Regulatory Networks that Direct the Development of Specialized Cell Types in the Drosophila Heart
Abstract
:1. Introduction
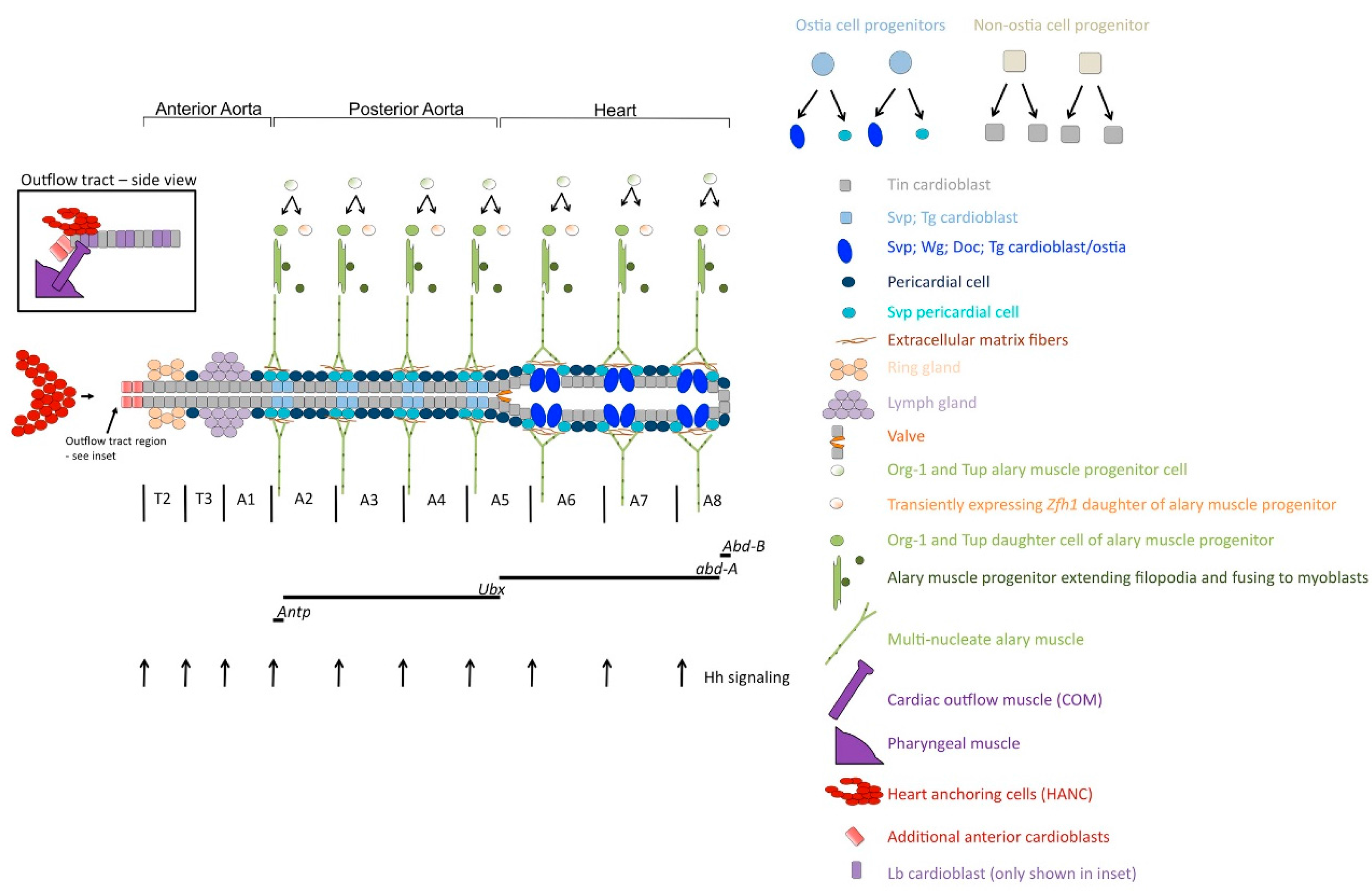
2. Development and Organization of the Drosophila Dorsal Vessel
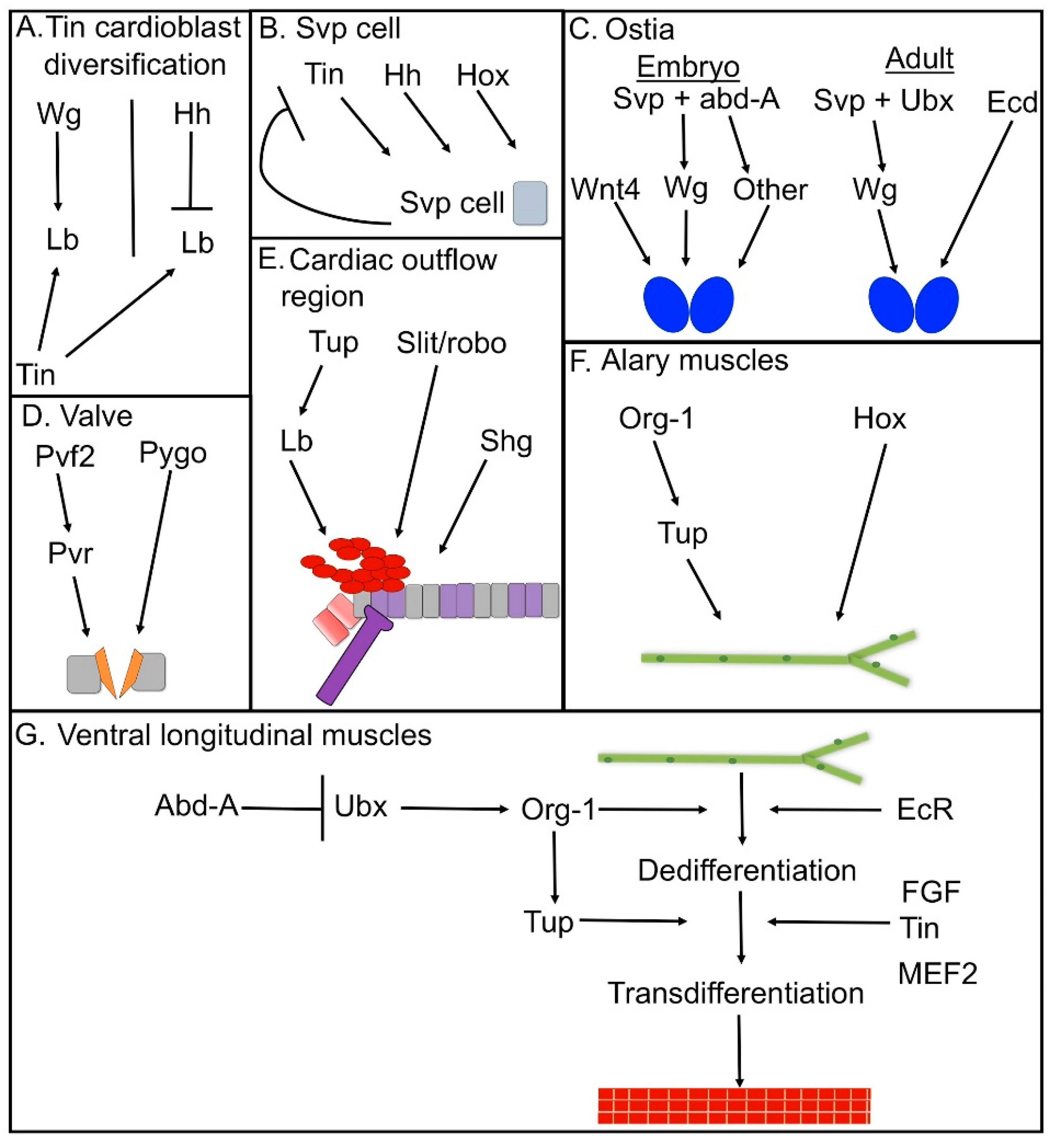
3. Cell-Type Diversification in the Dorsal Vessel
4. Genetic Control of Ostia Formation
5. Formation of the Adult Ostia
6. Regulatory Networks in Drosophila Valve Formation
7. Development and Specification of the Cardiac Outflow Tract
8. Regulatory Networks in Alary Muscle Formation
9. Regulatory Networks in the Formation of Ventral Longitudinal Muscle Fibers in the Adult Heart
10. Conclusions
Acknowledgments
Conflicts of Interest
References
- Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; de Ferranti, S.; Després, J.-P.; Fullerton, H.J.; Howard, V.J.; et al. Heart Disease and Stroke Statistics—2015 Update: A Report from the American Heart Association. Circulation 2015, 131, e29–e322. [Google Scholar] [CrossRef] [PubMed]
- Richards, A.A.; Garg, V. Genetics of congenital heart disease. Curr. Cardiol Rev. 2010, 6, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Anvari, M.S.; Boroumand, M.A.; Karimi, A.; Alidoosti, M.; Yazdanifard, P.; Shirzad, M.; Abbasi, S.H.; Soleymani, A. Aortic and mitral valve atherosclerosis: Predictive factors and associations with coronary atherosclerosis using Gensini score. Arch Med. Res. 2009, 40, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Libby, P.; Ridker, P.; Hansson, G. Progress and challenges in translating the biology of atherosclerosis. Nature 2011, 473, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Dirkx, E.; da Costa Martins, P.A.; de Windt, L.J. Regulation of fetal gene expression in heart failure. Biochim. Biophys. Acta 2013, 1832, 2414–2424. [Google Scholar] [CrossRef] [PubMed]
- Cripps, R.M.; Olson, E.N. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev. Biol. 2002, 246, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Olson, E.N. Gene regulatory networks in the evolution and development of the heart. Science 2006, 313, 1922–1927. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D. Making or Breaking the Heart: From Lineage Determination to Morphogenesis. Cell 2006, 6, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Chien, S.; Reiter, L.T.; Bier, E.; Gribskov, M. Homophila: human disease gene cognates in Drosophila. Nucleic Acids Res. 2002, 30, 149–151. [Google Scholar] [CrossRef] [PubMed]
- Reiter, L.T.; Potocki, L.; Chien, S.; Gribskov, M.; Bier, E. A systematic analysis of human disease-associated gene sequences in drosophila melanogaster. Genome Res. 2001, 11, 1114–1135. [Google Scholar] [CrossRef] [PubMed]
- Bier, E.; Bodmer, R. Drosophila, an emerging model for cardiac disease. Gene 2004, 342, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Schott, J.J.; Benson, D.W.; Basson, C.T.; Pease, W.; Silberbach, G.M.; Moak, J.P.; Maron, B.J.; Seidman, C.E.; Seidman, J.G. Congenital heart disease caused by mutations in the transcription factor NKX2–5. Science 1998, 281, 108–111. [Google Scholar] [CrossRef] [PubMed]
- Basson, C.T.; Bachinsky, D.R.; Lin, R.C.; Levi, T.; Elkins, J.A.; Soults, J.; Grayzel, D.; Droumpouzou, E.; Traill, T.A.; Leblanc-Straceski, J.; et al. Mutations in human TBX5 cause limb and cardiac malformation in Holt-Oram syndrome. Nat. Genet. 1997, 15, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Garg, V.; Kathiriya, I.S.; Barnes, R.; Schluterman, M.K.; King, I.N.; Butler, C.A.; Rothrock, C.R.; Eapen, R.S.; Hirayama-Yamada, K.; Joo, K.; et al. GATA4 muations cause human congenital heart defects and reveal an interaction with TBX5. Nature 2003, 424, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Bodmer, R.; Frasch, M. Genetic determination of Drosophila heart development. In Heart Development; Harvey, R.P., Rosenthal, N., Eds.; Academic Press: San Diego, CA, USA, 1999; Volume 1, pp. 65–90. [Google Scholar]
- Rizki, T.M. The circulatory system and associated cells and tissues. In The Genetics and Biology of Drosophila; Ashburner, M., Wright, T.R.F., Eds.; Academic Press: New York, NY, USA, 1978; Volume 2b, pp. 397–452. [Google Scholar]
- Zhang, F.; Zhao, Y.; Han, Z. An in vivo functional analysis system for renal gene discovery in Drosophila pericardial nephrocytes. J. Am. Soc. Nephrol. 2013, 24, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Sato, T.N. On the Mechanics of Cardiac Function of Drosophila Embryo. PLoS ONE 2008, 3, e4045. [Google Scholar] [CrossRef] [PubMed]
- LaBeau, E.M.; Trujillo, D.L.; Cripps, R.M. Bithorax Complex genes control alary muscle patterning along the cardiac tube of Drosophila. Mech. of Dev 2009, 126, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Lehmacher, C.; Bettina, A.; Paululat, A. The ultrastructure of Drosophila heart cells. Arthropod Struct. Dev. 2012, 41, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Boukhatmi, H.; Frendo, J.L.; Enriquez, J.; Crozatier, M.; Dubois, L.; Vincent, A. Tup/Islet1 integrates time and position to specify muscle identity in Drosophila. Development 2012, 139, 3572–3582. [Google Scholar] [CrossRef] [PubMed]
- Molina, M.R.; Cripps, R.M. Ostia, the inflow tracts of the Drosophila heart, develop from a genetically distinct subset of cardial cells. Mech. Dev. 2001, 109, 51–59. [Google Scholar] [CrossRef]
- Sellin, J.; Albrecht, S.; Kölsch, V.; Paululat, A. Dynamics of heart differentiation, visualized utilizing heart enhancer elements of the Drosophila melanogaster bHLH transcription factor Hand. Gene Expression Patterns 2006, 6, 360–375. [Google Scholar] [CrossRef] [PubMed]
- Monier, B.; Astier, M.; Semeriva, M.; Perrin, L. Steroid-dependent modification of Hox function drives myocyte reprogramming in the Drosophila heart. Development 2005, 132, 5283–5293. [Google Scholar] [CrossRef] [PubMed]
- Wasserthal, L.T. Drosophila flies combine periodic heartbeat reversal with a circulation in the anterior body mediated by a newly discovered anterior pair of ostial valves and venous channels. J. Exp. Biol. 2007, 210, 3707–3719. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.P.; Nongthomba, U.; Kelly Tanaka, K.K.; Denton, M.L.B.; Meadows, S.M.; Bancroft, N.; Molina, M.R.; Cripps, R.M. Cardiac remodeling in Drosophila arises from changes in actin gene expression and from a contribution of lymph gland-like cells to the heart musculature. Mech. Dev. 2011, 128, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Yuan, W.; Bodmer, R.; Wu, X.; Ocorr, K. The role of pygopus in the differentiation of intra-cardiac valves in Drosophila. Genesis 2014, 52, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Bodmer, R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development 1993, 118, 719–729. [Google Scholar] [PubMed]
- Azpiazu, N.; Frasch, M. tinman and bagpipe: Two homeobox genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993, 7, 1325–1340. [Google Scholar] [CrossRef] [PubMed]
- Jagla, K.; Frasch, M.; Jagla, T.; Dretzen, G.; Bellard, F.; Bellard, M. ladybird, a new component of the cardiogenic pathway in Drosophila required for diversification of heart precursors. Development 1997, 124, 3471–3479. [Google Scholar] [PubMed]
- Schafer, K.; Neuhaus, P.; Kruse, J.; Braun, T. The homeobox gene Lbx1 specifies a subpopulation of cardiac neural crest necessary for normal heart development. Circ. Res. 2003, 92, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Ward, E.J.; Skeath, J.B. Characterization of a novel subset of cardiac cells and their progenitors in the Drosophila embryo. Development 2000, 127, 4959–4969. [Google Scholar] [PubMed]
- Dye, C.A.; Lee, J.K.; Atkinson, R.C.; Brewster, R.; Han, P.L.; Bellen, H.J. The Drosophila sanpodo gene controls sibling cell fate and encodes a tropomodulin homolog, an actin/tropomyosin associated protein. Development 1998, 125, 1845–1856. [Google Scholar] [PubMed]
- Skeath, J.B.; Doe, C.Q. Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development 1998, 125, 1857–1865. [Google Scholar] [PubMed]
- Gajewski, K.; Choi, C.Y.; Kim, Y.; Schulz, R.A. Genetically Distinct Cardial Cells Within the Drosophila Heart. Genesis 2000, 28, 36–43. [Google Scholar] [CrossRef]
- Lovato, T.L.; Nguyen, T.P.; Molina, M.R.; Cripps, R.M. The Hox gene abdominal-A specifies heart cell fate in the Drosophila dorsal vessel. Development 2002, 129, 5019–5027. [Google Scholar] [PubMed]
- Ponzielli, R.; Astier, M.; Chartier, A.; Gallet, A.; Therond, P.; Semeriva, M. Heart tube patterning in Drosophila requires integration of axial and segmental information provided by the Bithorax Complex genes and hedgehog signaling. Development 2002, 129, 4509–4521. [Google Scholar] [PubMed]
- Lo, P.C.H.; Skeath, J.B.; Gajewski, K.; Schulz, R.A.; Frasch, M. Homeotic genes autonomously specify the anteroposterior subdivision of the Drosophila dorsal vessel into aorta and heart. Dev. Biol. 2002, 251, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Perrin, L.; Monier, B.; Ponzielli, R.; Astier, M.; Semeriva, M. Drosophila cardiac tube organogenesis requires multiple phases of Hox activity. Dev. Biol. 2004, 272, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Ryan, K.M.; Hoshizaki, D.K.; Cripps, R.M. Homeotic selector genes control the patterning of seven-up expressing cells in the Drosophila dorsal vessel. Mech. Dev. 2005, 122, 1023–1033. [Google Scholar] [CrossRef] [PubMed]
- Lo, P.C.H.; Frasch, M. A role for the COUP-TF-related gene seven-up in the diversification of cardioblast identities in the dorsal vessel of Drosophila. Mech. Dev. 2001, 104, 49–60. [Google Scholar] [CrossRef]
- Park, M.; Wu, X.; Golden, K.; Axelrod, J.D.; Bodmer, R. The wingless signaling pathway is directly involved in Drosophila heart development. Dev. Biol. 1996, 177, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Ryan, K.M.; Hendren, J.D.; Helander, L.A.; Cripps, R.M. The NK homeodomain transcription factor Tinman is a direct activator of seven-up in the Drosophila dorsal vessel. Dev. Biol. 2007, 302, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Reim, I.; Frasch, M. The Dorsocross T-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Development 2005, 132, 4911–4925. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, V.E. The T-box gene family: Emerging roles in development, stem cells, and cancer. Development 2014, 141, 3819–3833. [Google Scholar] [CrossRef] [PubMed]
- Reim, I.; Mohler, J.P.; Frasch, M. Tbx20-related genes, mid and H15, are required for tinman expression, proper patterning, and normal differentiation of cardioblasts in Drosophila. Mech. Dev. 2005, 122, 1056–1069. [Google Scholar] [CrossRef] [PubMed]
- Zaffran, S.; Ingolf, R.; Qian, L.; Lo, P.C.; Bodmer, R.; Frasch, M. Cardioblast-intrinsic Tinman activity controls proper diversification and differentiation of myocardial cells in Drosophila. Development 2006, 133, 4073–4083. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Wang, J.; Tokusumi, T.; Gajewski, K.; Schulz, R.A. Requirement of the LIM Homeodomain Transcription Factor Tailup for Normal Heart and Hematopoietic Organ Formation in Drosophila melanogaster. Mol. Cell. Biol. 2007, 27, 3962–3969. [Google Scholar] [CrossRef] [PubMed]
- Iklé, J.; Elwell, J.A.; Bryantsev, A.L.; Cripps, R.M. Cardiac expression of the Drosophila Transglutaminase (CG7356) gene is directly controlled by Myocyte enhancer factor-2. Dev. Dyn. 2008, 237, 2090–2099. [Google Scholar] [CrossRef] [PubMed]
- Tomancak, P.; Beaton, A.; Weiszmann, R.; Kwan, E.; Shu, S.; Lewis, S.E.; Richards, S.; Ashburner, M.; Hartenstein, V.; Celniker, S.E.; et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [Green Version]
- Trujillo, G.V.; Nodal, D.H.; Lovato, C.V.; Hendren, J.D.; Helander, L.A.; Bodmer, R.; Cripps, R.M. The canonical Wingless signaling pathway is required but not sufficient for inflow tract formation in the Drosophila melanogaster heart. Dev. Biol. 2016, in press. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhu, J.-Y.; Fu, Y.; Richman, A.; Han, Z. Wnt4 is required for ostia development in the Drosophila heart. Dev. Biol. 2016, in press. [Google Scholar] [CrossRef] [PubMed]
- Zeitouni, B.; Senatore, S.; Severac, D.; Aknin, C.; Semeriva, M.; Perrin, L. Signaling Pathways Involved in Adult Heart Formation Revealed by Gene Expression Profiling in Drosophila. PLoS Genet. 2007, 3, e174. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F.A.; Qiu, Y.; Zhou, G.; Tsai, M.J.; Tsai, S.Y. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999, 13, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.P.; Cheng, C.M.; Lanz, R.B.; Wang, T.; Respress, J.L.; Ather, S.; Chen, W.; Tsai, S.J.; Wehrens, X.H.; Tsai, M.J.; et al. Atrial identity is determined by a COUP-TFII regulatory network. Dev. Cell 2013, 25, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Tian, Y.; Boogerd, C.J.; Lu, M.M.; Kadzik, R.S.; Stewart, K.M.; Evans, S.M.; Morrisey, E.E. Coordination of heart and lung co-development by a multipotent cardiopulmonary progenitor. Nature 2013, 500, 589–592. [Google Scholar] [CrossRef] [PubMed]
- Moro, E.; Ozhan-Kizil, G.; Mongera, A.; Beis, D.; Wierzbicki, C.; Young, R.M.; Bournele, D.; Domenichini, A.; Valdivia, L.E.; Lum, L.; et al. In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev. Biol. 2012, 366, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Hurlstone, A.F.L.; Haramis, A.-P.G.; Wienholds, E.; Begthel, H.; Korving, J.; van Eeden, F.; Cuppen, E.; Zivkovic, D.; Plasterk, R.H.A.; Clevers, H. The Wnt/β-catenin pathway regulates cardiac valve formation. Nature 2003, 425, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Combs, M.D.; Yutzey, K.E. Heart Valve Development. Circ. Res. 2009, 105, 408–421. [Google Scholar] [CrossRef] [PubMed]
- Belenkaya, TY.; Han, C.; Sandley, H.J.; Lin, X.; Houston, D.W. pygopus Encodes a nuclear protein essential for wingless/Wnt signaling. Development 2002, 129, 4089–4101. [Google Scholar] [PubMed]
- Kramps, T.; Peter, O.; Brunner, E.; Nellen, D.; Froesch, B.; Chatterjee, S.; Murone, M.; Zullig, S.; Basler, K. Wnt/Wingless Signaling Requires BCL9/Legless-Mediated Recruitment of Pygopus to the Nuclear β-Catenin-TCF Complex. Cell 2002, 109, 47–60. [Google Scholar] [CrossRef] [Green Version]
- Parker, D.S.; Jemison, J.; Cadigan, K.M. Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development 2002, 129, 2565–2576. [Google Scholar] [PubMed]
- Thompson, B.; Townsley, F.; Rosin-Arbesfeld, R.; Musisi, H.; Bienz, M. A new nuclear component of the Wnt signally pathway. Nat. Cell Biol. 2002, 4, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Liebner, S.; Cattelino, A.; Gallini, R.; Rudini, N.; Iurlaro, M.; Piccolo, S.; Dejana, E. β-catenin is required for endothelial-mesenchymal transformation during heart cushion development in the mouse. J. Cell Biol. 2004, 166, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Stankunas, K.; Ma, G.K.; Kuhnert, F.J.; Kuo, C.J.; Chang, C.P. VEGF signaling has distinct spatiotemporal roles during heart valve development. Dev. Biol. 2010, 347, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Zikova, M.; Da Ponte, J.P.; Dastugue, B.; Jagla, K. Patterning of the cardiac outflow region in Drosophila. Proc. Natl. Acad. Sci. USA 2003, 100, 12189–12194. [Google Scholar] [CrossRef] [PubMed]
- Zmojdzian, M.; da Ponte, J.P.; Jagla, K. Cellular components and signals required for the cardiac outflow tract assembly in Drosophila. Proc. Natl. Acad. Sci. USA 2008, 105, 2475–2480. [Google Scholar] [CrossRef] [PubMed]
- Zmojdzian, M.; Jagla, K. Tailup plays multiple roles during cardiac outflow tract assembly in Drosophila. Cell Tissue Res. 2013, 354, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.G.; Buckingham, M.E. The anterior heart-forming field: Voyage to the arterial pole of the heart. Trends Genet. 2002, 18, 210–216. [Google Scholar] [CrossRef]
- Cai, C.L.; Liang, X.; Shi, Y.; Chu, P.H.; Pfaff, S.L.; Chen, J.; Evans, S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 2003, 5, 877–889. [Google Scholar] [CrossRef]
- Brown, C.B.; Baldwin, H.S. Neural crest contribution to the cardiovascular system. Adv. Exp. Med. Biol. 2006, 589, 134–154. [Google Scholar] [PubMed]
- Boukhatmi, H.; Schaub, C.; Bataille, L.; Reim, I.; Frendo, J.L.; Frasch, M.; Vincent, A. An Org-1-Tup transcriptional cascade reveals different types of alary muscles connecting internal organs in Drosophila. Development 2014, 141, 3761–3771. [Google Scholar] [CrossRef] [PubMed]
- Bate, M. The mesoderm and its derivatives. In The Development of Drosophila melanogaster; Bate, M., Martinez Arias, A., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1993; Volume 2, pp. 1013–1090. [Google Scholar]
- Dulcis, D.; Levine, R.B. Innervation of the Heart of the Adult Fruit Fly, Drosophila melanogaster. J. Comp. Neurol. 2003, 465, 560–578. [Google Scholar] [CrossRef] [PubMed]
- Standring, S. Gray’s Anatomy: The Anatomical Basis of Clinical Practice, 39th ed.; Elsevier Churchill Livingstone: New York, NY, USA, 2004; p. 1627. [Google Scholar]
- Schaub, C.; Nagaso, H.; Jin, H.; Frasch, M. Org-1, the Drosophila ortholog of Tbx1, is a direct activator of known identity genes during muscle specification. Development 2012, 139, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Schaub, C.; März, J.; Reim, I.; Frasch, M. Org-1-Dependent Lineage Reprogramming Generates the Ventral Longitudinal Musculature of the Drosophila heart. Curr. Biol. 2015, 25, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kelly, K.K.; Meadows, S.M.; Cripps, R.M. Drosophila MEF2 is an essential regulator of ACTIN57B transcription in cardiac, skeletal and visceral muscle lineages. Mec. Dev. 2002, 110, 39–50. [Google Scholar] [CrossRef]
- Kelly, R.G.; Evans, S.M. The Second Heart Field, 1st ed.; Rosenthal, N., Harvey, R.P., Eds.; Academic Press: London, UK, 2010; Volume 1, pp. 143–169. [Google Scholar]
- Barnes, R.M.; Harris, I.S.; Jaehnig, E.J.; Sauls, K.; Sinha, T.; Rojas, A.; Schachterle, W.; McCulley, D.J.; Norris, R.A.; Black, B.L. MEF2C regulates outflow tract alignment and transcriptional control of Tdgf1. Development 2016, 143, 774–779. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lovato, T.L.; Cripps, R.M. Regulatory Networks that Direct the Development of Specialized Cell Types in the Drosophila Heart. J. Cardiovasc. Dev. Dis. 2016, 3, 18. https://doi.org/10.3390/jcdd3020018
Lovato TL, Cripps RM. Regulatory Networks that Direct the Development of Specialized Cell Types in the Drosophila Heart. Journal of Cardiovascular Development and Disease. 2016; 3(2):18. https://doi.org/10.3390/jcdd3020018
Chicago/Turabian StyleLovato, TyAnna L., and Richard M. Cripps. 2016. "Regulatory Networks that Direct the Development of Specialized Cell Types in the Drosophila Heart" Journal of Cardiovascular Development and Disease 3, no. 2: 18. https://doi.org/10.3390/jcdd3020018




