Fungal Resistance to Echinocandins and the MDR Phenomenon in Candida glabrata
Abstract
:1. Epidemiology and Mechanisms of Resistance
2. Evolution of Echinocandin Resistance
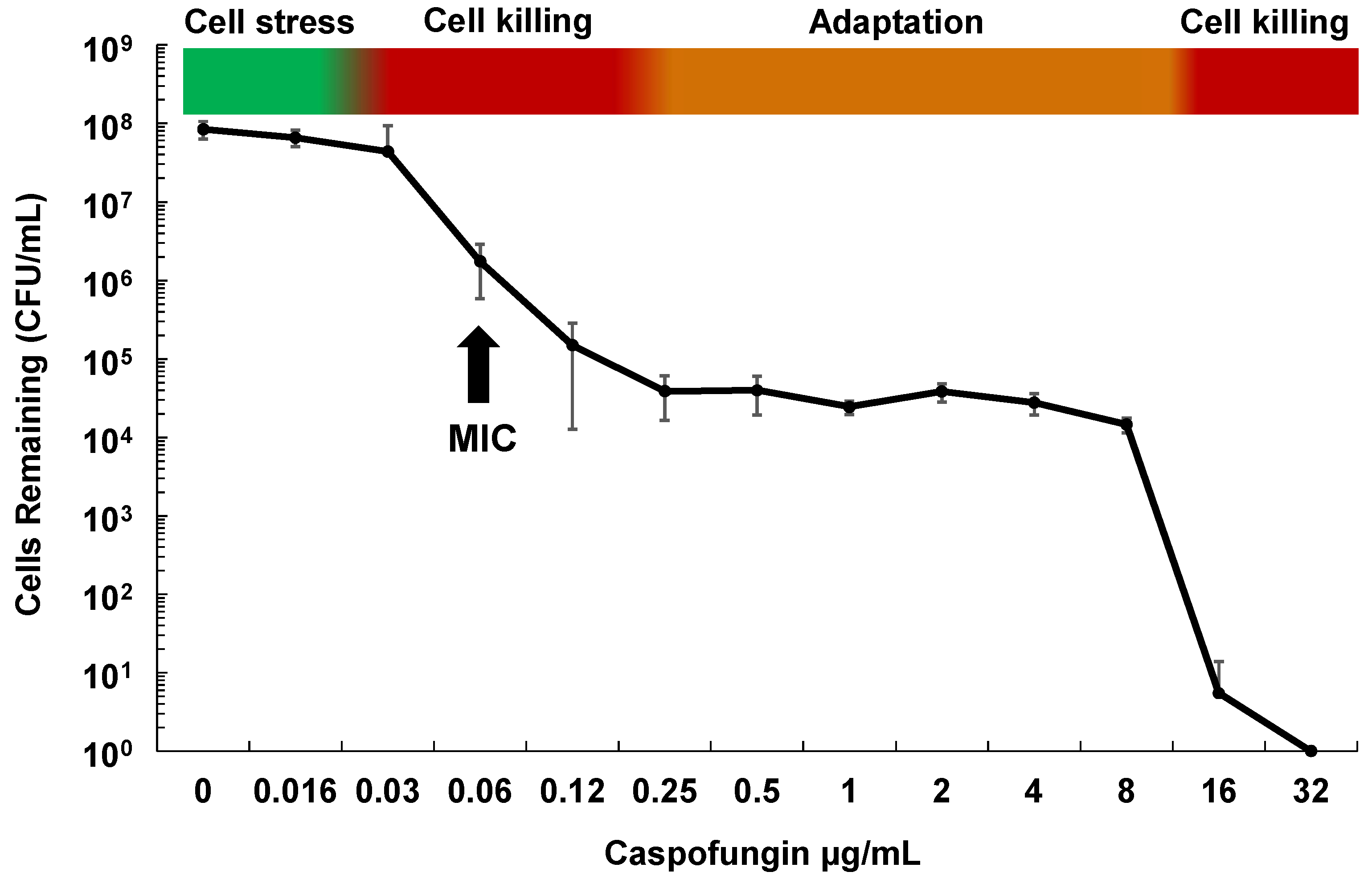
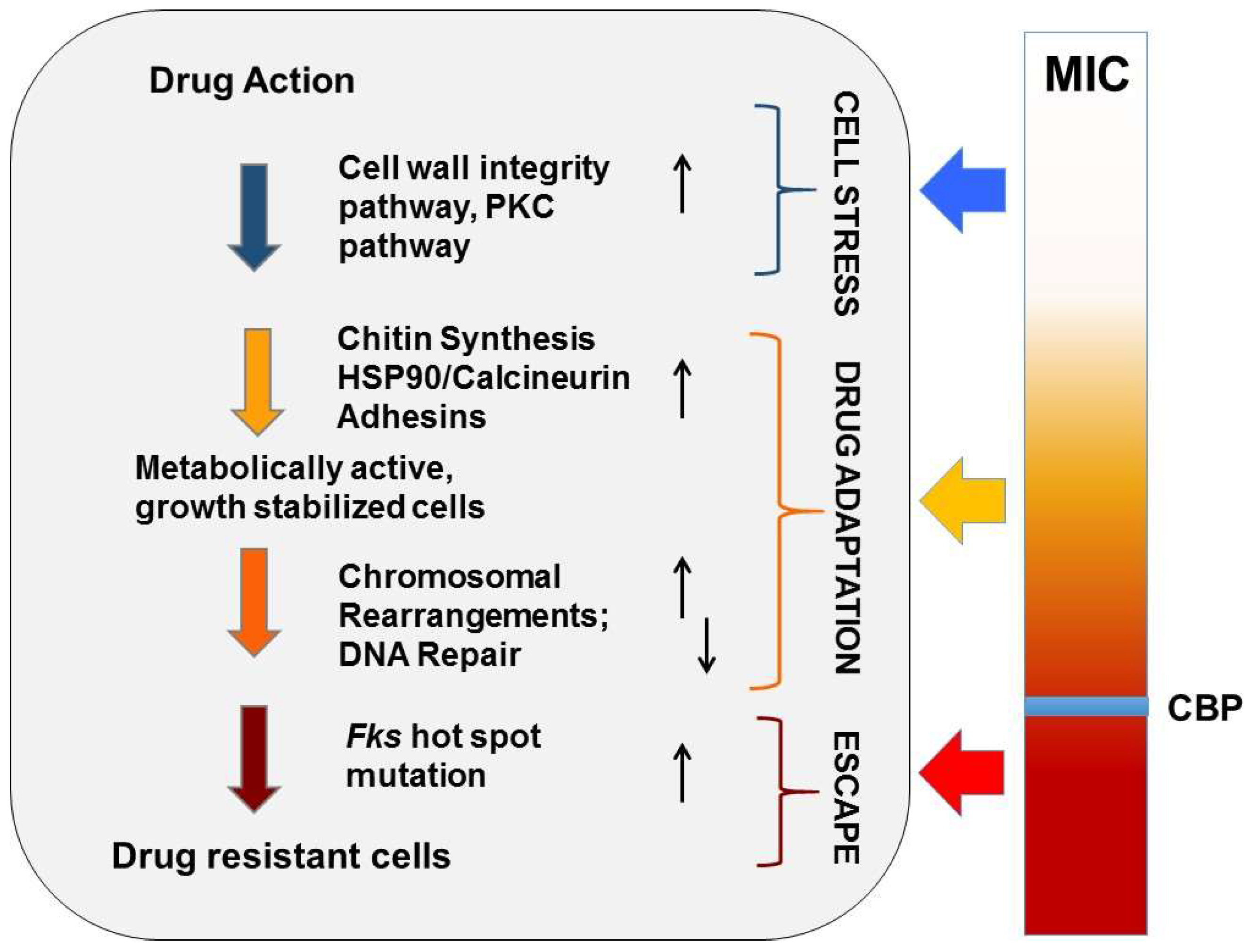
2.1. Drug Adaptation Is a Key Intermediate Leading to Echinocandin Resistance
2.2. Cellular Drivers of Echinocandin Adaptation
2.3. C. glabrata Specific Echinocandin Adaptation
2.4. Echinocandin- and FKS Gene- Specific Effects
2.5. MDR, PDR1 and Adhesins
2.6. Exploiting Genetic Diversity
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brown, G.D.; Denning, D.W.; Levitz, S.M. Tackling human fungal infections. Science 2012, 336, 647. [Google Scholar] [CrossRef] [PubMed]
- Huseyin, C.E.; O’Toole, P.W.; Cotter, P.D.; Scanlan, P.D. Forgotten fungi-the gut mycobiome in human health and disease. FEMS Microbiol. Rev. 2017, 41, 479–511. [Google Scholar] [CrossRef] [PubMed]
- Farmakiotis, D.; Tarrand, J.J.; Kontoyiannis, D.P. Drug-resistant Candida glabrata infection in cancer patients. Emerg. Infect. Dis. 2014, 20, 1833–1840. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Deshpande, L.M.; Davis, A.P.; Rhomberg, P.R.; Pfaller, M.A. Monitoring antifungal resistance in a global collection of invasive yeasts and molds: Application of clsi epidemiological cutoff values and whole-genome sequencing analysis for detection of azole resistance in Candida albicans. Antimicrob. Agents Chemother. 2017, 61, e00906-17. [Google Scholar] [CrossRef] [PubMed]
- Farmakiotis, D.; Kontoyiannis, D.P. Epidemiology of antifungal resistance in human pathogenic yeasts: Current viewpoint and practical recommendations for management. Int. J. Antimicrob. Agents 2017, 50, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Dannaoui, E.; Desnos-Ollivier, M.; Garcia-Hermoso, D.; Grenouillet, F.; Cassaing, S.; Baixench, M.T.; Bretagne, S.; Dromer, F.; Lortholary, O. Candida spp. With acquired echinocandin resistance, france, 2004–2010(1). Emerg. Infect. Dis. 2012, 18, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Woosley, L.N.; Diekema, D.J.; Messer, S.A.; Jones, R.N.; Pfaller, M.A. Low prevalence of FKS1 hot spot 1 mutations in a worldwide collection of Candida strains. Antimicrob. Agents Chemother. 2010, 54, 2655–2659. [Google Scholar] [CrossRef] [PubMed]
- Ostrosky-Zeichner, L. Candida glabrata and FKS mutations: Witnessing the emergence of the true multidrug-resistant Candida. Clin. Infect. Dis. 2013, 56, 1733–1734. [Google Scholar] [CrossRef] [PubMed]
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet Infect. Dis. 2017, 17, e383–e392. [Google Scholar] [CrossRef]
- Perlin, D.S. Mechanisms of echinocandin antifungal drug resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, B.D.; Johnson, M.D.; Pfeiffer, C.D.; Jimenez-Ortigosa, C.; Catania, J.; Booker, R.; Castanheira, M.; Messer, S.A.; Perlin, D.S.; Pfaller, M.A. Increasing echinocandin resistance in Candida glabrata: Clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin. Infect. Dis. 2013, 56, 1724–1732. [Google Scholar] [CrossRef] [PubMed]
- Pham, C.D.; Iqbal, N.; Bolden, C.B.; Kuykendall, R.J.; Harrison, L.H.; Farley, M.M.; Schaffner, W.; Beldavs, Z.G.; Chiller, T.M.; Park, B.J.; et al. Role of FKS mutations in Candida glabrata: Mic values, echinocandin resistance, and multidrug resistance. Antimicrob. Agents Chemother. 2014, 58, 4690–4696. [Google Scholar] [CrossRef] [PubMed]
- Cowen, L.E.; Sanglard, D.; Howard, S.J.; Rogers, P.D.; Perlin, D.S. Mechanisms of antifungal drug resistance. Cold Spring Harb. Perspect. Med. 2014. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.F.; Krol, A.A.; Sarti, K.E.; Bennett, J.E. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob. Agents Chemother. 2006, 50, 1384–1392. [Google Scholar] [CrossRef] [PubMed]
- Vermitsky, J.P.; Earhart, K.D.; Smith, W.L.; Homayouni, R.; Edlind, T.D.; Rogers, P.D. PDR1 regulates multidrug resistance in Candida glabrata: Gene disruption and genome-wide expression studies. Mol. Microbiol. 2006, 61, 704–722. [Google Scholar] [CrossRef] [PubMed]
- Niimi, K.; Maki, K.; Ikeda, F.; Holmes, A.R.; Lamping, E.; Niimi, M.; Monk, B.C.; Cannon, R.D. Overexpression of Candida albicans cdr1, cdr2, or mdr1 does not produce significant changes in echinocandin susceptibility. Antimicrob. Agents Chemother. 2006, 50, 1148–1155. [Google Scholar] [CrossRef] [PubMed]
- Perlin, D.S. Current perspectives on echinocandin class drugs. Future Microbiol. 2011, 6, 441–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazur, P.; Baginsky, W. In vitro activity of 1,3-β-d-glucan synthase requires the GTP-binding protein rho1. J. Biol. Chem. 1996, 271, 14604–14609. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kelly, R.; Kahn, J.N.; Robles, J.; Hsu, M.J.; Register, E.; Li, W.; Vyas, V.; Fan, H.; Abruzzo, G.; et al. Specific substitutions in the echinocandin target fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. Isolates. Antimicrob. Agents Chemother. 2005, 49, 3264–3273. [Google Scholar] [CrossRef] [PubMed]
- Katiyar, S.K.; Alastruey-Izquierdo, A.; Healey, K.R.; Johnson, M.E.; Perlin, D.S.; Edlind, T.D. FkS1 and FkS2 are functionally redundant but differentially regulated in Candida glabrata: Implications for echinocandin resistance. Antimicrob. Agents Chemother. 2012, 56, 6304–6309. [Google Scholar] [CrossRef] [PubMed]
- Arendrup, M.C.; Perlin, D.S. Echinocandin resistance: An emerging clinical problem? Curr. Opin. Infect. Dis. 2014, 27, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Effron, G.; Park, S.; Perlin, D.S. Correlating echinocandin mic and kinetic inhibition of FKS1 mutant glucan synthases for Candida albicans: Implications for interpretive breakpoints. Antimicrob. Agents Chemother. 2009, 53, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Shields, R.K.; Nguyen, M.H.; Press, E.G.; Kwa, A.L.; Cheng, S.; Du, C.; Clancy, C.J. The presence of an fks mutation rather than mic is an independent risk factor for failure of echinocandin therapy among patients with invasive candidiasis due to Candida glabrata. Antimicrob. Agents Chemother. 2012, 56, 4862–4869. [Google Scholar] [CrossRef] [PubMed]
- Pappas, P.G.; Kauffman, C.A.; Andes, D.R.; Clancy, C.J.; Marr, K.A.; Ostrosky-Zeichner, L.; Reboli, A.C.; Schuster, M.G.; Vazquez, J.A.; Walsh, T.J.; et al. Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of America. Clin. Infect. Dis. 2016, 62, e1–e50. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.S., 2nd; Wiederhold, N.P.; Wickes, B.L.; Patterson, T.F.; Jorgensen, J.H. Rapid emergence of echinocandin resistance in Candida glabrata resulting in clinical and microbiologic failure. Antimicrob. Agents Chemother. 2013, 57, 4559–4561. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Berkow, E.L.; Chow, N.; Welsh, R.M. Candida auris for the clinical microbiology laboratory: Not your grandfather’s Candida species. Clin. Microbiol. Newsl. 2017, 39, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Brunke, S.; Hube, B. Two unlike cousins: Candida albicans and C. glabrata infection strategies. Cell. Microbiol. 2013, 15, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Howard, S.J.; Livermore, J.; Sharp, A.; Goodwin, J.; Gregson, L.; Alastruey-Izquierdo, A.; Perlin, D.S.; Warn, P.A.; Hope, W.W. Pharmacodynamics of echinocandins against Candida glabrata: Requirement for dosage escalation to achieve maximal antifungal activity in neutropenic hosts. Antimicrob. Agents Chemother. 2011, 55, 4880–4887. [Google Scholar] [CrossRef] [PubMed]
- Lepak, A.; Castanheira, M.; Diekema, D.; Pfaller, M.; Andes, D. Optimizing echinocandin dosing and susceptibility breakpoint determination via in vivo pharmacodynamic evaluation against Candida glabrata with and without FKS mutations. Antimicrob. Agents Chemother. 2012, 56, 5875–5882. [Google Scholar] [CrossRef] [PubMed]
- Healey, K.R.; Nagasaki, Y.; Zimmerman, M.; Kordalewska, M.; Park, S.; Zhao, Y.; Perlin, D.S. The gastrointestinal tract is a major source of echinocandin drug resistance in a murine model of Candida glabrata colonization and systemic dissemination. Antimicrob. Agents Chemother. 2017, 61, e01412-17. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Prideaux, B.; Nagasaki, Y.; Lee, M.H.; Chen, P.Y.; Blanc, L.; Ho, H.; Clancy, C.J.; Nguyen, M.H.; Dartois, V.; et al. Unraveling drug penetration of echinocandin antifungals at the site of infection in an intra-abdominal abscess model. Antimicrob. Agents Chemother. 2017, 61, e01009-17. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, C.D.; Garcia-Effron, G.; Zaas, A.K.; Perfect, J.R.; Perlin, D.S.; Alexander, B.D. Breakthrough invasive candidiasis in patients on micafungin. J. Clin. Microbiol. 2010, 48, 2373–2380. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Rodrigues, C.F.; Araujo, D.; Rodrigues, M.E.; Henriques, M. Candida species biofilms’ antifungal resistance. J. Fungi 2017, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Bader, J.C.; Lakota, E.A.; Flanagan, S.; Ong, V.; Sandison, T.; Rubino, C.M.; Bhavnani, S.M.; Ambrose, P.G. Overcoming the resistance hurdle: Pharmacokinetic-pharmacodynamic target attainment analyses for rezafungin (cd101) against Candida albicans and Candida glabrata. Antimicrob. Agents Chemother. 2018, 62, e02614-17. [Google Scholar] [CrossRef] [PubMed]
- Lesage, G.; Bussey, H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006, 70, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.K.; Rogers, P.D.; Baerson, S.R.; Jacob, M.R.; Barker, K.S.; Cleary, J.D.; Walker, L.A.; Nagle, D.G.; Clark, A.M. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 2003, 278, 34998–35015. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.T.; Lee, R.E.; Barker, K.S.; Wei, L.; Homayouni, R.; Rogers, P.D. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 2005, 49, 2226–2236. [Google Scholar] [CrossRef] [PubMed]
- Rosenwald, A.G.; Arora, G.; Ferrandino, R.; Gerace, E.L.; Mohammednetej, M.; Nosair, W.; Rattila, S.; Subic, A.Z.; Rolfes, R. Identification of genes in Candida glabrata conferring altered responses to caspofungin, a cell wall synthesis inhibitor. G3 2016, 6, 2893–2907. [Google Scholar] [CrossRef] [PubMed]
- Cowen, L.E.; Steinbach, W.J. Stress, drugs, and evolution: The role of cellular signaling in fungal drug resistance. Eukaryot. Cell 2008, 7, 747–764. [Google Scholar] [CrossRef] [PubMed]
- Markovich, S.; Yekutiel, A.; Shalit, I.; Shadkchan, Y.; Osherov, N. Genomic approach to identification of mutations affecting caspofungin susceptibility in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 2004, 48, 3871–3876. [Google Scholar] [CrossRef] [PubMed]
- Reinoso-Martin, C.; Schuller, C.; Schuetzer-Muehlbauer, M.; Kuchler, K. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of slt2p mitogen-activated protein kinase signaling. Eukaryot. Cell 2003, 2, 1200–1210. [Google Scholar] [CrossRef] [PubMed]
- Wiederhold, N.P.; Kontoyiannis, D.P.; Prince, R.A.; Lewis, R.E. Attenuation of the activity of caspofungin at high concentrations against Candida albicans: Possible role of cell wall integrity and calcineurin pathways. Antimicrob. Agents Chemother. 2005, 49, 5146–5148. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Inamine, T.; Yamauchi, S.; Nagayoshi, Y.; Saijo, T.; Izumikawa, K.; Seki, M.; Kakeya, H.; Yamamoto, Y.; Yanagihara, K.; et al. Role of the slt2 mitogen-activated protein kinase pathway in cell wall integrity and virulence in Candida glabrata. FEMS Yeast Res. 2010, 10, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Schwarzmuller, T.; Ma, B.; Hiller, E.; Istel, F.; Tscherner, M.; Brunke, S.; Ames, L.; Firon, A.; Green, B.; Cabral, V.; et al. Systematic phenotyping of a large-scale Candida glabrata deletion collection reveals novel antifungal tolerance genes. PLoS Pathog. 2014, 10, e1004211. [Google Scholar] [CrossRef] [PubMed]
- Cota, J.M.; Grabinski, J.L.; Talbert, R.L.; Burgess, D.S.; Rogers, P.D.; Edlind, T.D.; Wiederhold, N.P. Increases in slt2 expression and chitin content are associated with incomplete killing of Candida glabrata by caspofungin. Antimicrob. Agents Chemother. 2008, 52, 1144–1146. [Google Scholar] [CrossRef] [PubMed]
- Singh-Babak, S.D.; Babak, T.; Diezmann, S.; Hill, J.A.; Xie, J.L.; Chen, Y.L.; Poutanen, S.M.; Rennie, R.P.; Heitman, J.; Cowen, L.E. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog. 2012, 8, e1002718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyazaki, T.; Yamauchi, S.; Inamine, T.; Nagayoshi, Y.; Saijo, T.; Izumikawa, K.; Seki, M.; Kakeya, H.; Yamamoto, Y.; Yanagihara, K.; et al. Roles of calcineurin and crz1 in antifungal susceptibility and virulence of Candida glabrata. Antimicrob. Agents Chemother. 2010, 54, 1639–1643. [Google Scholar] [CrossRef] [PubMed]
- Rai, M.N.; Balusu, S.; Gorityala, N.; Dandu, L.; Kaur, R. Functional genomic analysis of Candida glabrata-macrophage interaction: Role of chromatin remodeling in virulence. PLoS Pathog. 2012, 8, e1002863. [Google Scholar] [CrossRef] [PubMed]
- Garnaud, C.; Champleboux, M.; Maubon, D.; Cornet, M.; Govin, J. Histone deacetylases and their inhibition in Candida species. Front. Microbiol. 2016, 7, 1238. [Google Scholar] [CrossRef] [PubMed]
- Stevens, D.A.; Ichinomiya, M.; Koshi, Y.; Horiuchi, H. Escape of Candida from caspofungin inhibition at concentrations above the mic (paradoxical effect) accomplished by increased cell wall chitin; evidence for β-1,6-glucan synthesis inhibition by caspofungin. Antimicrob. Agents Chemother. 2006, 50, 3160–3161. [Google Scholar] [CrossRef] [PubMed]
- Munro, C.A.; Selvaggini, S.; de Bruijn, I.; Walker, L.; Lenardon, M.D.; Gerssen, B.; Milne, S.; Brown, A.J.; Gow, N.A. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol. Microbiol. 2007, 63, 1399–1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, L.A.; Gow, N.A.; Munro, C.A. Elevated chitin content reduces the susceptibility of Candida species to caspofungin. Antimicrob. Agents Chemother. 2013, 57, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Charlet, R.; Pruvost, Y.; Tumba, G.; Istel, F.; Poulain, D.; Kuchler, K.; Sendid, B.; Jawhara, S. Remodeling of the Candida glabrata cell wall in the gastrointestinal tract affects the gut microbiota and the immune response. Sci. Rep. 2018, 8, 3316. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.F.; Henriques, M. Portrait of matrix gene expression in Candida glabrata biofilms with stress induced by different drugs. Genes 2018, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.F.; Rodrigues, M.E.; Henriques, M. Susceptibility of Candida glabrata biofilms to echinocandins: Alterations in the matrix composition. Biofouling 2018, 34, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Healey, K.R.; Challa, K.K.; Edlind, T.D.; Katiyar, S.K. Sphingolipids mediate differential echinocandin susceptibility in Candida albicans and Aspergillus nidulans. Antimicrob. Agents Chemother. 2015, 59, 3377–3384. [Google Scholar] [CrossRef] [PubMed]
- Healey, K.R.; Katiyar, S.K.; Raj, S.; Edlind, T.D. Crs-mis in Candida glabrata: Sphingolipids modulate echinocandin-fks interaction. Mol. Microbiol. 2012, 86, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Ong, V.; James, K.D.; Smith, S.; Krishnan, B.R. Pharmacokinetics of the novel echinocandin cd101 in multiple animal species. Antimicrob. Agents Chemother. 2017, 61, e01626-16. [Google Scholar] [CrossRef] [PubMed]
- Sandison, T.; Ong, V.; Lee, J.; Thye, D. Safety and pharmacokinetics of cd101 IV, a novel echinocandin, in healthy adults. Antimicrob. Agents Chemother. 2017, 61, e01627-16. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Messer, S.A.; Rhomberg, P.R.; Borroto-Esoda, K.; Castanheira, M. Differential activity of the oral glucan synthase inhibitor scy-078 against wild-type and echinocandin-resistant strains of Candida species. Antimicrob. Agents Chemother. 2017, 61, e00161-17. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Ortigosa, C.; Perez, W.B.; Angulo, D.; Borroto-Esoda, K.; Perlin, D.S. De novo acquisition of resistance to scy-078 in Candida glabrata involves FKS mutations that both overlap and are distinct from those conferring echinocandin resistance. Antimicrob. Agents Chemother. 2017, 61, e00833-17. [Google Scholar] [CrossRef] [PubMed]
- Mazur, P.; Morin, N.; Baginsky, W.; el-Sherbeini, M.; Clemas, J.A.; Nielsen, J.B.; Foor, F. Differential expression and function of two homologous subunits of yeast 1,3-β-d-glucan synthase. Mol. Cell. Biol. 1995, 15, 5671–5681. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, S.; Hirata, A.; Nogami, S.; Beauvais, A.; Latge, J.P.; Ohya, Y. Homologous subunits of 1,3-β-glucan synthase are important for spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 2007, 6, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Suwunnakorn, S.; Wakabayashi, H.; Kordalewska, M.; Perlin, D.S.; Rustchenko, E. Fks2 and fks3 genes of opportunistic human pathogen Candida albicans influence echinocandin susceptibility. Antimicrob. Agents Chemother. 2018, 62, e02299-17. [Google Scholar] [CrossRef] [PubMed]
- Vale-Silva, L.; Ischer, F.; Leibundgut-Landmann, S.; Sanglard, D. Gain-of-function mutations in PDR1, a regulator of antifungal drug resistance in Candida glabrata, control adherence to host cells. Infect. Immun. 2013, 81, 1709–1720. [Google Scholar] [CrossRef] [PubMed]
- Vale-Silva, L.A.; Moeckli, B.; Torelli, R.; Posteraro, B.; Sanguinetti, M.; Sanglard, D. Upregulation of the adhesin gene epa1 mediated by PDR1 in Candida glabrata leads to enhanced host colonization. mSphere 2016, 1, e00065-15. [Google Scholar] [CrossRef] [PubMed]
- Ni, Q.; Wang, C.; Tian, Y.; Dong, D.; Jiang, C.; Mao, E.; Peng, Y. CgPDR1 gain-of-function mutations lead to azole-resistance and increased adhesion in clinical Candida glabrata strains. Mycoses 2018, 61, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Salazar, S.B.; Wang, C.; Munsterkotter, M.; Okamoto, M.; Takahashi-Nakaguchi, A.; Chibana, H.; Lopes, M.M.; Guldener, U.; Butler, G.; Mira, N.P. Comparative genomic and transcriptomic analyses unveil novel features of azole resistance and adaptation to the human host in Candida glabrata. FEMS Yeast Res. 2018, 18, fox079. [Google Scholar] [CrossRef] [PubMed]
- Cormack, B.P.; Ghori, N.; Falkow, S. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science 1999, 285, 578–582. [Google Scholar] [CrossRef] [PubMed]
- De Groot, P.W.; Kraneveld, E.A.; Yin, Q.Y.; Dekker, H.L.; Gross, U.; Crielaard, W.; de Koster, C.G.; Bader, O.; Klis, F.M.; Weig, M. The cell wall of the human pathogen Candida glabrata: Differential incorporation of novel adhesin-like wall proteins. Eukaryot. Cell 2008, 7, 1951–1964. [Google Scholar] [CrossRef] [PubMed]
- Vale-Silva, L.; Beaudoing, E.; Tran, V.D.T.; Sanglard, D. Comparative genomics of two sequential Candida glabrata clinical isolates. G3 2017, 7, 2413–2426. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Fuentes, E.; Gutierrez-Escobedo, G.; Timmermans, B.; Van Dijck, P.; De Las Penas, A.; Castano, I. Candida glabrata’s genome plasticity confers a unique pattern of expressed cell wall proteins. J. Fungi 2018, 4, 67. [Google Scholar] [CrossRef] [PubMed]
- Castano, I.; Pan, S.J.; Zupancic, M.; Hennequin, C.; Dujon, B.; Cormack, B.P. Telomere length control and transcriptional regulation of subtelomeric adhesins in Candida glabrata. Mol. Microbiol. 2005, 55, 1246–1258. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Sanguinetti, M.; Torelli, R.; Posteraro, B.; Sanglard, D. Contribution of cgpdr1-regulated genes in enhanced virulence of azole-resistant Candida glabrata. PLoS ONE 2011, 6, e17589. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Ischer, F.; Calabrese, D.; Posteraro, B.; Sanguinetti, M.; Fadda, G.; Rohde, B.; Bauser, C.; Bader, O.; Sanglard, D. Gain of function mutations in cgpdr1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog. 2009, 5, e1000268. [Google Scholar] [CrossRef] [PubMed]
- Vitenshtein, A.; Charpak-Amikam, Y.; Yamin, R.; Bauman, Y.; Isaacson, B.; Stein, N.; Berhani, O.; Dassa, L.; Gamliel, M.; Gur, C.; et al. NK cell recognition of Candida glabrata through binding of nkp46 and ncr1 to fungal ligands epa1, epa6, and epa7. Cell Host Microbe 2016, 20, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Galhardo, R.S.; Hastings, P.J.; Rosenberg, S.M. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 399–435. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, M.J.; Aponyi, I.; Rosenberg, S.M. General stress response regulator rpos in adaptive mutation and amplification in Escherichia coli. Genetics 2004, 166, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Ponder, R.G.; Fonville, N.C.; Rosenberg, S.M. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 2005, 19, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Shee, C.; Gibson, J.L.; Darrow, M.C.; Gonzalez, C.; Rosenberg, S.M. Impact of a stress-inducible switch to mutagenic repair of DNA breaks on mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA 2011, 108, 13659–13664. [Google Scholar] [CrossRef] [PubMed]
- Shor, E.; Fox, C.A.; Broach, J.R. The yeast environmental stress response regulates mutagenesis induced by proteotoxic stress. PLoS Genet. 2013, 9, e1003680. [Google Scholar] [CrossRef] [PubMed]
- MacLean, R.C.; Torres-Barcelo, C.; Moxon, R. Evaluating evolutionary models of stress-induced mutagenesis in bacteria. Nat. Rev. Genet. 2013, 14, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ami, R.; Zimmerman, O.; Finn, T.; Amit, S.; Novikov, A.; Wertheimer, N.; Lurie-Weinberger, M.; Berman, J. Heteroresistance to fluconazole is a continuously distributed phenotype among Candida glabrata clinical strains associated with in vivo persistence. mBio 2016, 7, e00655-16. [Google Scholar] [CrossRef] [PubMed]
- Mota, S.; Alves, R.; Carneiro, C.; Silva, S.; Brown, A.J.; Istel, F.; Kuchler, K.; Sampaio, P.; Casal, M.; Henriques, M.; et al. Candida glabrata susceptibility to antifungals and phagocytosis is modulated by acetate. Front. Microbiol. 2015, 6, 919. [Google Scholar] [CrossRef] [PubMed]
- Lott, T.J.; Frade, J.P.; Lockhart, S.R. Multilocus sequence type analysis reveals both clonality and recombination in populations of Candida glabrata bloodstream isolates from U.S. Surveillance studies. Eukaryot. Cell 2010, 9, 619–625. [Google Scholar] [CrossRef] [PubMed]
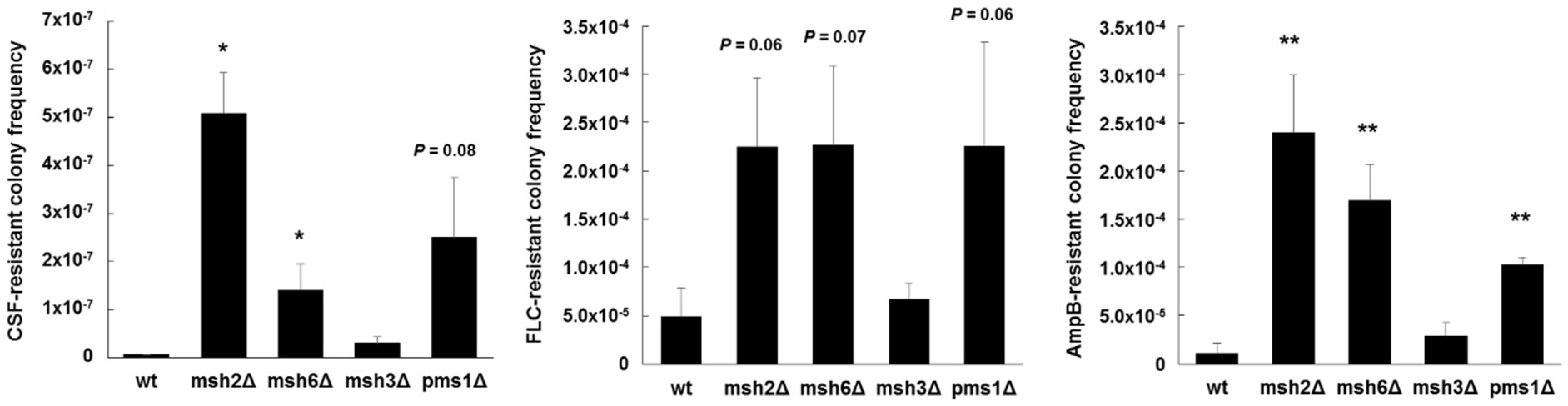
- Healey, K.R.; Jimenez Ortigosa, C.; Shor, E.; Perlin, D.S. Genetic drivers of multidrug resistance in Candida glabrata. Front. Microbiol. 2016, 7, 1995. [Google Scholar] [CrossRef] [PubMed]
- Healey, K.R.; Zhao, Y.; Perez, W.B.; Lockhart, S.R.; Sobel, J.D.; Farmakiotis, D.; Kontoyiannis, D.P.; Sanglard, D.; Taj-Aldeen, S.J.; Alexander, B.D.; et al. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat. Commun. 2016, 7, 11128. [Google Scholar] [CrossRef] [PubMed]
- Polakova, S.; Blume, C.; Zarate, J.A.; Mentel, M.; Jorck-Ramberg, D.; Stenderup, J.; Piskur, J. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc. Natl. Acad. Sci. USA 2009, 106, 2688–2693. [Google Scholar] [CrossRef] [PubMed]
- Dodgson, A.R.; Pujol, C.; Denning, D.W.; Soll, D.R.; Fox, A.J. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J. Clin. Microbiol. 2003, 41, 5709–5717. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.; Thierry, A.; Coppee, J.Y.; Gouyette, C.; Hennequin, C.; Sismeiro, O.; Talla, E.; Dujon, B.; Fairhead, C. Genomic polymorphism in the population of Candida glabrata: Gene copy-number variation and chromosomal translocations. Fungal Genet. Biol. 2009, 46, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Chen, Y.C.; Lo, H.J.; Chen, K.W.; Li, S.Y. Assessment of Candida glabrata strain relatedness by pulsed-field gel electrophoresis and multilocus sequence typing. J. Clin. Microbiol. 2007, 45, 2452–2459. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Chae, M.J.; Song, J.W.; Jung, S.I.; Cho, D.; Kee, S.J.; Kim, S.H.; Shin, M.G.; Suh, S.P.; Ryang, D.W. Changes in karyotype and azole susceptibility of sequential bloodstream isolates from patients with Candida glabrata candidemia. J. Clin. Microbiol. 2007, 45, 2385–2391. [Google Scholar] [CrossRef] [PubMed]
- Haw, R.; Yarragudi, A.D.; Uemura, H. Isolation of a Candida glabrata homologue of rap1, a regulator of transcription and telomere function in Saccharomyces cerevisiae. Yeast 2001, 18, 1277–1284. [Google Scholar] [CrossRef] [PubMed]
- Malkova, A.; Haber, J.E. Mutations arising during repair of chromosome breaks. Annu. Rev. Genet. 2012, 46, 455–473. [Google Scholar] [CrossRef] [PubMed]
- Sonoda, E.; Hochegger, H.; Saberi, A.; Taniguchi, Y.; Takeda, S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair 2006, 5, 1021–1029. [Google Scholar] [CrossRef] [PubMed]
- Boiteux, S.; Jinks-Robertson, S. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics 2013, 193, 1025–1064. [Google Scholar] [CrossRef] [PubMed]
- Hakem, R. DNA-damage repair; the good, the bad, and the ugly. EMBO J. 2008, 27, 589–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delliere, S.; Healey, K.; Gits-Muselli, M.; Carrara, B.; Barbaro, A.; Guigue, N.; Lecefel, C.; Touratier, S.; Desnos-Ollivier, M.; Perlin, D.S.; et al. Fluconazole and echinocandin resistance of Candida glabrata correlates better with antifungal drug exposure rather than with MSH2 mutator genotype in a french cohort of patients harboring low rates of resistance. Front. Microbiol. 2016, 7, 2038. [Google Scholar] [CrossRef] [PubMed]
- Byun, S.A.; Won, E.J.; Kim, M.N.; Lee, W.G.; Lee, K.; Lee, H.S.; Uh, Y.; Healey, K.R.; Perlin, D.S.; Choi, M.J.; et al. Multilocus sequence typing (mlst) genotypes of Candida glabrata bloodstream isolates in Korea: Association with antifungal resistance, mutations in mismatch repair gene (MSH2), and clinical outcomes. Front. Microbiol. 2018, 9, 1523. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Xiao, M.; Wang, H.; Yu, S.Y.; Zhang, G.; Zhao, Y.; Xu, Y.C. Profiling of PDR1 and MSH2 in Candida glabrata bloodstream isolates from a multicenter study in China. Antimicrob. Agents Chemother. 2018, 62, e00153-18. [Google Scholar] [CrossRef] [PubMed]
- Cleveland, A.A.; Farley, M.M.; Harrison, L.H.; Stein, B.; Hollick, R.; Lockhart, S.R.; Magill, S.S.; Derado, G.; Park, B.J.; Chiller, T.M. Changes in incidence and antifungal drug resistance in candidemia: Results from population-based laboratory surveillance in Atlanta and Baltimore, 2008–2011. Clin. Infect. Dis. 2012, 55, 1352–1361. [Google Scholar] [CrossRef] [PubMed]
- Lott, T.J.; Frade, J.P.; Lyon, G.M.; Iqbal, N.; Lockhart, S.R. Bloodstream and non-invasive isolates of candida glabrata have similar population structures and fluconazole susceptibilities. Med Mycol 2012, 50, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Healey, K.R.; Yadav, P.; Upadhyaya, G.; Sachdeva, N.; Sarma, S.; Kumar, A.; Tarai, B.; Perlin, D.S.; Chowdhary, A. Absence of azole or echinocandin resistance in Candida glabrata isolates in India despite background prevalence of strains with defects in the DNA mismatch repair pathway. Antimicrob. Agents Chemother. 2018, 62, e00195-18. [Google Scholar] [CrossRef] [PubMed]
- Healey, K.; Shor, E.; Perlin, D. Antifungal resistant isolates of Candida glabrata from the united states are enriched for specific sequence types with distinct MSH2 alleles. In Proceedings of the 20th Congress of the International Society for Human and Animal Mycology, ISHAM, Amsterdam, The Netherlands, 30 June–4 July 2018. Poster: P475. [Google Scholar]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Healey, K.R.; Perlin, D.S. Fungal Resistance to Echinocandins and the MDR Phenomenon in Candida glabrata. J. Fungi 2018, 4, 105. https://doi.org/10.3390/jof4030105
Healey KR, Perlin DS. Fungal Resistance to Echinocandins and the MDR Phenomenon in Candida glabrata. Journal of Fungi. 2018; 4(3):105. https://doi.org/10.3390/jof4030105
Chicago/Turabian StyleHealey, Kelley R., and David S. Perlin. 2018. "Fungal Resistance to Echinocandins and the MDR Phenomenon in Candida glabrata" Journal of Fungi 4, no. 3: 105. https://doi.org/10.3390/jof4030105




