Quantitative Assessment of Grapevine Wood Colonization by the Dieback Fungus Eutypa lata
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Fungal Material and Inoculation
2.3. Re-Isolation Method
2.4. Fungal Biomass Measurement by Quantitative PCR
2.5. Experimental Design
2.6. Data Analysis and Reproducible Research
3. Results
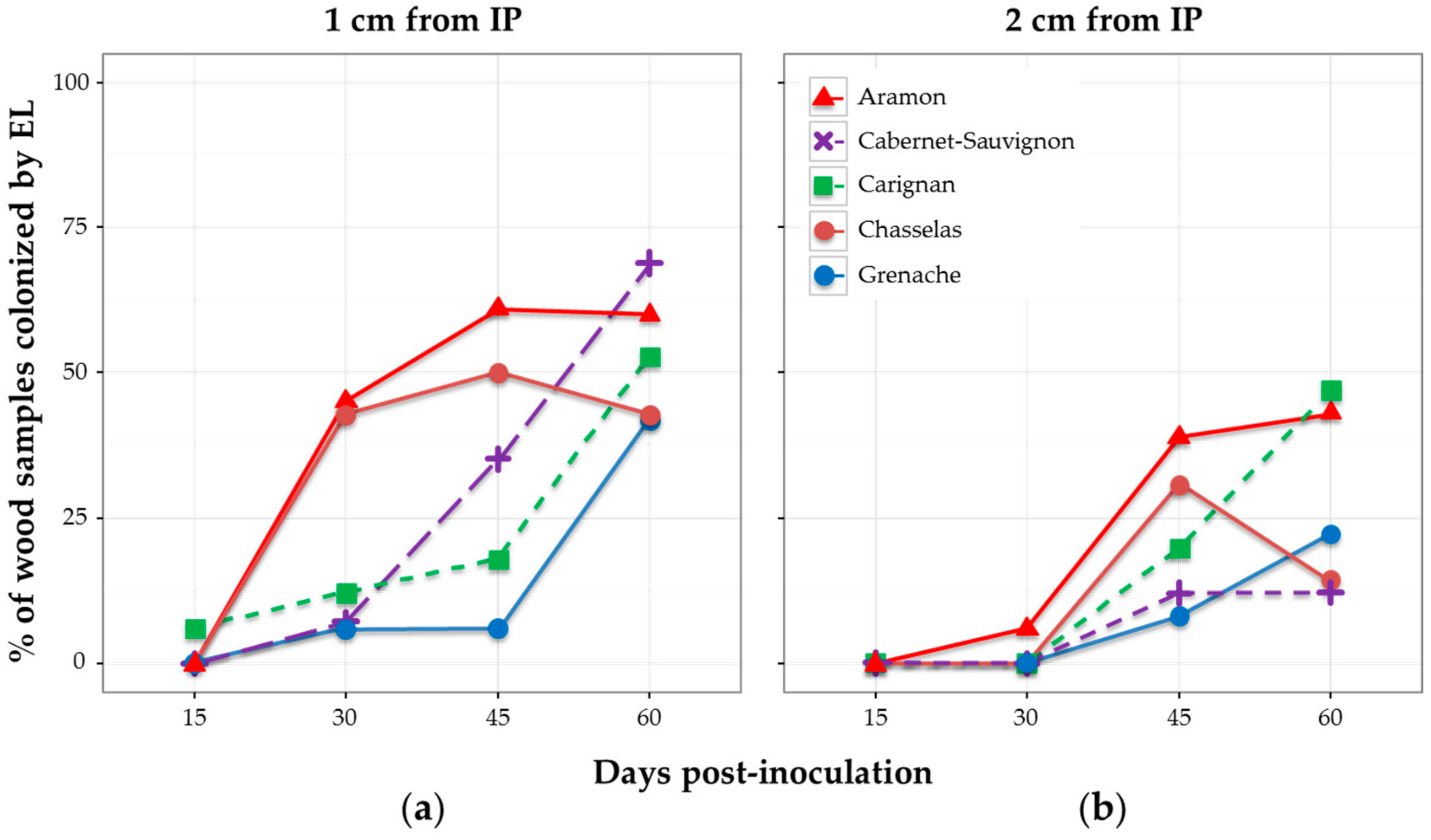
3.1. Wood Colonization in Relation to Aggressiveness and Distance from Inoculation Site (Experiment 1)
3.2. Comparison of Grapevine Cultivars for Their Tolerance to Wood Colonization by E. lata (Experiment 2)
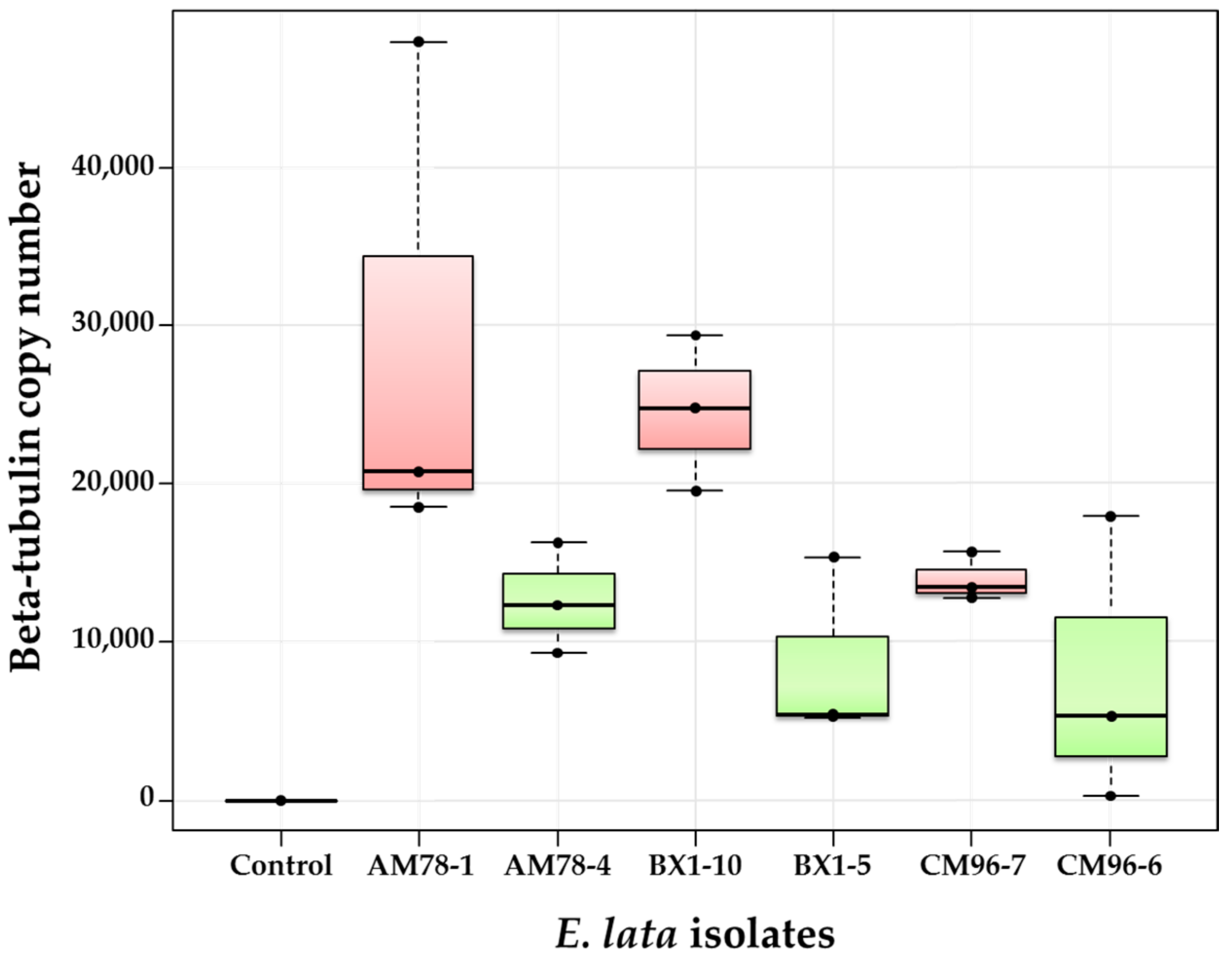
3.3. qPCR as a Tool to Evaluate Wood Colonization by E. lata (Experiment 3)
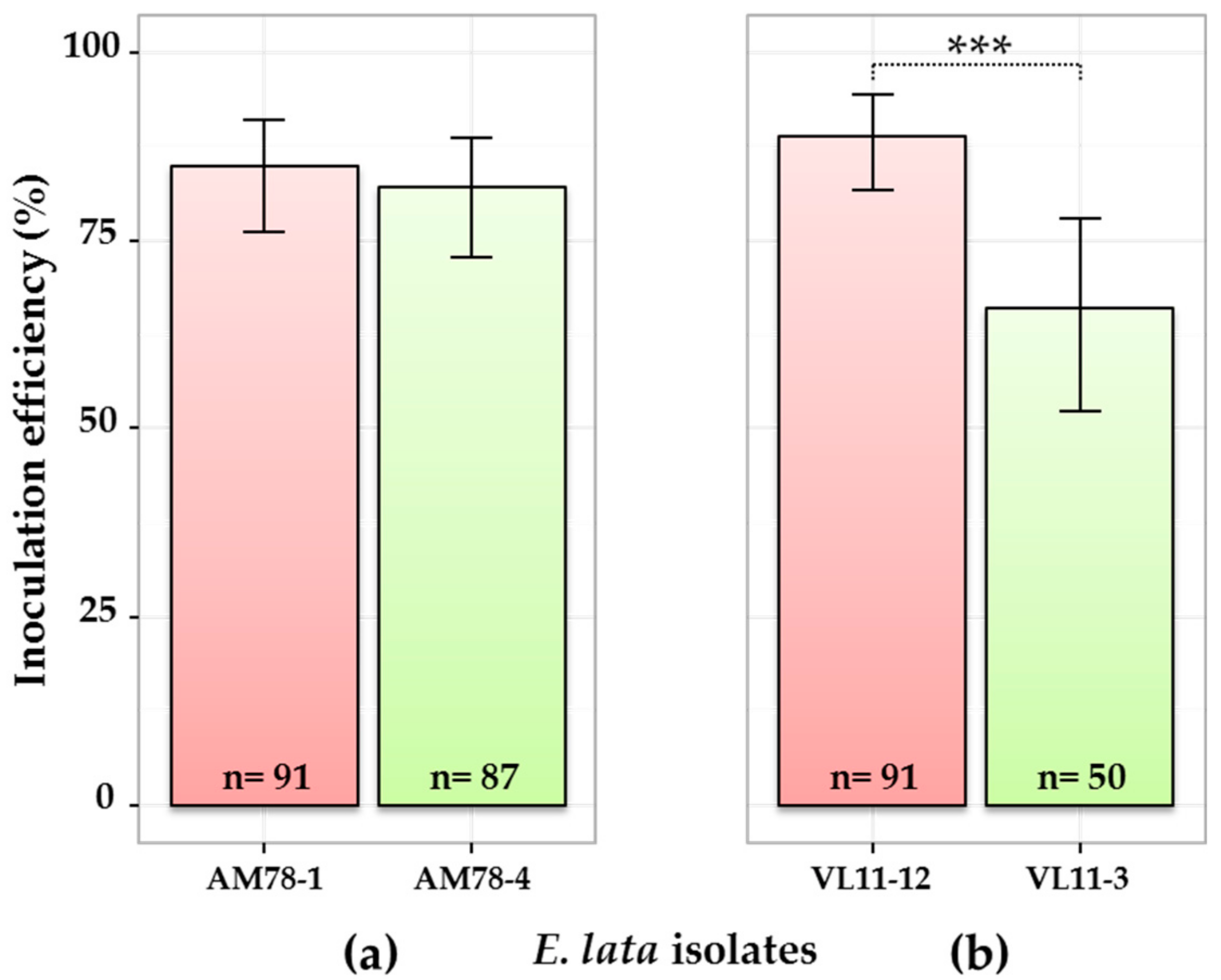
3.4. Comparison of E. lata Aggressiveness by qPCR (Experiment 4)
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bruez, E.; Lecomte, P.; Grosman, J.; Doublet, B.; Bertsch, C.; Fontaine, F.; Ugaglia, A.; Teissedre, P.L.; Da Costa, J.P.; Guerin-Dubrana, L.; et al. Overview of grapevine trunk diseases in France in the 2000s. Phytopathol. Mediterr. 2013, 52, 262–275. [Google Scholar]
- Mugnai, L.; Graniti, A.; Surico, G. Esca (Black Measles) and Brown Wood-Streaking: Two Old and Elusive Diseases of Grapevines. Plant Dis. 1999, 83, 404–418. [Google Scholar] [CrossRef]
- Reisenzein, H.; Berger, N.; Nieder, G. Esca in Austria. Phytopathol. Mediterr. 2000, 39, 26–34. [Google Scholar]
- Sosnowski, M.R.; Lardner, R.; Wicks, T.J.; Scott, E.S. The influence of grapevine cultivar and isolate of Eutypa lata on wood and foliar symptoms. Plant Dis. 2007, 91, 924–931. [Google Scholar] [CrossRef]
- Úrbez-Torres, J.R.; Leavitt, G.M.; Voegel, T.M.; Gubler, W.D. Identification and distribution of Botryosphaeria spp. associated with grapevine cankers in California. Plant Dis. 2006, 90, 1490–1503. [Google Scholar] [CrossRef]
- White, C.L.; Halleen, F.; Mostert, L. Symptoms and fungi associated with esca in South African vineyards. Phytopathol. Mediterr. 2011, 50, S236–S246. [Google Scholar]
- Bertsch, C.; Ramírez-Suero, M.; Magnin-Robert, M.; Larignon, P.; Chong, J.; Abou-Mansour, E.; Spagnolo, A.; Clément, C.; Fontaine, F. Grapevine trunk diseases: Complex and still poorly understood. Plant Pathol. 2013, 62, 243–265. [Google Scholar] [CrossRef]
- Rappaz, F. Sanctioned species of the genus Eutypa (Diatrypaceae, ascomycetes)—Taxonomic and nomenclatural study. Mycotaxon 1984, 20, 567–586. [Google Scholar]
- Péros, J.; Berger, G. Diversity within natural progenies of the grapevine dieback fungus Eutypa lata. Curr. Genet. 1999, 36, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.V.; Bolay, A.; Rappaz, F. An annotated host list and biblography of Eutypa armeniacae. Rev. Plant Pathol. 1983, 62, 251–258. [Google Scholar]
- Glawe, D.A.; Rogers, J.D. Observations on the anamorphs of six species of Eutypa and Eutypella. Mycotaxon 1982, 14, 334–346. [Google Scholar]
- Trouillas, F.P.; Gubler, W.D. Host range, biological variation, and phylogenetic diversity of Eutypa lata in California. Phytopathology 2010, 100, 1048–1056. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.V. The Status of Eutypa Lata as a Pathogen; Phytopathological Papers no. 32; International Mycological Institute: Kew, UK, 1991. [Google Scholar]
- Péros, J.P.; Berger, G.; Lahogue, F. Variation in Pathogenicity and Genetic Structure in the Eutypa lata Population of a Single Vineyard. Phytopathology 1997, 87, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Péros, J.P.; Berger, G. Genetic Structure and Variation in Aggressiveness in European and Australian Populations of the Grapevine Dieback Fungus, Eutypa lata. Eur. J. Plant Pathol. 2003, 109, 909–919. [Google Scholar] [CrossRef]
- Travadon, R.; Baumgartner, K.; Rolshausen, P.E.; Gubler, W.D.; Sosnowski, M.R.; Lecomte, P.; Halleen, F.; Péros, J.P. Genetic structure of the fungal grapevine pathogen Eutypa lata from four continents. Plant Pathol. 2012, 61, 85–95. [Google Scholar] [CrossRef]
- DeScenzo, R.A.; Engel, S.R.; Gomez, G.; Jackson, E.L.; Munkvold, G.P.; Weller, J.; Irelan, N.A. Genetic analysis of Eutypa strains from california supports the presence of two pathogenic species. Phytopathology 1999, 89, 884–893. [Google Scholar] [CrossRef] [PubMed]
- Molyneux, R.J.; Mahoney, N.; Bayman, P.; Wong, R.Y.; Meyer, K.; Irelan, N. Eutypa Dieback in Grapevines: Differential production of acetylenic phenol metabolites by strains of Eutypa lata. J. Agric. Food Chem. 2002, 50, 1393–1399. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, N.; Lardner, R.; Molyneux, R.J.; Scott, E.S.; Smith, L.R.; Schoch, T.K. Phenolic and heterocyclic metabolite profiles of the grapevine pathogen Eutypa lata. Phytochemistry 2003, 64, 475–484. [Google Scholar] [CrossRef]
- Dubos, B. Mise au point sur les maladies de dépérissement dans le vignoble francais. Compte-rendu de la réunion du groupe de travail sur les maladies de dépérissement de la vigne eutypiose (Eutypa Armenicae), esca (Stereum hirsutum, Phellinus sp.). Progrès Agric. Vitic. 1987, 104, 135–141. [Google Scholar]
- Chapuis, L.; Richard, L.; Dubos, B. Variation in susceptibility of grapevine pruning wound to infection by Eutypa lata in south-western France. Plant Pathol. 1998, 47, 463–472. [Google Scholar] [CrossRef]
- Travadon, R.; Rolshausen, P.E.; Gubler, W.D.; Cadle-Davidson, L.; Baumgartner, K. Susceptibility of cultivated and wild Vitis spp. to wood infection by fungal trunk pathogens. Plant Dis. 2013, 97, 1529–1536. [Google Scholar] [CrossRef]
- Octave, S.; Roblin, G.; Vachaud, M.; Fleurat-Lessard, P. Polypeptide metabolites secreted by the fungal pathogen Eutypa lata participate in Vitis vinifera cell structure damage observed in Eutypa dieback. Funct. Plant Biol. 2006, 33, 297–307. [Google Scholar] [CrossRef]
- Sosnowski, M.R.; Shtienberg, D.; Creaser, M.L.; Wicks, T.J.; Lardner, R.; Scott, E.S. The influence of climate on foliar symptoms of eutypa dieback in grapevines. Phytopathology 2007, 97, 1284–1289. [Google Scholar] [CrossRef] [PubMed]
- Dumot, V.; Ménard, E.; Courlit, Y.; Ouvrie, M.; Desache, F.; Boursier, N.; David, S.; Dubos, B.; Larignon, P. Eutypa canker in the Charentes region (France). Results of a 10-year study on Ugni blanc. Phytoma 2004, 568, 4–7. [Google Scholar]
- Creaser, M.; Wicks, T. Yearly variation in Eutypa dieback symptoms and the relationship to grapevine yield. Aust. Grapegrow. Winemak. 2001, 452, 50–52. [Google Scholar]
- Péros, J.-P.; Berger, G. A rapid method to assess the aggressiveness of Eutypa lata isolates and the susceptibility of grapevine cultivars to Eutypa dieback. Agronomie 1994, 14, 515–523. [Google Scholar] [CrossRef]
- Schena, L.; Nigro, F.; Ippolito, A.; Gallitelli, D. Real-time quantitative PCR: A new technology to detect and study phytopathogenic and antagonistic fungi. Eur. J. Plant Pathol. 2004, 110, 893–908. [Google Scholar] [CrossRef]
- Atallah, Z.K.; Bae, J.; Jansky, S.H.; Rouse, D.I.; Stevenson, W.R. Multiplex real-time quantitative PCR to detect and quantify Verticillium dahliae colonization in potato lines that differ in response to verticillium wilt. Phytopathology 2007, 97, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Lecomte, P.; Péros, J.P.; Blancard, D.; Bastien, N.; Delye, C. PCR assays that identify the grapevine dieback fungus Eutypa lata. Appl. Environ. Microbiol. 2000, 66, 4475–4480. [Google Scholar] [CrossRef] [PubMed]
- Camps, C.; Kappel, C.; Lecomte, P.; Leon, C.; Gomes, E.; Coutos-Thevenot, P.; Delrot, S. A transcriptomic study of grapevine (Vitis vinifera cv. Cabernet-Sauvignon) interaction with the vascular ascomycete fungus Eutypa lata. J. Exp. Bot. 2010, 61, 1719–1737. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.; Constable, F.; Wiechel, T.; Salib, S. Comparison of the molecular tests-single PCR, nested PCR and quantitative PCR (SYBR® Green and TaqMan®)-for detection of Phaeomoniella chlamydospora during grapevine nursery propagation. Phytopathol. Mediterr. 2007, 46, 58–72. [Google Scholar]
- Overton, B.E.; Stewart, E.L.; Qu, X.; Wenner, N.G.; Christ, B.J. Qualitative real-time PCR SYBR Green detection of Petri disease fungi. Phytopathol. Mediterr. 2004, 43, 403–410. [Google Scholar]
- Martin, M.T.; Cobos, R.; Martin, L.; Lopez-Enriquez, L. Real-Time PCR Detection of Phaeomoniella chlamydospora and Phaeoacremonium aleophilum. Appl. Environ. Microbiol. 2012, 78, 3985–3991. [Google Scholar] [CrossRef] [PubMed]
- Pouzoulet, J.; Mailhac, N.; Couderc, C.; Besson, X.; Daydé, J.; Lummerzheim, M.; Jacques, A. A method to detect and quantify Phaeomoniella chlamydospora and Phaeoacremonium aleophilum DNA in grapevine-wood samples. Appl. Microbiol. Biotechnol. 2013, 97, 10163–10175. [Google Scholar] [CrossRef] [PubMed]
- Gatto, P.; Vrhovsek, U.; Muth, J.; Segala, C.; Romualdi, C.; Fontana, P.; Pruefer, D.; Stefanini, M.; Moser, C.; Mattivi, F.; et al. Ripening and genotype control stilbene accumulation in healthy grapes. J. Agric. Food Chem. 2008, 56, 11773–11785. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Lee, D.; Schulman, A.H. FastPCR software for PCR primer and probe design and repeat search. Genes Genomes Genom. 2009, 3, 1–14. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2016. [Google Scholar]
- Goodman, S.N.; Fanelli, D.; Ioannidis, J.P.A. What does research reproducibility mean? Sci. Transl. Med. 2016, 8, 341ps12. [Google Scholar] [CrossRef] [PubMed]
- Bååth, R. Bayesian First Aid: A Package that Implements Bayesian Alternatives to the Classical * .test Functions in R. In Proceedings of the UseR! 2014-the International R User Conference, UCLA, Los Angeles, CA, USA, 3–30 July 2014. [Google Scholar]
- Surico, G.; Marchi, G.; Braccini, P.; Mugnai, L. Epidemiology of esca in some vineyards in Tuscany (Italy). Phytopathol. Mediterr. 2000, 39, 190–205. [Google Scholar]
- Munkvold, G.; Marois, J. Factors associated with variation in susceptibility of grapevine pruning wounds to infection by Eutypa lata. Phytopathol. Mediterr. 1995, 85, 249–256. [Google Scholar] [CrossRef]
- del Río, J.A.; Gómez, P.; Báidez, A.; Fuster, M.D.; Ortuño, A.; Frías, V. Phenolic compounds have a role in the defence mechanism protecting grapevine against the fungi involved in Petri disease. Phytopathol. Mediterr. 2004, 43, 87–94. [Google Scholar]
- Rudelle, J.; Octave, S.; Kaid-Harche, M.; Roblin, G.; Fleurat-Lessard, P. Structural modifications induced by Eutypa lata in the xylem of trunk and canes of Vitis vinifera. Funct. Plant Biol. 2005, 32, 537–547. [Google Scholar] [CrossRef]
- Yuan, H.Y.; Yao, L.L.; Jia, Z.Q.; Li, Y.; Li, Y.Z. Verticillium dahliae toxin induced alterations of cytoskeletons and nucleoli in Arabidopsis thaliana suspension cells. Protoplasma 2006, 229, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Pouzoulet, J.; Pivovaroff, A.L.; Santiago, L.S.; Rolshausen, P.E. Can vessel dimension explain tolerance toward fungal vascular wilt diseases in woody plants? Lessons from Dutch elm disease and esca disease in grapevine. Front. Plant Sci. 2014, 5, 253. [Google Scholar] [CrossRef] [PubMed]



| Stroma | Location | Year of Isolation | High-Aggressive Isolate | Low-Aggressive Isolate |
|---|---|---|---|---|
| AM78 | Aigues-Mortes | 1992 | AM78-4 | AM78-1 |
| BX1 | Bordeaux | 1990 | BX1-10 | BX1-5 |
| CM96 | Chambolle-Musigny | 1993 | CM96-7 | CM96-6 |
| VL11 | Villeneuve-lès-Maguelone | 1992 | VL11-12 | VL11-3 |
| Primers | Sequences (5’ -> 3’) | Length (nt) | Target | PCR Product Size | Organism | Source | |
|---|---|---|---|---|---|---|---|
| Act3_37_F | TCCTTGCCTTGCGTCATCTAT | Fwd | 21 | Actin | 72 bp | V. vinifera | Gatto et al. [36] |
| Act3_38_R | CACCAATCACTCTCCTGCTACAA | Rev | 23 | ||||
| EL_b-tub_33_F | CTGGCAATGCTAACTCGCCGCT | Fwd | 22 | β-tubulin | 90 bp | E. lata | This study |
| EL_b-tub_34_R | CGAGGAACATACTTGTTGCCGGAC | Rev | 24 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moisy, C.; Berger, G.; Flutre, T.; Le Cunff, L.; Péros, J.-P. Quantitative Assessment of Grapevine Wood Colonization by the Dieback Fungus Eutypa lata. J. Fungi 2017, 3, 21. https://doi.org/10.3390/jof3020021
Moisy C, Berger G, Flutre T, Le Cunff L, Péros J-P. Quantitative Assessment of Grapevine Wood Colonization by the Dieback Fungus Eutypa lata. Journal of Fungi. 2017; 3(2):21. https://doi.org/10.3390/jof3020021
Chicago/Turabian StyleMoisy, Cédric, Gilles Berger, Timothée Flutre, Loïc Le Cunff, and Jean-Pierre Péros. 2017. "Quantitative Assessment of Grapevine Wood Colonization by the Dieback Fungus Eutypa lata" Journal of Fungi 3, no. 2: 21. https://doi.org/10.3390/jof3020021






