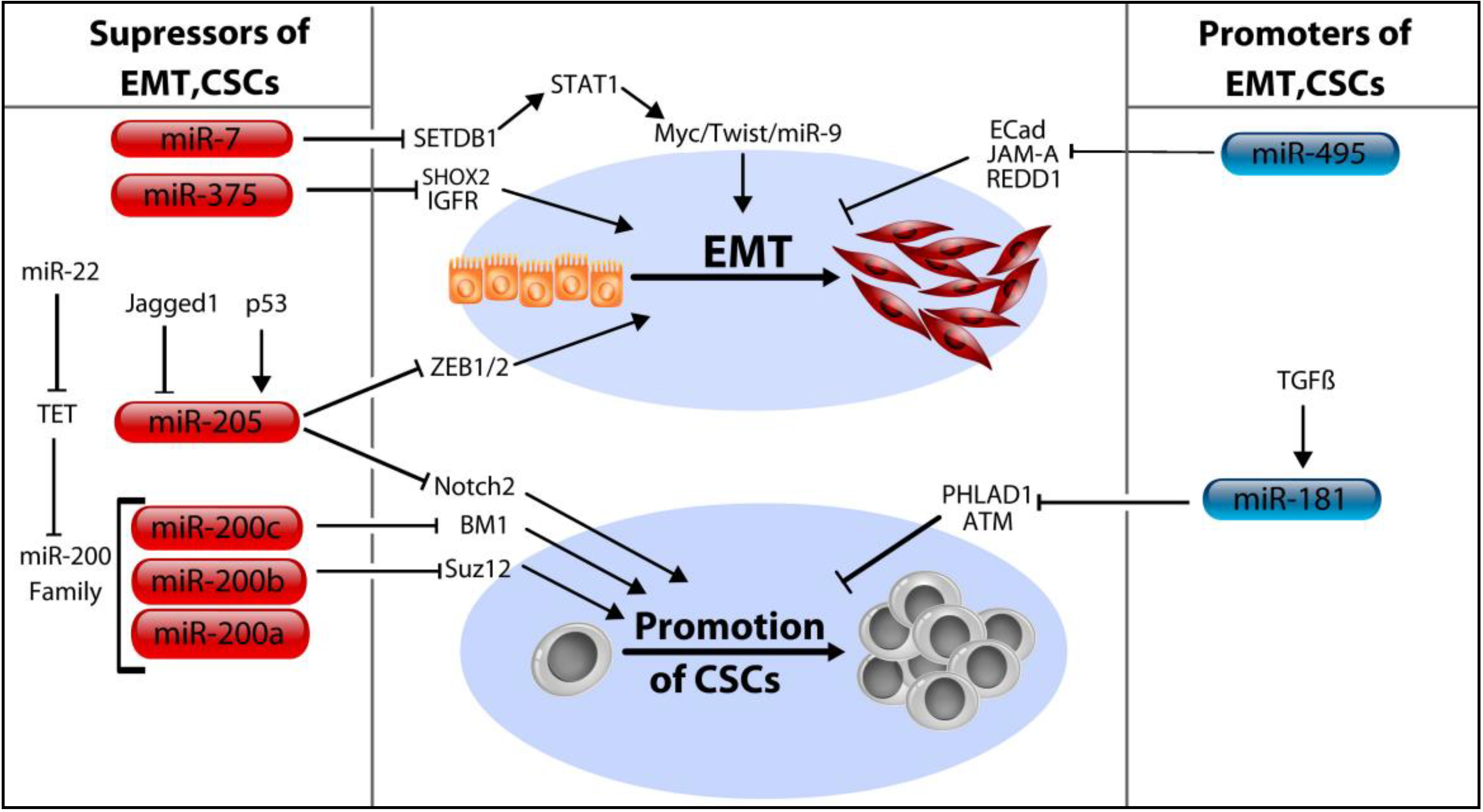
5. MicroRNA Control of Cancer Stem Cells
5.1. The MicroRNA-200 Family (miR-200a/b/c, miR-429, and miR-141)
5.2. MicroRNA-205
5.3. MicroRNA-7, miR-34a, and miR-375
5.4. MicroRNA-181a/b and miR-495
6. Modification of Cancer Cell Energy Metabolism by MicroRNAs
| MicroRNA | Primary Function | Target Genes | Reference |
|---|---|---|---|
| Let-7 Family | Tumor Suppressor | Ras, HMGA2 | [19] |
| miR-17-5p | Tumor Suppressor | AIB1, IGF1, IL-8, CcnD1, HBP-1 | [26,27,29] |
| miR-34 | Tumor Suppressor | CcnD1, CDK4, CDK6, SIRT1, Bcl-2, ZNF281, CD44, E2F3, SIRT1 | [33,35,36,100] |
| miR-379 | Tumor Suppressor | CcnB1, IL-11 | [37,38] |
| miR-497 | Tumor Suppressor | CcnD1, CcnE1 | [39,40] |
| miR-22 | Tumor Suppressor | ErbB3, CDC25C, EVI-1, ERα, CDK6, SIRT1, Sp1, CD147, TET Demethylases | [41,42,43,44,45] |
| miR-21 | Oncomir | PCDC4, PTEN, TPM1, MMP3, Smad7 | [48,49,50,51,52,54] |
| miR-23/27 | Oncomir | FOXO1, ZBTB10, ST14, Nisch, CYP1B1, PSAP | [57,58,59,60,61] |
| miR-155 | Oncomir | VHL, SOCS1, FOXO3, RhoA, TP53INP1 TRF1, Rad51, HK2 | [62,63,64,65,66,67,68,113] |
| miR-126 | Anti-metastatic | IRS,1 VEGF, PI3K , IGFBP2, PITPNC1, MERTK | [73,74,75] |
| miR-335 | Anti-metastatic | SOX4, TNC | [71] |
| miR-31 | Anti-metastatic | MIRP, MMP16, Radixin, FZD3, Rho, WAVE3 | [77,78,79] |
| miR-10b | Pro-metastatic | HoxD10 | [82] |
| miR-105 | Pro-metastatic | ZO-1, DNMT3A/B , ZFP36 | [83,85] |
| miR-200 | Inhibit EMT/CSCs | Suz12, BMI1 | [89,90] |
| miR-205 | Inhibit EMT/CSCs | Zeb1/2, ErbB2, ErbB3 Notch2 | [94,95,96,97] |
| miR-7 | Inhibit EMT/CSCs | SETDB1 | [99] |
| miR-375 | Inhibit EMT/CSCs | SHOX2, IGFR1 | [101,103,104] |
| miR-181a/b | Enhance EMT/CSCs | ATM, PHLDA1 | [105,106] |
| miR-495 | Enhance EMT/CSCs | E-Cadherin, REDD1 and JAM-A | [109,110] |
7. Circulating MicroRNAs as Biomarkers
8. MicroRNAs and Cancer Therapy: Agents of Drug Resistance
9. Conclusions
Acknowledgements
Conflicts of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014, 42, D68–D73. [Google Scholar]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar]
- Siegel, R.; Ma, J.; Zou, Z.; Jemal, A. Cancer Statistics, 2014. CA 2014, 64, 9–29. [Google Scholar]
- Blenkiron, C.; Goldstein, L.D.; Thorne, N.P.; Spiteri, I.; Chin, S.F.; Dunning, M.J.; Barbosa-Morais, N.L.; Teschendorff, A.E.; Green, A.R.; Ellis, I.O.; et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007, 8. [Google Scholar] [CrossRef]
- Dvinge, H.; Git, A.; Graf, S.; Salmon-Divon, M.; Curtis, C.; Sottoriva, A.; Zhao, Y.; Hirst, M.; Armisen, J.; Miska, E.A.; Chin, S.F.; et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 2013, 497, 378–382. [Google Scholar]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar]
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.N.; Hughes, S.M.; Cheng, H.H.; Arroyo, J.D.; Meredith, E.K.; Gallichotte, E.N.; et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893. [Google Scholar]
- Siomi, H.; Siomi, M.C. Posttranscriptional regulation of microRNA biogenesis in animals. Mol. Cell 2010, 38, 323–332. [Google Scholar]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar]
- Bartel, D.P. MicroRNAs: target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar]
- Hobert, O. Gene regulation by transcription factors and microRNAs. Science 2008, 319, 1785–1786. [Google Scholar]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar]
- Mulrane, L.; McGee, S.F.; Gallagher, W.M.; O’Connor, D.P. miRNA dysregulation in breast cancer. Cancer Res. 2013, 73, 6554–6562. [Google Scholar]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Muller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109–1123. [Google Scholar]
- Liu, Y.; Li, H.; Feng, J.; Cui, X.; Huang, W.; Li, Y.; Su, F.; Liu, Q.; Zhu, J.; Lv, X. Lin28 induces epithelial-to-mesenchymal transition and stemness via downregulation of Let-7a in breast cancer cells. PLoS ONE 2013, 8, e83083. [Google Scholar]
- Yang, M.; Li, S.N.; Anjum, K.M.; Gui, L.X.; Zhu, S.S.; Liu, J.; Chen, J.K.; Liu, Q.F.; Ye, G.D.; Wang, W.J.; et al. A double-negative feedback loop between Wnt-beta-catenin signaling and HNF4alpha regulates epithelial-mesenchymal transition in hepatocellular carcinoma. J. Cell Sci. 2013, 126, 5692–5703. [Google Scholar]
- Hu, X.; Guo, J.; Zheng, L.; Li, C.; Zheng, T.M.; Tanyi, J.L.; Liang, S.; Benedetto, C.; Mitidieri, M.; Katsaros, D.; et al. The heterochronic microRNA let-7 inhibits cell motility by regulating the genes in the actin cytoskeleton pathway in breast cancer. Mol. Cancer Res. 2013, 11, 240–250. [Google Scholar]
- Erturk, E.; Cecener, G.; Egeli, U.; Tunca, B.; Tezcan, G.; Gokgoz, S.; Tolunay, S.; Tasdelen, I. Expression status of let-7a and miR-335 among breast tumors in patients with and without germ-line BRCA mutations. Mol. Cell Biochem. 2014, 395, 77–88. [Google Scholar]
- Nair, V.S.; Maeda, L.S.; Ioannidis, J.P. Clinical outcome prediction by microRNAs in human cancer: A systematic review. J. Natl. Cancer Inst. 2012, 104, 528–540. [Google Scholar]
- Hossain, A.; Kuo, M.T.; Saunders, G.F. Mir-17–5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol. Cell. Biol. 2006, 26, 8191–8201. [Google Scholar]
- Yu, Z.; Willmarth, N.E.; Zhou, J.; Katiyar, S.; Wang, M.; Liu, Y.; McCue, P.A.; Quong, A.A.; Lisanti, M.P.; Pestell, R.G. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 8231–8236. [Google Scholar]
- Fan, M.; Sethuraman, A.; Brown, M.; Sun, W.; Pfeffer, L.M. Systematic analysis of metastasis-associated genes identifies miR-17–5p as a metastatic suppressor of basal-like breast cancer. Breast Cancer Res. Treat. 2014, 146, 487–502. [Google Scholar]
- Li, H.; Bian, C.; Liao, L.; Li, J.; Zhao, R.C. miR-17–5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res. Treat. 2011, 126, 565–75. [Google Scholar]
- Cloonan, N.; Brown, M.K.; Steptoe, A.L.; Wani, S.; Chan, W.L.; Forrest, A.R.; Kolle, G.; Gabrielli, B.; Grimmond, S.M. The miR-17–5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008, 9, R127. [Google Scholar] [CrossRef]
- Liu, S.; Clouthier, S.G.; Wicha, M.S. Role of microRNAs in the regulation of breast cancer stem cells. J. Mammary Gland Biol. Neoplasia 2012, 17, 15–21. [Google Scholar]
- Liu, S.; Patel, S.H.; Ginestier, C.; Ibarra, I.; Martin-Trevino, R.; Bai, S.; McDermott, S.P.; Shang, L.; Ke, J.; Ou, S.J. MicroRNA93 regulates proliferation and differentiation of normal and malignant breast stem cells. PLoS Genet. 2012, 8, e1002751. [Google Scholar]
- Achari, C.; Winslow, S.; Ceder, Y.; Larsson, C. Expression of miR-34c induces G2/M cell cycle arrest in breast cancer cells. BMC Cancer 2014, 14, 1471–1407. [Google Scholar]
- Kato, M.; Paranjape, T.; Ullrich, R.; Nallur, S.; Gillespie, E.; Keane, K.; Esquela-Kerscher, A.; Weidhaas, J.; Slack, F. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene 2009, 28, 2419–2424. [Google Scholar]
- Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13421–13426. [Google Scholar]
- Hahn, S.; Jackstadt, R.; Siemens, H.; Hunten, S.; Hermeking, H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013, 32, 3079–3095. [Google Scholar]
- Khan, S.; Brougham, C.L.; Ryan, J.; Sahrudin, A.; O’Neill, G.; Wall, D.; Curran, C.; Newell, J.; Kerin, M.J.; Dwyer, R.M. miR-379 regulates cyclin B1 expression and is decreased in breast cancer. PLoS ONE 2013, 8, e68753. [Google Scholar]
- Pollari, S.; Leivonen, S.K.; Perala, M.; Fey, V.; Kakonen, S.M.; Kallioniemi, O. Identification of microRNAs inhibiting TGF-beta-induced IL-11 production in bone metastatic breast cancer cells. PLoS ONE 2012, 7, e37361. [Google Scholar]
- Luo, Q.; Li, X.; Gao, Y.; Long, Y.; Chen, L.; Huang, Y.; Fang, L. MiRNA-497 regulates cell growth and invasion by targeting cyclin E1 in breast cancer. Cancer Cell Int. 2013, 13, 95. [Google Scholar] [CrossRef]
- Li, D.; Zhao, Y.; Liu, C.; Chen, X.; Qi, Y.; Jiang, Y.; Zou, C.; Zhang, X.; Liu, S.; Wang, X.; et al. Analysis of MiR-195 and MiR-497 expression, regulation and role in breast cancer. Clin. Cancer Res. 2011, 17, 1722–1730. [Google Scholar]
- Patel, J.; Appaiah, H.; Burnett, R.; Bhat-Nakshatri, P.; Wang, G.; Mehta, R.; Badve, S.; Thomson, M.; Hammond, S.; Steeg, P. Control of EVI-1 oncogene expression in metastatic breast cancer cells through microRNA miR-22. Oncogene 2011, 30, 1290–1301. [Google Scholar]
- Pandey, D.P.; Picard, D. miR-22 inhibits estrogen signaling by directly targeting the estrogen receptor alpha mRNA. Mol. Cell. Biol. 2009, 29, 3783–3790. [Google Scholar]
- Kong, L.M.; Liao, C.G.; Zhang, Y.; Xu, J.; Li, Y.; Huang, W.; Zhang, Y.; Bian, H.; Chen, Z.N. A regulatory loop involving miR-22, Sp1 and c-Myc modulates CD147 expression in breast cancer invasion and metastasis. Cancer Res. 2014, 74, 3764–3778. [Google Scholar]
- Xu, D.; Takeshita, F.; Hino, Y.; Fukunaga, S.; Kudo, Y.; Tamaki, A.; Matsunaga, J.; Takahashi, R.U.; Takata, T.; Shimamoto, A. miR-22 represses cancer progression by inducing cellular senescence. J. Cell Biol. 2011, 193, 409–424. [Google Scholar]
- Song, S.J.; Ito, K.; Ala, U.; Kats, L.; Webster, K.; Sun, S.M.; Jongen-Lavrencic, M.; Manova-Todorova, K.; Teruya-Feldstein, J.; Avigan, D.E. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell 2013, 13, 87–101. [Google Scholar]
- Song, S.J.; Pandolfi, P.P. miR-22 in tumorigenesis. Cell Cycle 2014, 13, 11–12. [Google Scholar]
- Chen, J.; Wang, X. MicroRNA-21 in breast cancer: diagnostic and prognostic potential. Clin. Transl. Oncol. 2014, 16, 225–233. [Google Scholar]
- Lu, Z.; Liu, M.; Stribinskis, V.; Klinge, C.; Ramos, K.; Colburn, N.; Li, Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 2008, 27, 4373–4379. [Google Scholar]
- Frankel, L.B.; Christoffersen, N.R.; Jacobsen, A.; Lindow, M.; Krogh, A.; Lund, A.H. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008, 283, 1026–1033. [Google Scholar]
- Qi, L.; Bart, J.; Tan, L.P.; Platteel, I.; van der Sluis, T.; Huitema, S.; Harms, G.; Fu, L.; Hollema, H.; van den Berg, A. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer 2009, 9, 163. [Google Scholar] [CrossRef]
- Song, B.; Wang, C.; Liu, J.; Wang, X.; Lv, L.; Wei, L.; Xie, L.; Zheng, Y.; Song, X. MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J. Exp. Clin. Cancer Res. 2010, 29, 29. [Google Scholar] [CrossRef]
- Zhu, S.; Si, M.L.; Wu, H.; Mo, Y.Y. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J. Biol. Chem. 2007, 282, 14328–14336. [Google Scholar]
- Soria-Valles, C.; Gutierrez-Fernandez, A.; Guiu, M.; Mari, B.; Fueyo, A.; Gomis, R.R.; Lopez-Otin, C. The anti-metastatic activity of collagenase-2 in breast cancer cells is mediated by a signaling pathway involving decorin and miR-21. Oncogene 2014, 33, 3054–3063. [Google Scholar]
- Gong, C.; Nie, Y.; Qu, S.; Liao, J.Y.; Cui, X.; Yao, H.; Zeng, Y.; Su, F.; Song, E.; Liu, Q. miR-21 induces myofibroblast differentiation and promotes the malignant progression of breast phyllodes tumors. Cancer Res. 2014, 74, 4341–4352. [Google Scholar]
- Mertens-Talcott, S.U.; Chintharlapalli, S.; Li, X.; Safe, S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007, 67, 11001–11011. [Google Scholar]
- Chhabra, R.; Dubey, R.; Saini, N. Cooperative and individualistic functions of the microRNAs in the miR-23a~ 27a~ 24–2 cluster and its implication in human diseases. Mol. Cancer 2010, 9, 232. [Google Scholar] [CrossRef]
- Guttilla, I.K.; White, B.A. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J. Biol. Chem. 2009, 284, 23204–23216. [Google Scholar]
- Wang, Y.; Rathinam, R.; Walch, A.; Alahari, S.K. ST14 (suppression of tumorigenicity 14) gene is a target for miR-27b, and the inhibitory effect of ST14 on cell growth is independent of miR-27b regulation. J. Biol. Chem. 2009, 284, 23094–23106. [Google Scholar]
- Jin, L.; Wessely, O.; Marcusson, E.G.; Ivan, C.; Calin, G.A.; Alahari, S.K. Prooncogenic factors miR-23b and miR-27b are regulated by Her2/Neu, EGF, and TNF-α in breast cancer. Cancer Res. 2013, 73, 2884–2896. [Google Scholar]
- Tsuchiya, Y.; Nakajima, M.; Takagi, S.; Taniya, T.; Yokoi, T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006, 66, 9090–9098. [Google Scholar]
- Ell, B.; Qiu, Q.; Wei, Y.; Mercatali, L.; Ibrahim, T.; Amadori, D.; Kang, Y. The microRNA-23b/27b/24 cluster promotes breast cancer lung metastasis by targeting metastasis-suppressive gene prosaposin. J. Biol. Chem. 2014, 289, 21888–21895. [Google Scholar]
- Kong, W.; He, L.; Richards, E.; Challa, S.; Xu, C.; Permuth-Wey, J.; Lancaster, J.; Coppola, D.; Sellers, T.; Djeu, J. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene 2014, 33, 679–689. [Google Scholar]
- Jiang, S.; Zhang, H.W.; Lu, M.H.; He, X.H.; Li, Y.; Gu, H.; Liu, M.F.; Wang, E.D. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010, 70, 3119–3127. [Google Scholar]
- Kong, W.; Yang, H.; He, L.; Zhao, J.J.; Coppola, D.; Dalton, W.S.; Cheng, J.Q. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell. Biol. 2008, 28, 6773–6784. [Google Scholar]
- Kong, W.; He, L.; Coppola, M.; Guo, J.; Esposito, N.N.; Coppola, D.; Cheng, J.Q. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J. Biol. Chem 2010, 285, 17869–17879. [Google Scholar]
- Zhang, C.M.; Zhao, J.; Deng, H.Y. MiR-155 promotes proliferation of human breast cancer MCF-7 cells through targeting tumor protein 53-induced nuclear protein 1. J. Biomed. Sci. 2013, 20. [Google Scholar] [CrossRef]
- Dinami, R.; Ercolani, C.; Petti, E.; Piazza, S.; Ciani, Y.; Sestito, R.; Sacconi, A.; Biagioni, F.; le Sage, C.; Agami, R.; et al. miR-155 drives telomere fragility in human breast cancer by targeting TRF1. Cancer Res. 2014, 74, 4145–4156. [Google Scholar]
- Sochor, M.; Basova, P.; Pesta, M.; Dusilkova, N.; Bartos, J.; Burda, P.; Pospisil, V.; Stopka, T. Oncogenic MicroRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer 2014, 14, 448. [Google Scholar] [CrossRef]
- Gasparini, P.; Lovat, F.; Fassan, M.; Casadei, L.; Cascione, L.; Jacob, N.K.; Carasi, S.; Palmieri, D.; Costinean, S.; Shapiro, C.L. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc. Natl. Acad. Sci. USA 2014, 111, 4536–4541. [Google Scholar]
- Vanharanta, S.; Massague, J. Origins of metastatic traits. Cancer Cell 2013, 24, 410–421. [Google Scholar]
- Tavazoie, S.F.; Alarcon, C.; Oskarsson, T.; Padua, D.; Wang, Q.; Bos, P.D.; Gerald, W.L.; Massague, J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008, 451, 147–152. [Google Scholar]
- Zhang, Y.; Yang, P.; Sun, T.; Li, D.; Xu, X.; Rui, Y.; Li, C.; Chong, M.; Ibrahim, T.; Mercatali, L. miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat. Cell Biol. 2013, 15, 284–294. [Google Scholar]
- Zhang, J.; Du, Y.Y.; Lin, Y.F.; Chen, Y.T.; Yang, L.; Wang, H.J.; Ma, D. The cell growth suppressor, mir-126, targets IRS-1. Biochem. Biophys. Res. Commun. 2008, 377, 136–140. [Google Scholar]
- Zhu, N.; Zhang, D.; Xie, H.; Zhou, Z.; Chen, H.; Hu, T.; Bai, Y.; Shen, Y.; Yuan, W.; Jing, Q.; Qin, Y. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol. Cell. Biochem. 2011, 351, 157–164. [Google Scholar]
- Png, K.J.; Halberg, N.; Yoshida, M.; Tavazoie, S.F. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature 2012, 481, 190–194. [Google Scholar]
- Heyn, H.; Engelmann, M.; Schreek, S.; Ahrens, P.; Lehmann, U.; Kreipe, H.; Schlegelberger, B.; Beger, C. MicroRNA miR-335 is crucial for the BRCA1 regulatory cascade in breast cancer development. Int. J. Cancer 2011, 129, 2797–806. [Google Scholar]
- Valastyan, S.; Reinhardt, F.; Benaich, N.; Calogrias, D.; Szasz, A.M.; Wang, Z.C.; Brock, J.E.; Richardson, A.L.; Weinberg, R.A. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 2009, 137, 1032–1046. [Google Scholar]
- Sossey-Alaoui, K.; Downs-Kelly, E.; Das, M.; Izem, L.; Tubbs, R.; Plow, E.F. WAVE3, an actin remodeling protein, is regulated by the metastasis suppressor microRNA, miR-31, during the invasion-metastasis cascade. Int. J. Cancer 2011, 129, 1331–1343. [Google Scholar]
- Augoff, K.; Das, M.; Bialkowska, K.; McCue, B.; Plow, E.F.; Sossey-Alaoui, K. miR-31 is a broad regulator of b-integrin expression and function in cancer cells. Mol. Cancer Res. 2011, 9, 1500–1508. [Google Scholar]
- Chan, Y.T.; Lin, Y.C.; Lin, R.J.; Kuo, H.H.; Thang, W.C.; Chiu, K.P.; Alice, L.Y. Concordant and discordant regulation of target genes by miR-31 and its isoforms. PLoS ONE 2013, 8, e58169. [Google Scholar]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar]
- Ma, L.; Reinhardt, F.; Pan, E.; Soutschek, J.R.; Bhat, B.; Marcusson, E.G.; Teruya-Feldstein, J.; Bell, G.W.; Weinberg, R.A. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 2010, 28, 341–347. [Google Scholar]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar]
- Gebeshuber, C.A.; Zatloukal, K.; Martinez, J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009, 10, 400–405. [Google Scholar]
- Fabbri, M.; Garzon, R.; Cimmino, A.; Liu, Z.; Zanesi, N.; Callegari, E.; Liu, S.; Alder, H.; Costinean, S.; Fernandez-Cymering, C. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 2007, 104, 15805–15810. [Google Scholar]
- Jordan, C.T.; Guzman, M.L.; Noble, M. Cancer stem cells. N. Engl. J. Med. 2006, 355, 1253–1261. [Google Scholar]
- Pece, S.; Tosoni, D.; Confalonieri, S.; Mazzarol, G.; Vecchi, M.; Ronzoni, S.; Bernard, L.; Viale, G.; Pelicci, P.G.; di Fiore, P.P. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 2010, 140, 62–73. [Google Scholar]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar]
- Iliopoulos, D.; Jaeger, S.A.; Hirsch, H.A.; Bulyk, M.L.; Struhl, K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol. Cell 2010, 39, 493–506. [Google Scholar]
- Shimono, Y.; Zabala, M.; Cho, R.W.; Lobo, N.; Dalerba, P.; Qian, D.; Diehn, M.; Liu, H.; Panula, S.P.; Chiao, E. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009, 138, 592–603. [Google Scholar]
- Le, M.T.; Hamar, P.; Guo, C.; Basar, E.; Perdigao-Henriques, R.; Balaj, L.; Lieberman, J. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J. Clin. Investig. 2014, 124, 5109–5128. [Google Scholar]
- Sempere, L.F.; Christensen, M.; Silahtaroglu, A.; Bak, M.; Heath, C.V.; Schwartz, G.; Wells, W.; Kauppinen, S.; Cole, C.N. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007, 67, 11612–11620. [Google Scholar]
- Qin, A.Y.; Zhang, X.W.; Liu, L.; Yu, J.P.; Li, H.; Emily Wang, S.Z.; Ren, X.B.; Cao, S. MiR-205 in cancer: An angel or a devil? Eur. J. Cell Biol. 2013, 92, 54–60. [Google Scholar]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar]
- Wu, H.; Zhu, S.; Mo, Y.Y. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009, 19, 439–448. [Google Scholar]
- Iorio, M.V.; Casalini, P.; Piovan, C.; di Leva, G.; Merlo, A.; Triulzi, T.; Ménard, S.; Croce, C.M.; Tagliabue, E. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009, 69, 2195–2200. [Google Scholar]
- Chao, C.H.; Chang, C.C.; Wu, M.J.; Ko, H.W.; Wang, D.; Hung, M.C.; Yang, J.Y.; Chang, C.J. MicroRNA-205 signaling regulates mammary stem cell fate and tumorigenesis. J. Clin. Investig. 2014, 124, 3093–3106. [Google Scholar]
- Piovan, C.; Palmieri, D.; di Leva, G.; Braccioli, L.; Casalini, P.; Nuovo, G.; Tortoreto, M.; Sasso, M.; Plantamura, I.; Triulzi, T.; et al. Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol. Oncol. 2012, 6, 458–472. [Google Scholar]
- Zhang, H.; Cai, K.; Wang, J.; Wang, X.; Cheng, K.; Shi, F.; Jiang, L.; Zhang, Y.; Dou, J. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells 2014, 32, 2858–2868. [Google Scholar]
- Li, L.; Xie, X.; Luo, J.; Liu, M.; Xi, S.; Guo, J.; Kong, Y.; Wu, M.; Gao, J.; Xie, Z. Targeted expression of miR-34a using the T-VISA system suppresses breast cancer cell growth and invasion. Mol. Ther. 2012, 20, 2326–2334. [Google Scholar]
- Hong, S.; Noh, H.; Teng, Y.; Shao, J.; Rehmani, H.; Ding, H.F.; Dong, Z.; Su, S.B.; Shi, H.; Kim, J. SHOX2 Is a Direct miR-375 Target and a Novel Epithelial-to-Mesenchymal Transition Inducer in Breast Cancer Cells. Neoplasia 2014, 16, 279–290. e5. [Google Scholar]
- Ward, A.; Balwierz, A.; Zhang, J.D.; Kublbeck, M.; Pawitan, Y.; Hielscher, T.; Wiemann, S.; Sahin, O. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene 2013, 32, 1173–1182. [Google Scholar]
- Giricz, O.; Reynolds, P.A.; Ramnauth, A.; Liu, C.; Wang, T.; Stead, L.; Childs, G.; Rohan, T.; Shapiro, N.; Fineberg, S.; et al. Hsa-miR-375 is differentially expressed during breast lobular neoplasia and promotes loss of mammary acinar polarity. J. Pathol. 2012, 226, 108–119. [Google Scholar]
- Ye, X.M.; Zhu, H.Y.; Bai, W.D.; Wang, T.; Wang, L.; Chen, Y.; Yang, A.G.; Jia, L.T. Epigenetic silencing of miR-375 induces trastuzumab resistance in HER2-positive breast cancer by targeting IGF1R. BMC Cancer 2014, 14, 134. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.; Tsuyada, A.; Ren, X.; Wu, X.; Stubblefield, K.; Rankin-Gee, E.K.; Wang, S.E. Transforming growth factor-beta regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene 2011, 30, 1470–1480. [Google Scholar]
- Mansueto, G.; Forzati, F.; Ferraro, A.; Pallante, P.; Bianco, M.; Esposito, F.; Iaccarino, A.; Troncone, G.; Fusco, A. Identification of a New Pathway for Tumor Progression: MicroRNA-181b Up-Regulation and CBX7 Down-Regulation by HMGA1 Protein. Genes Cancer 2010, 1, 210–224. [Google Scholar]
- Neel, J.C.; Lebrun, J.J. Activin and TGFb regulate expression of the microRNA-181 family to promote cell migration and invasion in breast cancer cells. Cell. Signal. 2013, 25, 1556–1566. [Google Scholar]
- Taylor, M.A.; Sossey-Alaoui, K.; Thompson, C.L.; Danielpour, D.; Schiemann, W.P. TGF-b upregulates miR-181a expression to promote breast cancer metastasis. J. Clin. Investig. 2013, 123, 150–163. [Google Scholar]
- Cao, M.; Nie, W.; Li, J.; Zhang, Y.; Yan, X.; Guan, X.; Chen, X.; Zen, K.; Zhang, C.Y.; Jiang, X. MicroRNA-495 induces breast cancer cell migration by targeting JAM-A. Protein Cell 2014, 5, 862–872. [Google Scholar]
- Hwang-Verslues, W.; Chang, P.; Wei, P.; Yang, C.; Huang, C.; Kuo, W.; Shew, J.; Chang, K.; Lee, E.Y.P.; Lee, W. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene 2011, 30, 2463–2474. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: the next generation. Cell 2011, 144, 646–674. [Google Scholar]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar]
- Jiang, S.; Zhang, L.F.; Zhang, H.W.; Hu, S.; Lu, M.H.; Liang, S.; Li, B.; Li, Y.; Li, D.; Wang, E.D.; Liu, M.F. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 2012, 31, 1985–1998. [Google Scholar]
- Kim, S.; Rhee, J.; Yu, H.J.; Lee, H.J.; Lee, E.J.; Lee, J.W.; Yu, J.H.; Son, B.H.; Gong, G.; Kim, S.B. Bioinformatic and metabolomic analysis reveals miR-155 regulates thiamine level in breast cancer. Cancer Lett. 2015, 357, 488–497. [Google Scholar]
- Eichner, L.J.; Perry, M.C.; Dufour, C.R.; Bertos, N.; Park, M.; St-Pierre, J.; Giguere, V. miR-378(*) mediates metabolic shift in breast cancer cells via the PGC-1beta/ERRgamma transcriptional pathway. Cell Metab. 2010, 12, 352–361. [Google Scholar]
- Gao, P.; Tchernyshyov, I.; Chang, T.C.; Lee, Y.S.; Kita, K.; Ochi, T.; Zeller, K.I.; de Marzo, A.M.; van Eyk, J.E.; Mendell, J.T. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–765. [Google Scholar]
- Khatcheressian, J.L.; Hurley, P.; Bantug, E.; Esserman, L.J.; Grunfeld, E.; Halberg, F.; Hantel, A.; Henry, N.L.; Muss, H.B.; Smith, T.J. Breast cancer follow-up and management after primary treatment: American Society of Clinical Oncology clinical practice guideline update. J. Clin. Oncol. 2013, 31, 961–965. [Google Scholar]
- Turchinovich, A.; Samatov, T.R.; Tonevitsky, A.G.; Burwinkel, B. Circulating miRNAs: Cell-cell communication function? Front. Genet. 2013, 4. [Google Scholar] [CrossRef]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A. Cancer Exosomes Perform Cell-Independent MicroRNA Biogenesis and Promote Tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell. Biol. 2007, 9, 654–659. [Google Scholar]
- Foekens, J.A.; Sieuwerts, A.M.; Smid, M.; Look, M.P.; de Weerd, V.; Boersma, A.W.; Klijn, J.G.; Wiemer, E.A.; Martens, J.W. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc. Natl. Acad. Sci. USA 2008, 105, 13021–13026. [Google Scholar]
- Eichelser, C.; Flesch-Janys, D.; Chang-Claude, J.; Pantel, K.; Schwarzenbach, H. Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clin. Chem. 2013, 59, 1489–1496. [Google Scholar]
- Chen, W.; Cai, F.; Zhang, B.; Barekati, Z.; Zhong, X.Y. The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: potential biomarkers. Tumour Biol. 2013, 34, 455–462. [Google Scholar]
- Godfrey, A.C.; Xu, Z.; Weinberg, C.R.; Getts, R.C.; Wade, P.A.; DeRoo, L.A.; Sandler, D.P.; Taylor, J.A. Serum microRNA expression as an early marker for breast cancer risk in prospectively collected samples from the Sister Study cohort. Breast Cancer Res. 2013, 15, R42. [Google Scholar] [CrossRef]
- Guo, L.J.; Zhang, Q.Y. Decreased serum miR-181a is a potential new tool for breast cancer screening. Int. J. Mol. Med. 2012, 30, 680–686. [Google Scholar]
- Kodahl, A.R.; Zeuthen, P.; Binder, H.; Knoop, A.S.; Ditzel, H.J. Alterations in circulating miRNA levels following early-stage estrogen receptor-positive breast cancer resection in post-menopausal women. PLoS ONE 2014, 9, e101950. [Google Scholar]
- Zhao, H.; Shen, J.; Medico, L.; Wang, D.; Ambrosone, C.B.; Liu, S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS ONE 2010, 5, 0013735. [Google Scholar]
- Wang, F.; Zheng, Z.; Guo, J.; Ding, X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol. Oncol. 2010, 119, 586–593. [Google Scholar]
- Heneghan, H.M.; Miller, N.; Lowery, A.J.; Sweeney, K.J.; Newell, J.; Kerin, M.J. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann. Surg. 2010, 251, 499–505. [Google Scholar]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012, 5, 492–497. [Google Scholar]
- Cuk, K.; Zucknick, M.; Heil, J.; Madhavan, D.; Schott, S.; Turchinovich, A.; Arlt, D.; Rath, M.; Sohn, C.; Benner, A.; Junkermann, H.; Schneeweiss, A.; Burwinkel, B. Circulating microRNAs in plasma as early detection markers for breast cancer. Int. J. Cancer 2013, 132, 1602–1612. [Google Scholar]
- Witwer, K.W. Circulating microRNA biomarker studies: Pitfalls and potential solutions. Clin. Chem. 2015, 61, 56–63. [Google Scholar]
- Lagana, A.; Acunzo, M.; Romano, G.; Pulvirenti, A.; Veneziano, D.; Cascione, L.; Giugno, R.; Gasparini, P.; Shasha, D.; Ferro, A. miR-Synth: A computational resource for the design of multi-site multi-target synthetic miRNAs. Nucleic Acids Res. 2014. [Google Scholar] [CrossRef]
- Masri, S.; Liu, Z.; Phung, S.; Wang, E.; Yuan, Y.C.; Chen, S. The role of microRNA-128a in regulating TGFbeta signaling in letrozole-resistant breast cancer cells. Breast Cancer Res. Treat. 2010, 124, 89–99. [Google Scholar]
- Ring, A.; Dowsett, M. Mechanisms of tamoxifen resistance. Endocr. -Relat. Cancer 2004, 11, 643–658. [Google Scholar]
- Miller, T.E.; Ghoshal, K.; Ramaswamy, B.; Roy, S.; Datta, J.; Shapiro, C.L.; Jacob, S.; Majumder, S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J. Biol. Chem. 2008, 283, 29897–29903. [Google Scholar]
- Ward, A.; Shukla, K.; Balwierz, A.; Soons, Z.; Konig, R.; Sahin, O.; Wiemann, S. MicroRNA-519a is a novel oncomir conferring tamoxifen resistance by targeting a network of tumour-suppressor genes in ER+ breast cancer. J. Pathol. 2014, 233, 368–379. [Google Scholar]
- Adams, B.D.; Furneaux, H.; White, B.A. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol. Endocrinol. 2007, 21, 1132–1147. [Google Scholar]
- Cittelly, D.M.; Das, P.M.; Spoelstra, N.S.; Edgerton, S.M.; Richer, J.K.; Thor, A.D.; Jones, F.E. Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol. Cancer 2010, 9, 317. [Google Scholar]
- Si, M.; Zhu, S.; Wu, H.; Lu, Z.; Wu, F.; Mo, Y. miR-21-mediated tumor growth. Oncogene 2006, 26, 2799–2803. [Google Scholar]
- Mei, M.; Ren, Y.; Zhou, X.; Yuan, X.B.; Han, L.; Wang, G.X.; Jia, Z.; Pu, P.Y.; Kang, C.S.; Yao, Z. Downregulation of miR-21 enhances chemotherapeutic effect of taxol in breast carcinoma cells. Technol. Cancer Res. Treat. 2010, 9, 77–86. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eastlack, S.C.; Alahari, S.K. MicroRNA and Breast Cancer: Understanding Pathogenesis, Improving Management. Non-Coding RNA 2015, 1, 17-43. https://doi.org/10.3390/ncrna1010017
Eastlack SC, Alahari SK. MicroRNA and Breast Cancer: Understanding Pathogenesis, Improving Management. Non-Coding RNA. 2015; 1(1):17-43. https://doi.org/10.3390/ncrna1010017
Chicago/Turabian StyleEastlack, Steven C., and Suresh K. Alahari. 2015. "MicroRNA and Breast Cancer: Understanding Pathogenesis, Improving Management" Non-Coding RNA 1, no. 1: 17-43. https://doi.org/10.3390/ncrna1010017





