Function and Biosynthesis of Cell Wall α-1,3-Glucan in Fungi
Abstract
:1. Introduction
2. Brief History of Studies on α-1,3-Glucan in Fungi
2.1. Functional Analysis of α-1,3-Glucan in Aspergillus nidulans before Whole-Genome Sequencing
2.2. Functional Analysis of α-1,3-Glucan in Aspergillus Species after Complete Genome Sequences Became Available
3. Biosynthesis of α-1,3-Glucan
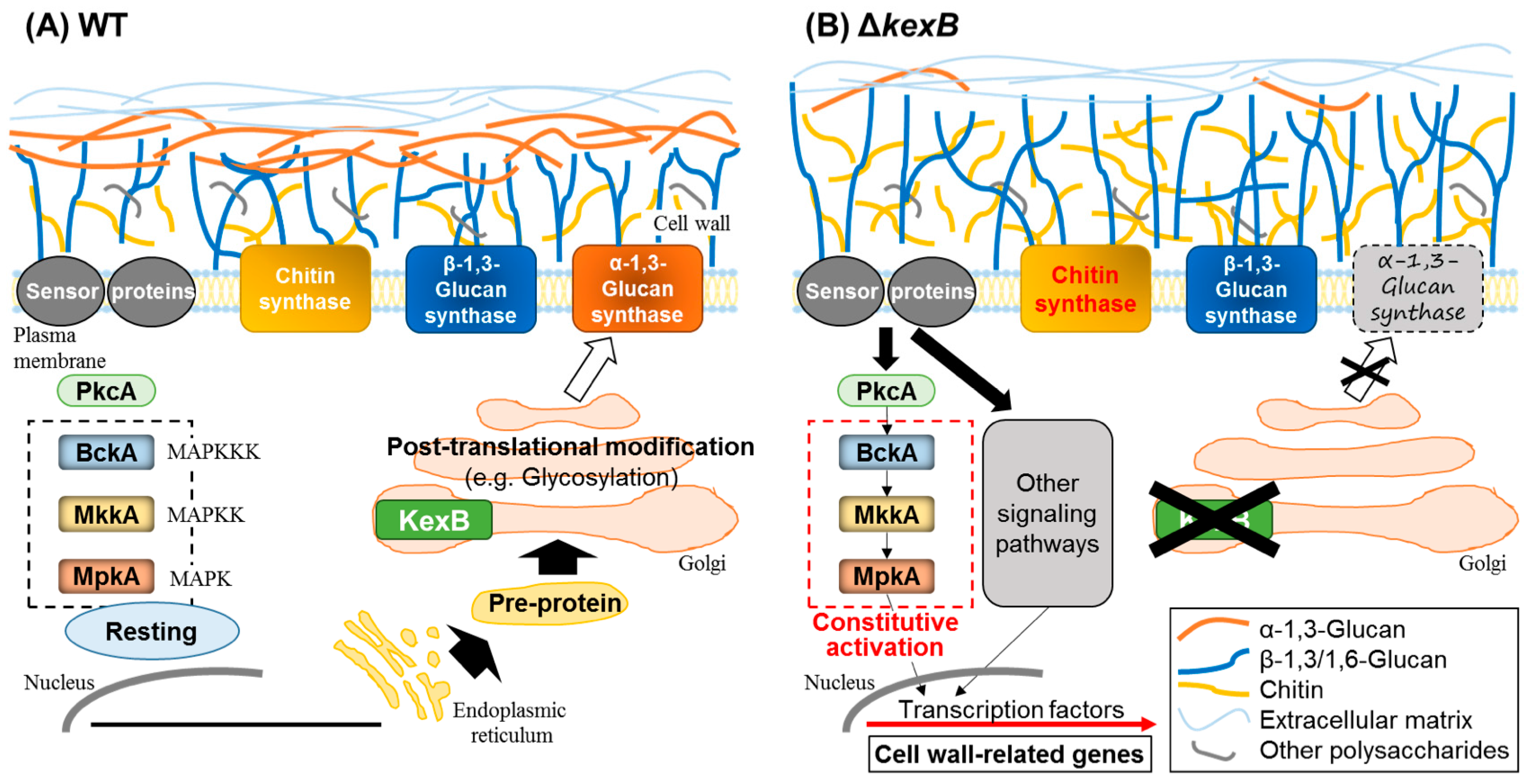
3.1. Regulation of α-1,3-Glucan Biosynthesis in Filamentous Fungi: Cell Wall Integrity Signaling
3.2. Biosynthesis of α-1,3-Glucan in Schizosaccharomyces pombe
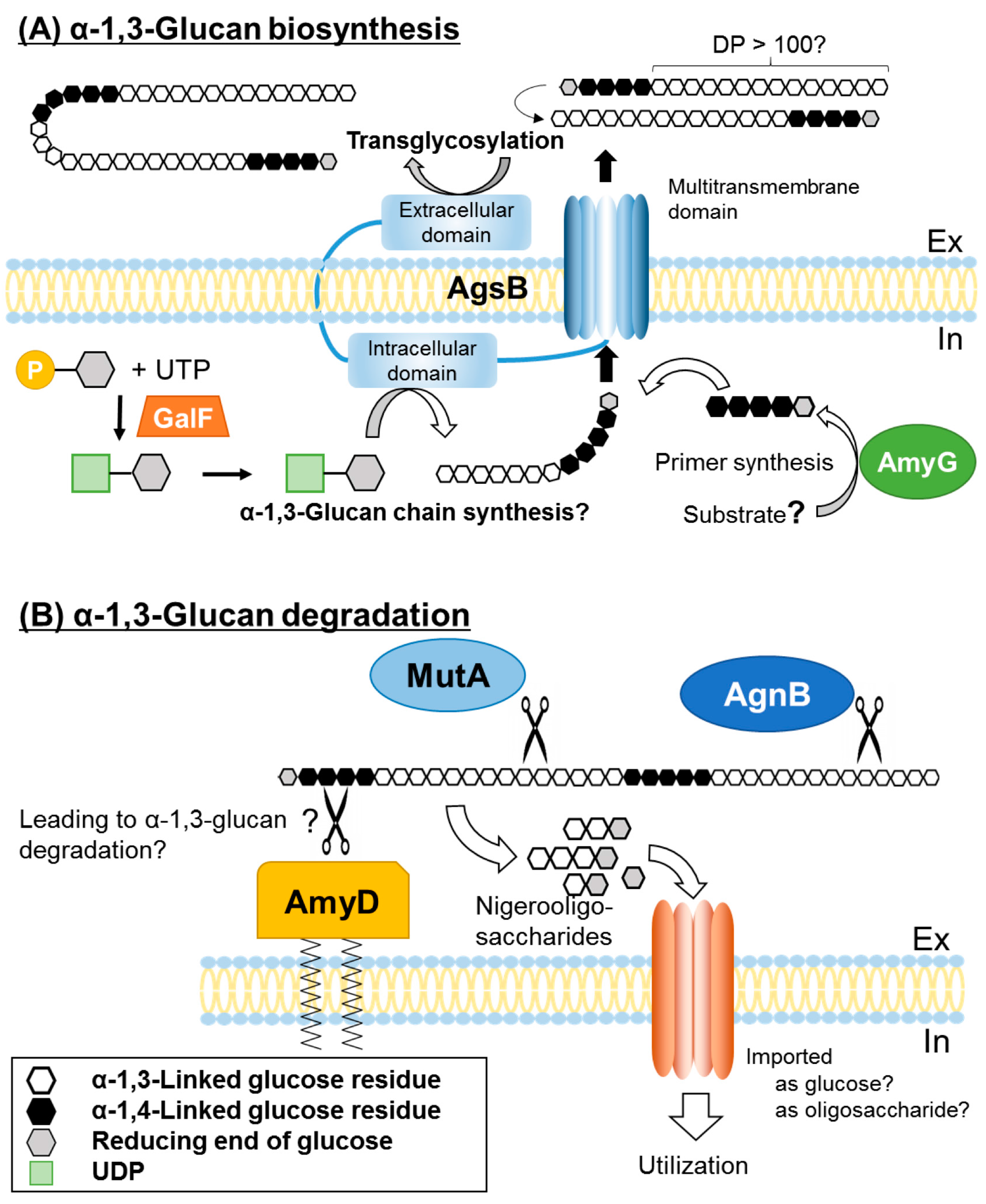
3.3. Intracellular Amylase
3.4. Other Enzymes Involved in α-1,3-Glucan Synthesis
4. Biological Functions of α-1,3-Glucan in Fungi
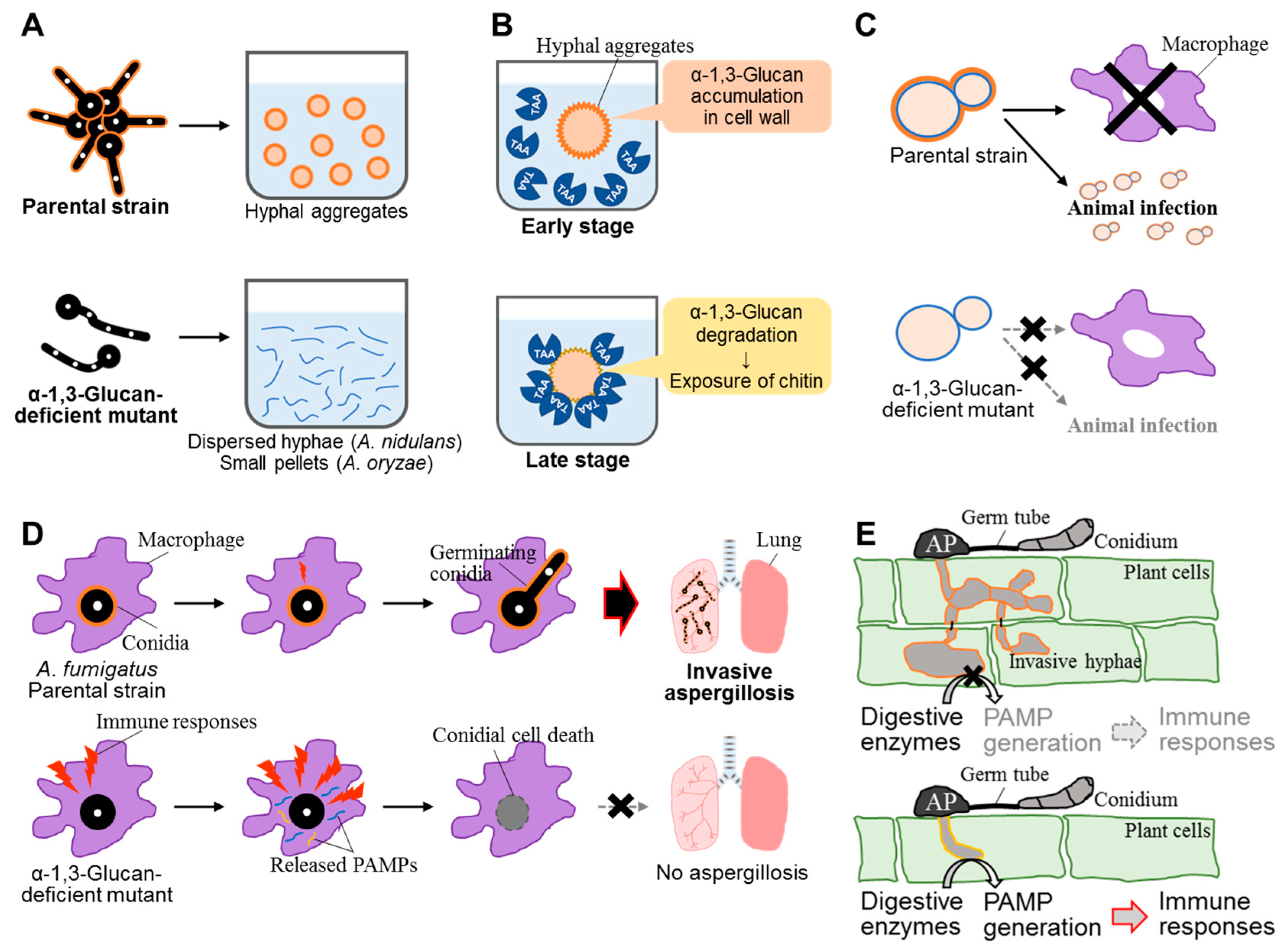
4.1. Function as an Aggregation Factor
4.2. Influence on Adsorption of α-Amylase onto the Cell Surface
4.3. Function as a Virulence Factor
4.3.1. Relationships between α-1,3-Glucan and Virulence in Pathogenic Yeasts
4.3.2. α-1,3-Glucan Is a Virulence Factor in Aspergillus fumigatus
4.3.3. α-1,3-Glucan Is a Virulence Factor in Magnaporthe grisea
5. Industrial Applications of α-1,3-Glucan Mutants
6. Conclusions and Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Latgè, J.P. Tasting the Fungal cell wall. Cell. Microbiol. 2010, 12, 863–872. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, A.; Miyazawa, K.; Abe, K. Cell wall structure and biogenesis in Aspergillus species. Biosci. Biotechnol. Biochem. 2016, 80, 1700–1711. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Inoue, K.; Kitagawa, H.; Meguro, H.; Shimoi, S.; Park, P. Extracellular matrix (ECM) in phytopathogenic fungi: Its role and potential target for disease protection. In Plant Pathology; Cumagun, C.J., Ed.; InTechOpen Access Publisher: Rijeka, Croatia, 2012; pp. 131–150. ISBN 978-953-51-0489-6. [Google Scholar]
- Sheppard, D.C.; Howell, P.L. Biofilm exopolysaccharides of pathogenic fungi: Lessons from bacteria. J. Biol. Chem. 2016, 291, 12529–12537. [Google Scholar] [CrossRef] [PubMed]
- Bernard, M.; Latgè, J.P. Aspergillus fumigatus cell wall: Composition and biosynthesis. Med. Mycol. 2001, 39, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Johnston, I.R. The composition of the cell wall of Aspergillus niger. Biochem. J. 1965, 96, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Zonneveld, B.J.M. Biochemical analysis of the cell wall of Aspergillus nidulans. Biochim. Biophys. Acta 1971, 249, 506–514. [Google Scholar] [CrossRef]
- Zonneveld, B.J.M. The Significance of α-1,3-glucan of the cell wall and α-1,3-glucanase for cleistothecium development. Biochim. Biophys. Acta 1972, 273, 174–187. [Google Scholar] [CrossRef]
- Rappleye, C.A.; Engle, J.T.; Goldman, W.E. RNA interference in Histoplasma capsulatum demonstrates a role for α-(1,3)-glucan in virulence. Mol. Microbiol. 2004, 53, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Rappleye, C.A.; Eissenberg, L.G.; Goldman, W.E. Histoplasma capsulatum α-(1,3)-glucan blocks innate immune recognition by the β-glucan receptor. Proc. Natl. Acad. Sci. USA 2007, 104, 1366–1370. [Google Scholar] [CrossRef] [PubMed]
- Beauvais, A.; Bozza, S.; Kniemeyer, O.; Formosa, C.; Balloy, V.; Henry, C.; Roberson, R.W.; Dague, E.; Chignard, M.; Brakhage, A.A.; et al. Deletion of the α-(1,3)-glucan synthase genes induces a restructuring of the conidial cell wall responsible for the avirulence of Aspergillus fumigatus. PLoS Pathog. 2013, 9, e1003716. [Google Scholar] [CrossRef]
- Fujikawa, T.; Kuga, Y.; Yano, S.; Yoshimi, A.; Tachiki, T.; Abe, K.; Nishimura, M. Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol. Microbiol. 2009, 73, 553–570. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa, T.; Sakaguchi, A.; Nishizawa, Y.; Kouzai, Y.; Minami, E.; Yano, S.; Koga, H.; Meshi, T.; Nishimiura, M. Surface α-1,3-glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PLoS Pathog. 2012, 8, e1002882. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, T.; Beauvais, A.; Loussert, C.; Thevenard, B.; Fulgsang, C.C.; Ohno, N.; Clavaud, C.; Prevost, M.C.; Latgé, J.P. Cell wall α1-3glucans induce the aggregation of germinating conidia of Aspergillus fumigatus. Fungal Genet. Biol. 2010, 47, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.; Latgé, J.P.; Beauvais, A. α1,3 Glucans are dispensable in Aspergillus fumigatus. Eukaryot. Cell 2012, 11, 26–29. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, A.; Sano, M.; Inaba, A.; Kokubun, Y.; Fujioka, T.; Mizutani, O.; Hagiwara, D.; Fujikawa, T.; Nishimura, M.; Yano, S.; et al. Functional Analysis of the α-1,3-glucan synthase genes agsA and agsB in Aspergillus nidulans: AgsB is the major α-1,3-glucan synthase in this fungus. PLoS ONE 2013, 8, e54893. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, K.; Yoshimi, A.; Zhang, S.; Sano, M.; Nakayama, M.; Gomi, K.; Abe, K. Increased enzyme production under liquid culture conditions in the industrial fungus Aspergillus oryzae by disruption of the genes encoding cell wall α-1,3-glucan synthase. Biosci. Biotechnol. Biochem. 2016, 80, 1853–1863. [Google Scholar] [CrossRef] [PubMed]
- Puanglek, S.; Kimura, S.; Enomoto-Rogers, Y.; Kabe, T.; Yoshida, M.; Wada, M.; Iwata, T. In vitro synthesis of linear α-1,3-glucan and chemical modification to ester derivatives exhibiting outstanding thermal properties. Sci. Rep. 2016, 6, 30479. [Google Scholar] [CrossRef] [PubMed]
- Zonneveld, B.J.M. New type of enzyme, an exo-splitting α-1,3-glucanase from non-induced cultures of Aspergillus nidulans. Biochim. Biophys. Acta 1972, 258, 541. [Google Scholar] [CrossRef]
- Zonneveld, B.J.M. Inhibitory Effect of 2-deozyglucose on cell-wall α-1,3-glucan synthesis and cleistothecium development in Aspergillus nidulans. Dev. Biol. 1973, 34, 1–8. [Google Scholar] [CrossRef]
- Zonneveld, B.J.M. α-1,3-Glucan synthesis correlated with α-1,3-glucanase synthesis, conidiation and fructification in morphogenetic mutants of Aspergillus nidulans. J. Gen. Microbiol. 1974, 81, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Zonneveld, B.J.M. Sexual differentiation in Aspergillus nidulans—Requirement for manganese and its effect on α-1,3-glucan synthesis and degradation. Arch. Microbiol. 1975, 105, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Polacheck, I.; Rosenberger, R.F. Aspergillus nidulans mutant lacking α-(1,3)-glucan, melanin, and cleistothecia. J. Bacteriol. 1977, 132, 650–656. [Google Scholar] [PubMed]
- Wei, H.; Scherner, M.; Singh, A.; Liese, R.; Fischer, R. Aspergillus nidulans α-1,3 glucanase (mutanase), mutA, is expressed during sexual development and mobilizes mutan. Fungal Genet. Biol. 2001, 34, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Galagan, J.E.; Calvo, S.E.; Cuomo, C.; Ma, L.J.; Wortman, J.R.; Batzoglou, S.; Lee, S.I.; Baştürkmen, M.; Spevak, C.C.; Kapitonov, V.; et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 2005, 438, 1105–1115. [Google Scholar]
- Machida, M.; Asai, K.; Sano, M.; Tanaka, T.; Kumagai, T.; Terai, G.; Kusumoto, K.; Arima, T.; Akita, O.; Kashiwagi, Y.; et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 2005, 438, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Nierman, W.C.; Pain, A.; Anderson, M.J.; Wortman, J.R.; Kim, H.S.; Arroyo, J.; Berriman, M.; Abe, K.; Archer, D.B.; Bermejo, C.; et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 2005, 438, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Pel, H.J.; de Winde, J.H.; Archer, D.B.; Dyer, P.S.; Hofmann, G.; Schaap, P.J.; Turner, G.; de Vries, R.P.; Albang, R.; Albermann, K.; et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 2007, 25, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Beauvais, A.; Maubon, D.; Park, S.; Morelle, W.; Tanguy, M.; Huerre, M.; Perlin, D.S.; Latgé, J.P. Two α(1-3)glucan synthases with different functions in Aspergillus fumigatus. Appl. Environ. Microbiol. 2005, 71, 1531–1538. [Google Scholar] [CrossRef] [PubMed]
- Maubon, D.; Park, S.; Tanguy, M.; Huerre, M.; Schmitt, C.; Prévost, M.C.; Perlin, D.S.; Latgé, J.P.; Beauvais, A. AGS3, an α(1-3)glucan synthase gene family member of Aspergillus fumigatus, modulates mycelium growth in the lung of experimentally infected mice. Fungal Genet. Biol. 2006, 43, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Damveld, R.A.; van Kuyk, P.A.; Arentshorst, M.; Klis, F.M.; van den Hondel, C.A.; Ram, A.F.J. Expression of agsA, one of five 1,3-α-D-glucan synthase-encoding genes in Aspergillus niger, is induced in response to cell wall stress. Fungal Genet. Biol. 2005, 42, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, T.; Mizutani, O.; Furukawa, K.; Sato, N.; Yoshimi, A.; Yamagata, Y.; Nakajima, T.; Abe, K. MpkA-dependent and -independent cell wall integrity signaling in Aspergillus nidulans. Eukaryot. Cell 2007, 6, 1497–1510. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Li, S.; Kaminskyj, S.G.W. Characterization of Aspergillus nidulans α-glucan synthesis: Roles for two synthases and two amylases. Mol. Microbiol. 2014, 91, 579–595. [Google Scholar] [CrossRef] [PubMed]
- Van der Kaaij, R.M.; Yuan, X.L.; Franken, A.; Ram, A.F.J.; Punt, P.J.; van der Maarel, M.J.E.C.; Dijkhuizen, L. Two novel, putatively cell wall-associated and glycosylphosphatidylinositol-anchored α-glucanotransferase enzymes of Aspergillus niger. Eukaryot. Cell 2007, 6, 1178–1188. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, K.; Yoshimi, A.; Abe, K. Laboratory of Applied Microbiology, Department of Microbial Biotechnology, Graduate School of Agricultural Sciences, Tohoku University, Sendai, Japan. Unpublised work. 2017. [Google Scholar]
- Zhang, S.; Sato, H.; Ichinose, S.; Tanaka, M.; Miyazawa, K.; Yoshimi, A.; Abe, K.; Shintani, T.; Gomi, K. Cell wall α-1,3-glucan prevents α-amylase adsorption onto fungal cell in submerged culture of Aspergillus oryzae. J. Biosci. Bioeng. 2017, 124, 47–53. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Li, S.; Kaminskyj, S.G.W. An amylase-like protein, AmyD, is the major negative regulator for α-glucan synthesis in Aspergillus nidulans during the asexual life cycle. Int. J. Mol. Sci. 2017, 18, 695. [Google Scholar] [CrossRef] [PubMed]
- Van der Kaaij, R.M.; Janecek, S.; van der Maarel, M.J.E.C.; Dijkhuizen, L. Phylogenetic and biochemical characterization of a novel cluster of intracellular fungal α-amylase enzymes. Microbiology 2007, 153, 4003–4015. [Google Scholar] [CrossRef] [PubMed]
- Van Munster, J.M.; Dobruchowska, J.M.; Veloo, R.; Dijkhuizen, L.; van der Maarel, M.J.E.C. Characterization of the starvation-induced chitinase and α-1,3-glucanase AgsB of Aspergillus niger. Appl. Microbiol. Biotechnol. 2015, 99, 2209–2223. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [PubMed]
- Verna, J.; Lodders, A.; Lee, K.H.; Vagts, A.; Ballester, R. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1997, 94, 13804–13809. [Google Scholar] [CrossRef] [PubMed]
- Lodders, A.L.; Lee, T.K.; Ballester, R. Characterization of the Wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics 1999, 152, 1489–1499. [Google Scholar]
- Levin, D.E.; Fields, F.O.; Kunisawa, R.; Bishop, J.M.; Thorner, J. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell 1990, 62, 213–224. [Google Scholar] [PubMed]
- Levin, D.E. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005, 69, 262–291. [Google Scholar] [CrossRef] [PubMed]
- Philip, B.; Levin, D.E. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that ant through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 2001, 21, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Kamada, Y.; Qadota, H.; Python, C.P.; Anraku, Y.; Ohya, Y.; Levin, D.E. Activation of yeast protein kinase C by Rho1 GTPase. J. Biol. Chem. 1996, 271, 9193–9196. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; Levin, D.E. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol. Cell. Biol. 1992, 12, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Irie, K.; Takase, M.; Lee, K.S.; Levin, D.E.; Araki, H.; Matsumoto, K.; Oshima, Y. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol. Cell. Biol. 1993, 13, 3076–3083. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.; Arroyo, J.; Sánchez, M.; Molina, M.; Nombela, C. Activity of the yeast MAP kinase homologue Slt2 is critically required for cell integrity at 37 °C. Mol. Gen. Genet. 1993, 241, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Jung, U.S.; Levin, D.E. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 1999, 34, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, O.; Nojima, A.; Yamamoto, M.; Furukawa, K.; Fujioka, T.; Yamagata, Y.; Abe, K.; Nakajima, T. Disordered cell integrity signaling caused by disruption of the kexB gene in Aspergillus oryzae. Eukaryot. Cell 2004, 3, 1036–1048. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, O.; Shiina, M.; Yoshimi, A.; Sano, M.; Watanabe, T.; Yamagata, Y.; Nakajima, T.; Gomi, K.; Abe, K. Substantial decrease in cell wall α-1,3-glucan caused by disruption of the kexB gene encoding a subtilisin-like processing protease in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2016, 80, 1781–1791. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Nakamura, T.; Ohshima, T.; Tanaka, S.; Matsuo, H. Yeast KEX2 genes encodes an endopeptidase homologous to subtilisin-like serine proteases. Biochem. Biophys. Res. Commun. 1988, 156, 246–254. [Google Scholar] [CrossRef]
- Fuller, R.S.; Brake, A.; Thorner, J. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc. Natl. Acad. Sci. USA 1989, 86, 1434–1438. [Google Scholar] [CrossRef] [PubMed]
- Hochstenbach, F.; Klis, F.M.; van den Ende, H.; van Donselaar, E.; Peters, P.J.; Klausner, R.D. Identification of a putative α-glucan synthase essential for cell wall construction and morphogenesis in fission yeast. Proc. Natl. Acad. Sci. USA 1998, 95, 9161–9166. [Google Scholar] [CrossRef] [PubMed]
- Vos, A.; Dekker, N.; Distel, B.; Leunissen, J.A.; Hochstenbach, F. Role of the synthase domain of Ags1p in cell wall α-glucan biosynthesis in fission yeast. J. Biol. Chem. 2007, 282, 18969–18979. [Google Scholar] [CrossRef] [PubMed]
- Grün, C.H.; Hochstenbach, F.; Humbel, B.M.; Verkleij, A.J.; Sietsma, J.H.; Klis, F.M.; Kamerling, J.P.; Vliegenthart, J.F.G. The structure of cell wall α-glucan from fission yeast. Glycobiology 2005, 15, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Lomako, J.; Lomako, W.M.; Whelan, W.J. Glycogenin: The primer for mammalian and yeast glycogen synthesis. Biochim. Biophys. Acta 2004, 1673, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Marion, C.L.; Rappleye, C.A.; Engle, J.T.; Goldman, W.E. An α-(1,4)-amylase is essential for α-(1,3)-glucan production and virulence in Histoplasma capsulatum. Mol. Microbiol. 2006, 62, 970–983. [Google Scholar] [CrossRef] [PubMed]
- Camacho, E.; Sepulveda, V.E.; Goldman, W.E.; San-Blas, G.; Niño-Vega, G.A. Expression of Paracoccidioides brasiliensis AMY1 in a Histoplasma capsulatum amy1 mutant, relates an α-(1,4)-amylase to cell wall α-(1,3)-glucan synthesis. PLoS ONE 2012, 7, e50201. [Google Scholar] [CrossRef] [PubMed]
- Choma, A.; Wiater, A.; Komaniecka, I.; Paduch, R.; Pleszczyñska, M.; Szczodrak, J. Chemical characterization of a water insoluble (1→3)-α-d-glucan from an alkaline extract of Aspergillus wentii. Carbohydr. Polym. 2013, 91, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Wiater, A.; Paduch, R.; Choma, A.; Sylwia, S.; Pleszczynska, M.; Tomczyk, M.; Locatelli, M.; Szczodrak, J. (1→3)-α-D-Glucans from Aspergillus spp.: Structural characterization and biological study on their carboxymethylated derivatives. Curr. Drug Targets 2015, 16, 1488–1494. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Maeda, Y.; Tanoue, N.; Makita, T.; Kato, M.; Kobayashi, T. Expression profile of amylolytic genes in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2006, 70, 2363–2370. [Google Scholar] [CrossRef] [PubMed]
- Nitsche, B.M.; Jorgensen, T.R.; Akeroyd, M.; Meyer, V.; Ram, A.F.J. The carbon starvation response of Aspergillus niger during submerged cultivation: Insight from the transcriptome and secretome. BMC Genomics 2012, 13, 380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klimpel, K.R.; Goldman, W.E. Isolation and characterization of spontaneous avirulent variants of Histoplasma capsulatum. Infect. Immun. 1987, 55, 528–533. [Google Scholar] [PubMed]
- Sato, H.; Toyoshima, Y.; Shintani, T.; Gomi, K. Identification of potential cell wall component that allows Taka-amylase A adsorption in submerged cultures of Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2011, 92, 961–969. [Google Scholar] [CrossRef] [PubMed]
- San-Blas, F.; San-Blas, G.; Cova, L.J. A morphological mutant of Paracoccidioides brasiliensis strain IVIC Pb9. Isolation and wall characterization. J. Gen. Microbiol. 1976, 93, 209–218. [Google Scholar] [CrossRef] [PubMed]
- San-Blas, F.; Vernet, D. Induction of the synthesis of cell wall α-1,3-glucan in the yeastlike form of Paracoccidioides brasiliensis strain IVIC Pb9 by fetal calf serum. Infect. Immun. 1977, 15, 897–902. [Google Scholar] [PubMed]
- San-Blas, G.; San-Blas, F.; Serrano, L.E. Host-parasite relationships in the yeast form of Paracoccidioides brasiliensis strain IVIC Pb9. Infect. Immun. 1977, 15, 343–346. [Google Scholar] [PubMed]
- San-Blas, G.; San-Blas, F.; Ormaechea, E.; Serrano, L.E. Cell wall analysis of an adenine-requiring mutant of the yeast-like form of Paracoccidioides brasiliensis strain IVIC Pb9. Sabouraudia 1977, 15, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Klimpel, K.R.; Goldman, W.E. Cell walls from avirulent variants of Histoplasma capsulatum lack α-(1,3)-glucan. Infect. Immun. 1988, 56, 2997–3000. [Google Scholar] [PubMed]
- Hogan, L.H.; Klein, B.S. Altered expression of surface α-1,3-glucan in genetically related strains of Blastomyces dermatitidis that differ in virulence. Infect. Immun. 1994, 62, 3543–3546. [Google Scholar] [PubMed]
- Domer, J.E. Monosaccharide and chitin content of cell walls of Histoplasma capsulatum and Blastomyces dermatitidis. J. Bacteriol. 1971, 107, 870–877. [Google Scholar] [PubMed]
- Edwards, J.A.; Alore, E.A.; Rappleye, C.A. The yeast-phase requirement for α-glucan synthase differs among Histoplasma capsulatum chemotypes. Eukaryot. Cell 2011, 10, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Reese, A.J.; Doering, T.L. Cell wall α-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol. Microbiol. 2003, 50, 1401–1409. [Google Scholar] [CrossRef] [PubMed]
- Reese, A.J.; Yoneda, A.; Breger, J.A.; Beauvais, A.; Liu, H.; Griffith, C.L.; Bose, I.; Kim, M.J.; Skau, C.; Yang, S.; et al. Loss of cell wall α(1-3) glucan affects Cryptococcus neoformans from ultrastructure to virulence. Mol. Microbiol. 2007, 63, 1385–1398. [Google Scholar] [CrossRef] [PubMed]
- Stephen-Victor, E.; Karnam, A.; Fontaine, T.; Beauvais, A.; Das, M.; Hegde, P.; Prakhae, P.; Holla, S.; Balaji, K.N.; Kaveri, S.V.; et al. Aspergillus fumigatus cell wall α-(1,3)-glucan stimulates regulatory T-cell polarization by inducing PD-L1 expression on human dendritic cells. J. Infect. Dis. 2017. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.S.; Possos, C.P.; Madureira, P.; Vilanova, M.; Coimbra, M.A. Structure-function relationships of immunostimulatory polysaccharides. Carbohydr. Polym. 2015, 132, 378–396. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, A.; Hirama, M.; Tsubota, Y.; Kawakami, K.; Zhang, S.; Gomi, K.; Abe, K. Characterization of cell wall α-1,3-glucan-deficient mutants in Aspergillus oryzae isolated by a screening method based on their sensitivity to Congo Red or Lysing Enzyme. J. Appl. Glycobiol. 2017, 64, 77–86. [Google Scholar] [CrossRef]
| A. nidulans | A. fumigatus | A. oryzae | A. niger | ||||
|---|---|---|---|---|---|---|---|
| Gene | Phenotypes | Gene | Phenotypes | Gene | Phenotypes | Gene | Phenotypes |
| α-1,3-Glucan synthases | |||||||
| agsA | Expressed at a very low level [16] High expression level in ΔmpkA strain [32] | AGS2 | No change in α-1,3-glucan content in Δags2 strain [29] | agsA | Expressed at a very low level [36] | agsD | Not reported |
| agsB | Main α-1,3-glucan synthase [16] Hyphal aggregation [16] Upregulated by micafungin treatment [32] | AGS1 | 50% reduction in α-1,3-glucan content in Δags1 strain [29] | agsB | Major α-1,3-glucan synthase [36] | agsE | Upregulated by CFW treatment [31] |
| AGS3 | Hypervirulence in Δags3 strain [30] | agsC | Not directly involved in α-1,3-glucan synthesis [36] | agsA | Upregulated by CFW treatment [31] | ||
| agsB | Not reported | ||||||
| agsC | Downregulated by CFW treatment [31] | ||||||
| Intracellular α-amylases | |||||||
| amyG | Important for α-1,3-glucan synthesis [33,37] | 1 | Not reported | 2 | Not reported | amyD | Starch hydrolysis [38] |
| 1 | Not reported | ||||||
| GPI-anchored α-amylases | |||||||
| amyD | Degradation of α-1,3-glucan [33,37] | 3 | Not reported | 2 | Not reported | agtA | α-1,4-Glucano-transferase [34] |
| agtB | α-1,4-Glucano-transferase [34] | ||||||
| agtC | Not reported | ||||||
| α-1,3-Glucanases | |||||||
| mutA | Hülle cell localization [24] Hydrolysis of α-1,3-glucan [37] | Not annotated | Not annotated | agnB | α-1,3-Glucan hydrolysis [39] | ||
| agnB | Hydrolysis of α-1,3-glucan [37] | agnE | Not reported | ||||
| agnE | No α-1,3-glucan hydrolysis [37] | ||||||
| 2 | Not reported | ||||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshimi, A.; Miyazawa, K.; Abe, K. Function and Biosynthesis of Cell Wall α-1,3-Glucan in Fungi. J. Fungi 2017, 3, 63. https://doi.org/10.3390/jof3040063
Yoshimi A, Miyazawa K, Abe K. Function and Biosynthesis of Cell Wall α-1,3-Glucan in Fungi. Journal of Fungi. 2017; 3(4):63. https://doi.org/10.3390/jof3040063
Chicago/Turabian StyleYoshimi, Akira, Ken Miyazawa, and Keietsu Abe. 2017. "Function and Biosynthesis of Cell Wall α-1,3-Glucan in Fungi" Journal of Fungi 3, no. 4: 63. https://doi.org/10.3390/jof3040063




