Morphology Changes in Human Fungal Pathogens upon Interaction with the Host
Abstract
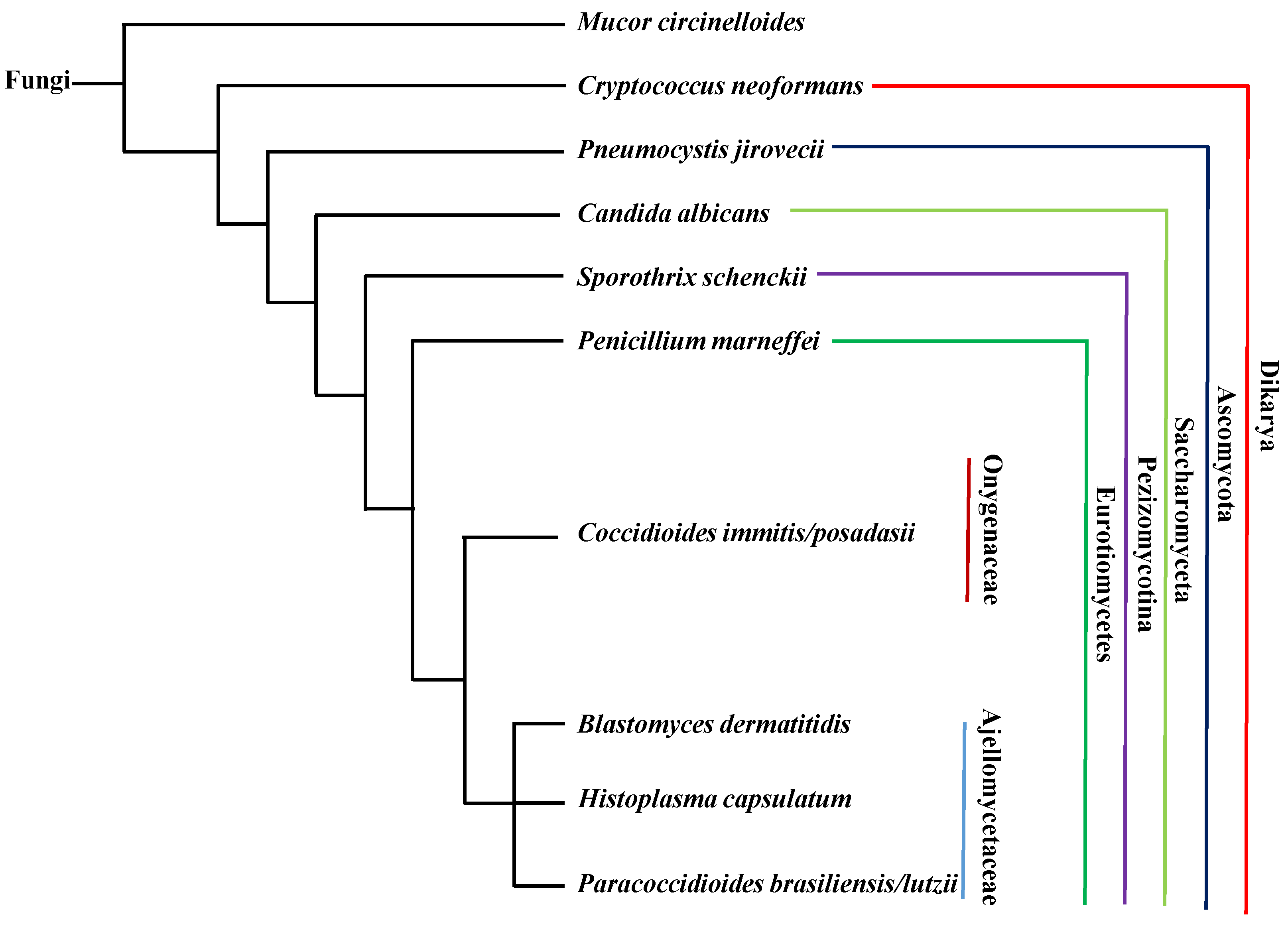
:1. Introduction
2. C. albicans—A Commensal Fungal Pathogen
3. Thermally Dimorphic Fungal Pathogens
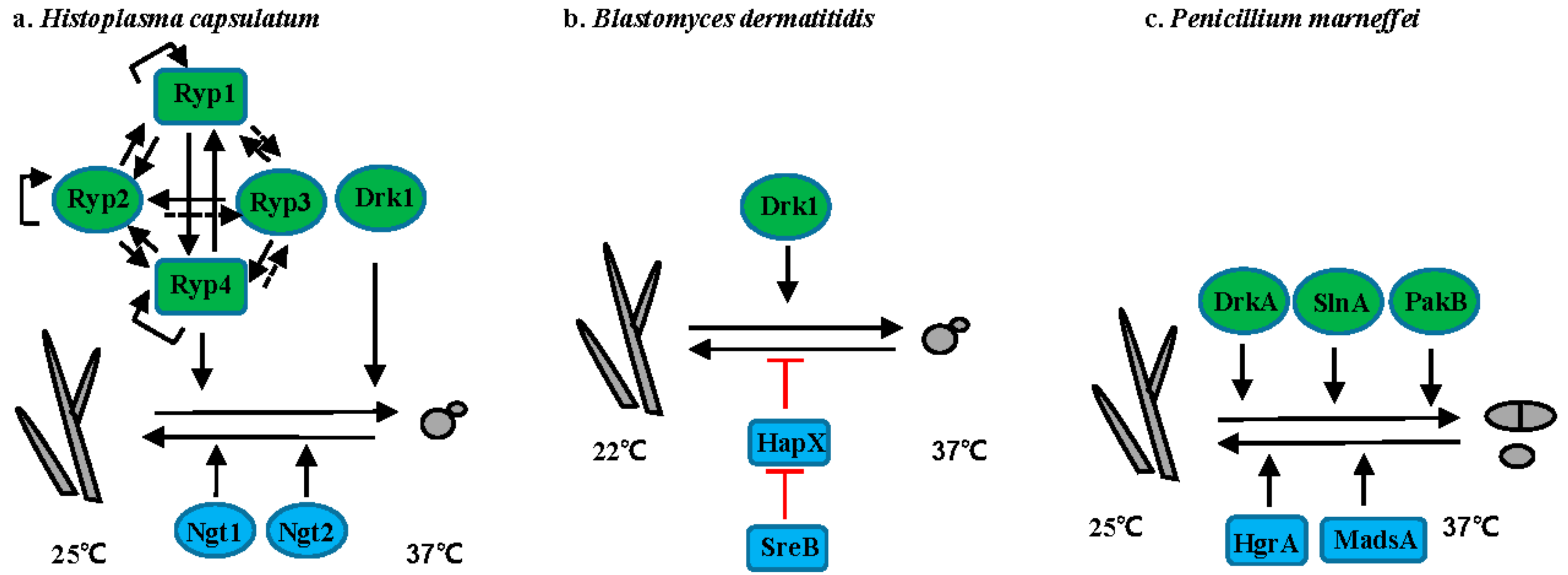
3.1. Histoplasma Species
3.2. Blastomyces Species
3.3. T. marneffei
3.4. Paracoccidioides Species
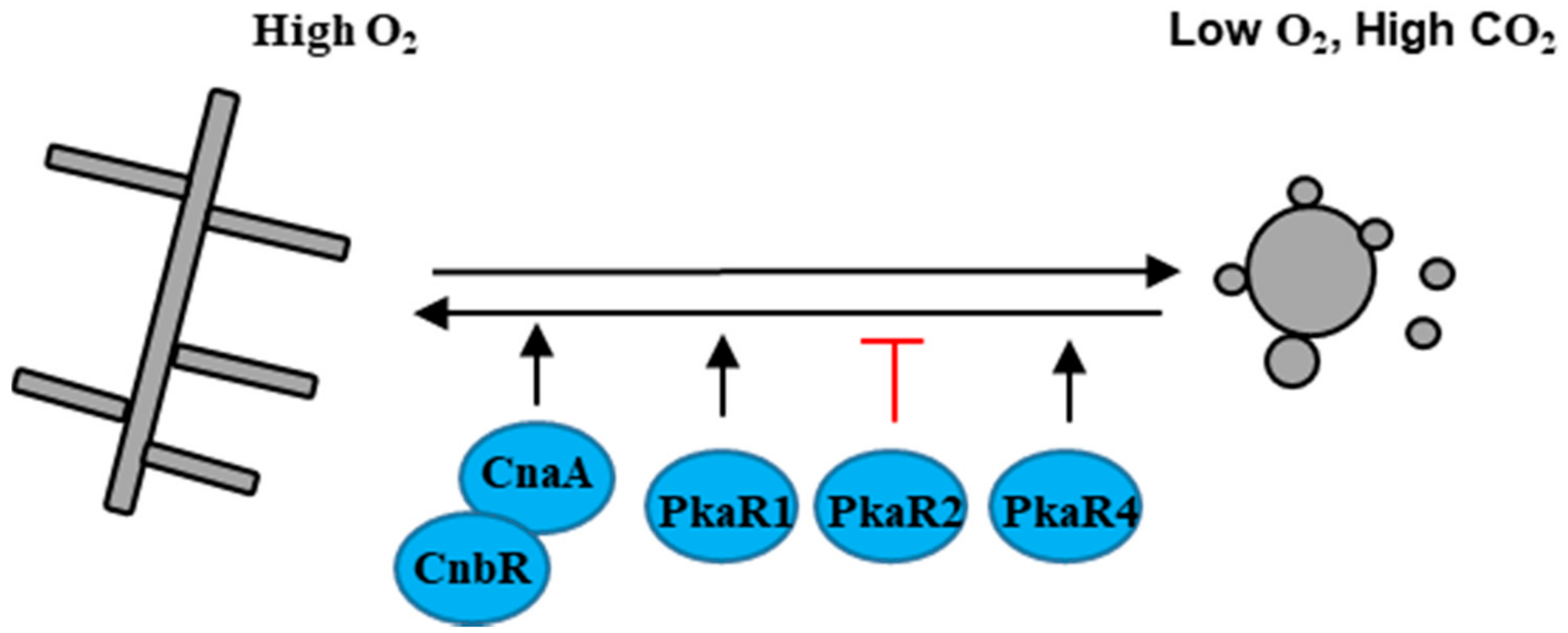
3.5. S. schenckii
3.6. Summary
4. Aerobically/Anaerobically Dimorphic Fungal Pathogen
5. Fungal Pathogens That Exhibit Cell Size Variation
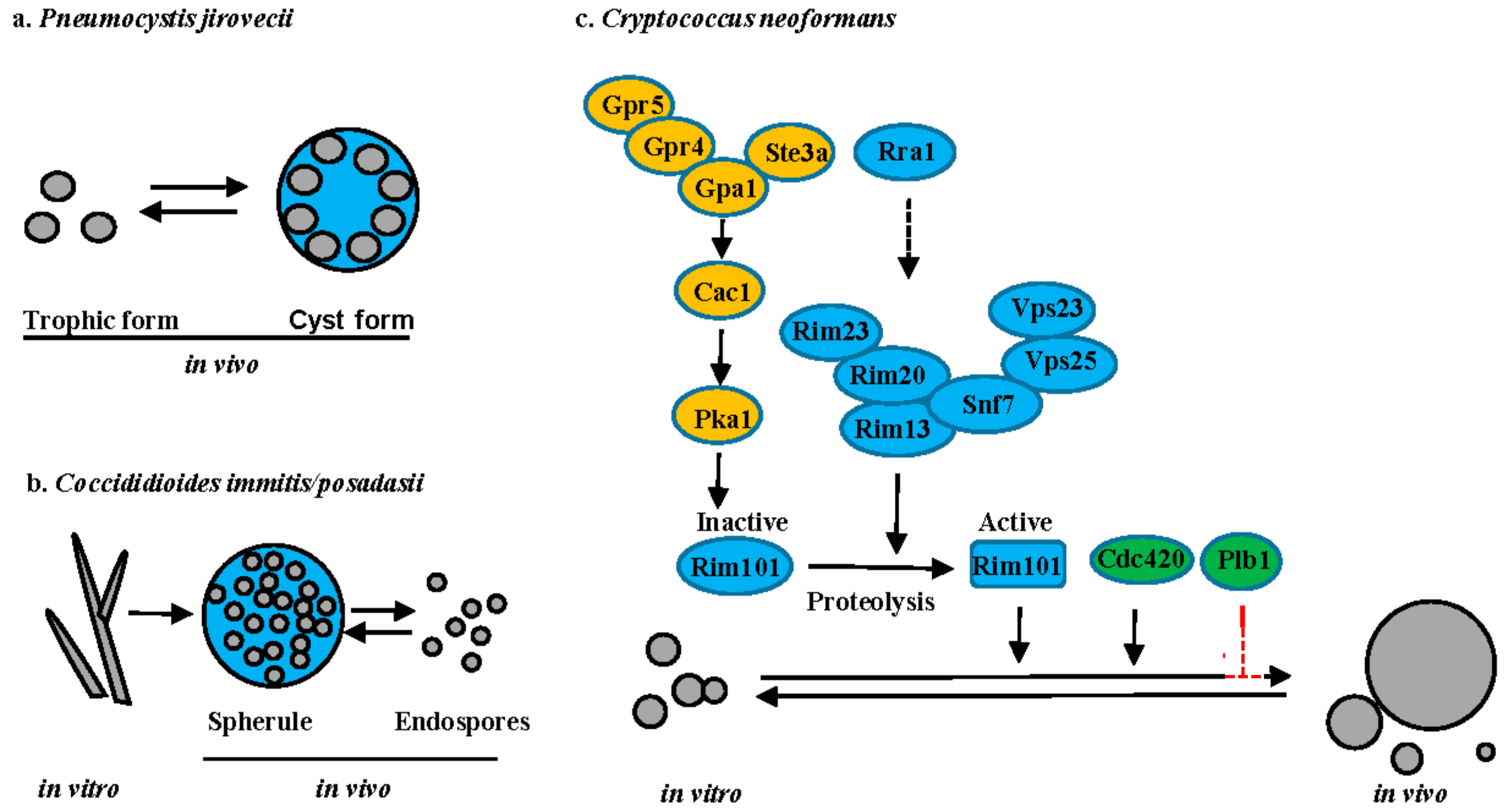
5.1. Pneumocystis Species
5.2. Coccidioides Species
5.3. Cryptococcus Species
5.4. Summary
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hawksworth, D.L. The magnitude of fungal diversity: The 1.5 million species estimate revisited. Mycol. Res. 2001, 105, 1422–1432. [Google Scholar] [CrossRef]
- Blackwell, M. The fungi: 1, 2, 3... 5.1 million species? Am. J. Bot. 2011, 98, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.H.; Latham, S.M.; Woolhouse, M.E. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 983–998. [Google Scholar] [CrossRef] [PubMed]
- Havlickova, B.; Czaika, V.A.; Friedrich, M. Epidemiological trends in skin mycoses worldwide. Mycoses 2008, 4, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Sobel, J.D. Vulvovaginal candidosis. Lancet 2007, 369, 1961–1971. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef] [PubMed]
- Saville, S.P.; Lazzell, A.L.; Monteagudo, C.; Lopez-Ribot, J.L. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell 2003, 2, 1053–1060. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.L.; Lee, R.T.; Fang, H.M.; Wang, Y.M.; Li, R.; Zou, H.; Zhu, Y.; Wang, Y. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe 2008, 4, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Klengel, T.; Liang, W.J.; Chaloupka, J.; Ruoff, C.; Schröppel, K.; Naglik, J.R.; Eckert, S.E.; Mogensen, E.G.; Haynes, K.; Tuite, M.F.; et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr. Biol. 2005, 15, 2021–2026. [Google Scholar] [CrossRef] [PubMed]
- Sonneborn, A.; Bockmühl, D.P.; Gerads, M.; Kurpanek, K.; Sanglard, D.; Ernst, J.F. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol. Microbiol. 2000, 35, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Stoldt, V.R.; Sonneborn, A.; Leuker, C.E.; Ernst, J.F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997, 16, 1982–1991. [Google Scholar] [CrossRef] [PubMed]
- Bockmühl, D.P.; Ernst, J.F. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics 2001, 157, 1523–1530. [Google Scholar] [PubMed]
- Lassak, T.; Schneider, E.; Bussmann, M.; Kurtz, D.; Manak, J.R.; Srikantha, T.; Soll, D.R.; Ernst, J.F. Target specificity of the Candida albicans Efg1 regulator. Mol. Microbiol. 2011, 82, 602–618. [Google Scholar] [CrossRef] [PubMed]
- Znaidi, S.; Nesseir, A.; Chauvel, M.; Rossignol, T.; d’Enfert, C. A comprehensive functional portrait of two heat shock factor-type transcriptional regulators involved in Candida albicans morphogenesis and virulence. PLoS Pathog. 2013, 9, e1003519. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Lane, S.; Raniga, P.P.; Lu, Y.; Zhou, Z.; Ramon, K.; Chen, J.; Liu, H. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol. Biol. Cell 2006, 17, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Guan, G.; Xie, J.; Cottier, F.; Sun, Y.; Jia, W.; Mühlschlegel, F.A.; Huang, G. The transcription factor Flo8 mediates CO2 sensing in the human fungal pathogen Candida albicans. Mol. Biol. Cell 2012, 23, 2692–2701. [Google Scholar] [CrossRef] [PubMed]
- Cleary, I.A.; Lazzell, A.L.; Monteagudo, C.; Thomas, D.P.; Saville, S.P. BRG1 and NRG1 form a novel feedback circuit regulating Candida albicans hypha formation and virulence. Mol. Microbiol. 2012, 85, 557–573. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, M.; Thompson, D.S.; Lazzell, A.; Carlisle, P.L.; Pierce, C.; Monteagudo, C.; López-Ribot, J.L.; Kadosh, D. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell 2008, 19, 1354–1365. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, A.; Rupp, S.; Taylor, B.N.; Röllinghoff, M.; Schröppel, K. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 2000, 38, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Hwang, C.S.; Oh, J.H.; Huh, W.K.; Yim, H.S.; Kang, S.O. Ssn6, an important factor of morphological conversion and virulence in Candida albicans. Mol. Microbiol. 2003, 47, 1029–1043. [Google Scholar] [CrossRef] [PubMed]
- Murad, A.M.; Leng, P.; Straffon, M.; Wishart, J.; Macaskill, S.; MacCallum, D.; Schnell, N.; Talibi, D.; Marechal, D.; Tekaia, F.; et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001, 20, 4742–4752. [Google Scholar] [CrossRef] [PubMed]
- Braun, B.R.; Kadosh, D.; Johnson, A.D. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 2001, 20, 4753–4761. [Google Scholar] [CrossRef] [PubMed]
- Kadosh, D.; Johnson, A.D. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 2001, 21, 2496–2505. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Su, C.; Liu, H. A GATA transcription factor recruits Hda1 in response to reduced Tor1 signaling to establish a hyphal chromatin state in Candida albicans. PLoS Pathog. 2012, 8, e1002663. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Wang, Y.; Wang, Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 2004, 23, 1845–1856. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.D.; Lee, R.T.; Wang, Y.M.; Lin, Q.S.; Wang, Y. Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 2007, 26, 3760–3769. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, R.S.; Zaas, A.K.; Betancourt-Quiroz, M.; Perfect, J.R.; Cowen, L.E. The Hsp90 co-chaperone Sgt1 governs Candida albicans morphogenesis and drug resistance. PLoS ONE 2012, 7, e44734. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, R.S.; Uppuluri, P.; Zaas, A.K.; Collins, C.; Senn, H.; Perfect, J.R.; Heitman, J.; Cowen, L.E. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr. Biol. 2009, 19, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Summers, E.; Guo, B.; Fink, G. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol. 1999, 181, 6339–6346. [Google Scholar] [PubMed]
- Fang, H.M.; Wang, Y. RA domain-mediated interaction of Cdc35 with Ras1 is essential for increasing cellular cAMP level for Candida albicans hyphal development. Mol. Microbiol. 2006, 61, 484–496. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, R.S.; Sellam, A.; Tebbji, F.; Whiteway, M.; Nantel, A.; Cowen, L.E. Pho85, Pcl1, and Hms1 signaling governs Candida albicans morphogenesis induced by high temperature or Hsp90 compromise. Curr. Biol. 2012, 22, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Raja, J.; Davis, D.A. The β-arrestin-like protein Rim8 is hyperphosphorylated and complexes with Rim21 and Rim101 to promote adaptation to neutral-alkaline pH. Eukaryot Cell 2012, 11, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Smith, F.J., Jr.; Subaran, R.; Mitchell, A.P. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 2004, 15, 5528–5537. [Google Scholar] [CrossRef] [PubMed]
- Kullas, A.L.; Li, M.; Davis, D.A. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot. Cell 2004, 3, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.M.; Davis, D.A. Mutational analysis of Candida albicans SNF7 reveals genetically separable Rim101 and ESCRT functions and demonstrates divergence in bro1-domain protein interactions. Genetics 2010, 184, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.; Wilson, R.B.; Mitchell, A.P. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 2000, 20, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Martin, S.J.; Bruno, V.M.; Mitchell, A.P.; Davis, D.A. Candida albicans Rim13p, a protease required for Rim101p processing at acidic and alkaline pHs. Eukaryot. Cell 2004, 3, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Bensen, E.S.; Martin, S.J.; Li, M.; Berman, J.; Davis, D.A. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol. Microbiol. 2004, 54, 1335–1351. [Google Scholar] [CrossRef] [PubMed]
- Vylkova, S.; Lorenz, M.C. Modulation of phagosomal pH by Candida albicans promotes hyphal morphogenesis and requires Stp2p, a regulator of amino acid transport. PLoS Pathog. 2014, 10, e1003995. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Cowen, L.E.; Griffin, A.M.; Chan, L.; Köhler, J.R. The Candida albicans pescadillo homolog is required for normal hypha-to-yeast morphogenesis and yeast proliferation. Proc. Natl. Acad. Sci. USA 2008, 105, 20918–22023. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, T.R.; Veri, A.O.; Ketela, T.; Jiang, B.; Roemer, T.; Cowen, L.E. Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Retallack, D.M.; Woods, J.P. Molecular epidemiology, pathogenesis, and genetics of the dimorphic fungus Histoplasma capsulatum. Microbes Infect. 1999, 1, 817–825. [Google Scholar] [CrossRef]
- Nemecek, J.C.; Wüthrich, M.; Klein, B.S. Global control of dimorphism and virulence in fungi. Science 2006, 312, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.Q.; Sil, A. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc. Natl. Acad. Sci. USA 2008, 105, 4880–4885. [Google Scholar] [CrossRef] [PubMed]
- Webster, R.H.; Sil, A. Conserved factors Ryp2 and Ryp3 control cell morphology and infectious spore formation in the fungal pathogen Histoplasma capsulatum. Proc. Natl. Acad. Sci. USA 2008, 105, 14573–14578. [Google Scholar] [CrossRef] [PubMed]
- Beyhan, S.; Gutierrez, M.; Voorhies, M.; Sil, A. A temperature-responsive network links cell shape and virulence traits in a primary fungal pathogen. PLoS Biol. 2013, 11, e1001614. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, S.A.; Naseem, S.; Konopka, J.B.; Sil, A. N-acetylglucosamine (GlcNAc) triggers a rapid, temperature-responsive morphogenetic program in thermally dimorphic fungi. PLoS Genet. 2013, 9, e1003799. [Google Scholar] [CrossRef] [PubMed]
- López-Martínez, R.; Méndéz-Tovar, L.J. Blastomycosis. Clin. Dermatol. 2012, 30, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, G.M.; Sullivan, T.D.; Gallardo, S.S.; Brandhorst, T.T.; Vanden Wymelenberg, A.J.; Cuomo, C.A.; Suen, G.; Currie, C.R.; Klein, B.S. SREB, a GATA transcription factor that directs disparate fates in Blastomyces dermatitidis including morphogenesis and siderophore biosynthesis. PLoS Pathog. 2010, 6, e1000846. [Google Scholar] [CrossRef] [PubMed]
- Marty, A.J.; Broman, A.T.; Zarnowski, R.; Dwyer, T.G.; Bond, L.M.; Lounes-Hadj Sahraoui, A.; Fontaine, J.; Ntambi, J.M.; Keleş, S.; Kendziorski, C.; et al. Fungal Morphology, Iron Homeostasis, and Lipid Metabolism Regulated by a GATA Transcription Factor in Blastomyces dermatitidis. PLoS Pathog. 2015, 11, e1004959. [Google Scholar] [CrossRef] [PubMed]
- Boyce, K.J.; Schreider, L.; Kirszenblat, L.; Andrianopoulos, A. The two-component histidine kinases DrkA and SlnA are required for in vivo growth in the human pathogen Penicillium marneffei. Mol. Microbiol. 2011, 82, 1164–1184. [Google Scholar] [CrossRef] [PubMed]
- Boyce, K.J.; Schreider, L.; Andrianopoulos, A. In vivo yeast cell morphogenesis is regulated by a p21-activated kinase in the human pathogen Penicillium marneffei. PLoS Pathog. 2009, 5, e1000678. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Chow, W.N.; Wang, G.; Woo, P.C.; Lau, S.K.; Yuen, K.Y.; Lin, X.; Cai, J.J. Signature gene expression reveals novel clues to the molecular mechanisms of dimorphic transition in Penicillium marneffei. PLoS Genet. 2014, 10, e1004662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bugeja, H.E.; Hynes, M.J.; Andrianopoulos, A. HgrA is necessary and sufficient to drive hyphal growth in the dimorphic pathogen Penicillium marneffei. Mol. Microbiol. 2013, 88, 998–1014. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, M.M.; Theodoro, R.C.; Nino-Vega, G.; Bagagli, E.; Felipe, M.S. Paracoccidioides Species Complex: Ecology, Phylogeny, Sexual Reproduction, and Virulence. PLoS Pathog. 2014, 10, e1004397. [Google Scholar] [CrossRef] [PubMed]
- Campos, C.B.; Di Benedette, J.P.; Morais, F.V.; Ovalle, R.; Nobrega, M.P. Evidence for the role of calcineurin in morphogenesis and calcium homeostasis during mycelium-to-yeast dimorphism of Paracoccidioides brasiliensis. Eukaryot. Cell 2008, 7, 1856–1864. [Google Scholar] [CrossRef] [PubMed]
- Matos, T.G.; Morais, F.V.; Campos, C.B. Hsp90 regulates Paracoccidioides brasiliensis proliferation and ROS levels under thermal stress and cooperates with calcineurin to control yeast to mycelium dimorphism. Med. Mycol. 2013, 51, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Téllez, M.D.; Batista-Duharte, A.; Portuondo, D.; Quinello, C.; Bonne-Hernández, R.; Carlos, I.Z. Sporothrix schenckii complex biology: Environment and fungal pathogenicity. Microbiology 2014, 160, 2352–2365. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Caban, J.; Gonzalez-Velazquez, W.; Perez-Sanchez, L.; Gonzalez-Mendez, R.; Rodriguez-del Valle, N. Calcium/calmodulin kinase1 and its relation to thermotolerance and HSP90 in Sporothrix schenckii: An RNAi and yeast two-hybrid study. BMC Microbiol. 2011, 11, 162. [Google Scholar] [CrossRef] [PubMed]
- Orlowski, M. Mucor dimorphism. Microbiol. Rev. 1991, 55, 234–258. [Google Scholar] [PubMed]
- Wolff, A.M.; Appel, K.F.; Petersen, J.B.; Poulsen, U.; Arnau, J. Identification and analysis of genes involved in the control of dimorphism in Mucor circinelloides (syn. racemosus). FEMS Yeast Res. 2002, 2, 203–213. [Google Scholar] [PubMed]
- Ocampo, J.; Nuñez, L.F.; Silva, F.; Pereyra, E.; Moreno, S.; Garre, V.; Rossi, S. A subunit of protein kinase A regulates growth and differentiation in the fungus Mucor circinelloides. Eukaryot. Cell 2009, 8, 933–944. [Google Scholar] [CrossRef] [PubMed]
- Ocampo, J.; McCormack, B.; Navarro, E.; Moreno, S.; Garre, V.; Rossi, S. Protein kinase a regulatory subunit isoforms regulate growth and differentiation in Mucor circinelloides: Essential role of PKAR4. Eukaryot. Cell. 2012, 11, 989–1002. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Li, A.; Calo, S.; Heitman, J. Calcineurin plays key roles in the dimorphic transition and virulence of the human pathogenic zygomycete Mucor circinelloides. PLoS Pathog. 2013, 9, e1003625. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.F., Jr.; Limper, A.H. Current insights into the biology and pathogenesis of Pneumocystis pneumonia. Nat. Rev. Microbiol. 2007, 5, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Kutty, G.; Davis, A.S.; Ma, L.; Taubenberger, J.K.; Kovacs, J.A. Pneumocystis encodes a functional endo-β-1,3-glucanase that is expressed exclusively in cysts. J. Infect. Dis. 2015, 211, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Cushion, M.T.; Linke, M.J.; Ashbaugh, A.; Sesterhenn, T.; Collins, M.S.; Lynch, K.; Brubaker, R.; Walzer, P.D. Echinocandin treatment of pneumocystis pneumonia in rodent models depletes cysts leaving trophic burdens that cannot transmit the infection. PLoS ONE 2010, 5, e8524. [Google Scholar] [CrossRef] [PubMed]
- Whiston, E.; Zhang Wise, H.; Sharpton, T.J.; Jui, G.; Cole, G.T.; Taylor, J.W. Comparative transcriptomics of the saprobic and parasitic growth phases in Coccidioides spp. PLoS ONE 2012, 7, e41034. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Hernández, B.; Palma-Cortés, G.; Cabello-Gutiérrez, C.; Martínez-Rivera, M.A. Parasitic polymorphism of Coccidioides spp. BMC Infect. Dis. 2014, 14, 213. [Google Scholar] [CrossRef] [PubMed]
- Cruickshank, J.G.; Cavill, R.; Jelbert, M. Cryptococcus neoformans of Unusual Morphology. Appl. Microbiol. 1973, 25, 309–312. [Google Scholar] [PubMed]
- Wang, J.M.; Zhou, Q.; Cai, H.R.; Zhuang, Y.; Zhang, Y.F.; Xin, X.Y.; Meng, F.Q.; Wang, Y.P. Clinicopathological features of pulmonary cryptococcosis with cryptococcal titan cells: A comparative analysis of 27 cases. Int. J. Clin. Exp. Pathol. 2014, 7, 4837–4846. [Google Scholar] [PubMed]
- Zaragoza, O. Multiple Disguises for the Same Party: The Concepts of Morphogenesis and Phenotypic Variations in Cryptococcus neoformans. Front. Microbiol. 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza, O.; García-Rodas, R.; Nosanchuk, J.D.; Cuenca-Estrella, M.; Rodríguez-Tudela, J.L.; Casadevall, A. Fungal cell gigantism during mammalian infection. PLoS Pathog. 2010, 6, e1000945. [Google Scholar] [CrossRef]
- Okagaki, L.H.; Strain, A.K.; Nielsen, J.N.; Charlier, C.; Baltes, N.J.; Chrétien, F.; Heitman, J.; Dromer, F.; Nielsen, K. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 2010, 6, e1000953. [Google Scholar] [CrossRef]
- Alanio, A.; Vernel-Pauillac, F.; Sturny-Leclère, A.; Dromer, F. Cryptococcus neoformans host adaptation: Toward biological evidence of dormancy. mBio 2015, 6, e02580–14. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, D.L.; Specht, C.A.; Lee, C.K.; Smith, K.D.; Mukaremera, L.; Lee, S.T.; Lee, C.G.; Elias, J.A.; Nielsen, J.N.; Boulware, D.R.; et al. Chitin recognition via chitotriosidase promotes pathologic type-2 helper T cell responses to cryptococcal infection. PLoS Pathog. 2015, 11, e1004701. [Google Scholar] [CrossRef] [PubMed]
- Okagaki, L.H.; Nielsen, K. Titan cells confer protection from phagocytosis in Cryptococcus neoformans infections. Eukaryot. Cell 2012, 11, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Gerstein, A.C.; Fu, M.S.; Mukaremera, L.; Li, Z.; Ormerod, K.L.; Fraser, J.A.; Berman, J.; Nielsen, K. Polyploid Titan Cells Produce Haploid and Aneuploid Progeny To Promote Stress Adaptation. mBio 2015, 6, e01340–15. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Vogl, A.W.; Kronstad, J.W. Regulated expression of cyclic AMP-dependent protein kinase A reveals an influence on cell size and the secretion of virulence factors in Cryptococcus neoformans. Mol. Microbiol. 2012, 85, 700–715. [Google Scholar] [CrossRef] [PubMed]
- Okagaki, L.H.; Wang, Y.; Ballou, E.R.; O’Meara, T.R.; Bahn, Y.S.; Alspaugh, J.A.; Xue, C.; Nielsen, K. Cryptococcal titan cell formation is regulated by G-protein signaling in response to multiple stimuli. Eukaryot. Cell 2011, 10, 1306–1316. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, T.R.; Norton, D.; Price, M.S.; Hay, C.; Clements, M.F.; Nichols, C.B.; Alspaugh, J.A. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog. 2010, 6, e1000776. [Google Scholar] [CrossRef] [PubMed]
- Ost, K.S.; O’Meara, T.R.; Huda, N.; Esher, S.K.; Alspaugh, J.A. The Cryptococcus neoformans alkaline response pathway: Identification of a novel rim pathway activator. PLoS Genet. 2015, 11, e1005159. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.J.; Li, Z.; Hughes, W.S.; Djordjevic, J.T.; Nielsen, K.; May, R.C. Cryptococcal phospholipase B1 is required for intracellular proliferation and control of titan cell morphology during macrophage infection. Infect Immun. 2015, 83, 1296–1304. [Google Scholar] [CrossRef] [PubMed]
- Edgar, B.A.; Zielke, N.; Gutierrez, C. Endocycles: A recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell. Biol. 2014, 15, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Kottom, T.J.; Thomas, C.F., Jr.; Mubarak, K.K.; Leof, E.B.; Limper, A.H. Pneumocystis carinii uses a functional cdc13 B-type cyclin complex during its life cycle. Am. J. Respir. Cell. Mol. Biol. 2000, 22, 722–731. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Nielsen, K. Morphology Changes in Human Fungal Pathogens upon Interaction with the Host. J. Fungi 2017, 3, 66. https://doi.org/10.3390/jof3040066
Li Z, Nielsen K. Morphology Changes in Human Fungal Pathogens upon Interaction with the Host. Journal of Fungi. 2017; 3(4):66. https://doi.org/10.3390/jof3040066
Chicago/Turabian StyleLi, Zhongming, and Kirsten Nielsen. 2017. "Morphology Changes in Human Fungal Pathogens upon Interaction with the Host" Journal of Fungi 3, no. 4: 66. https://doi.org/10.3390/jof3040066



