Transcriptional Regulation of Heart Development in Zebrafish
Abstract
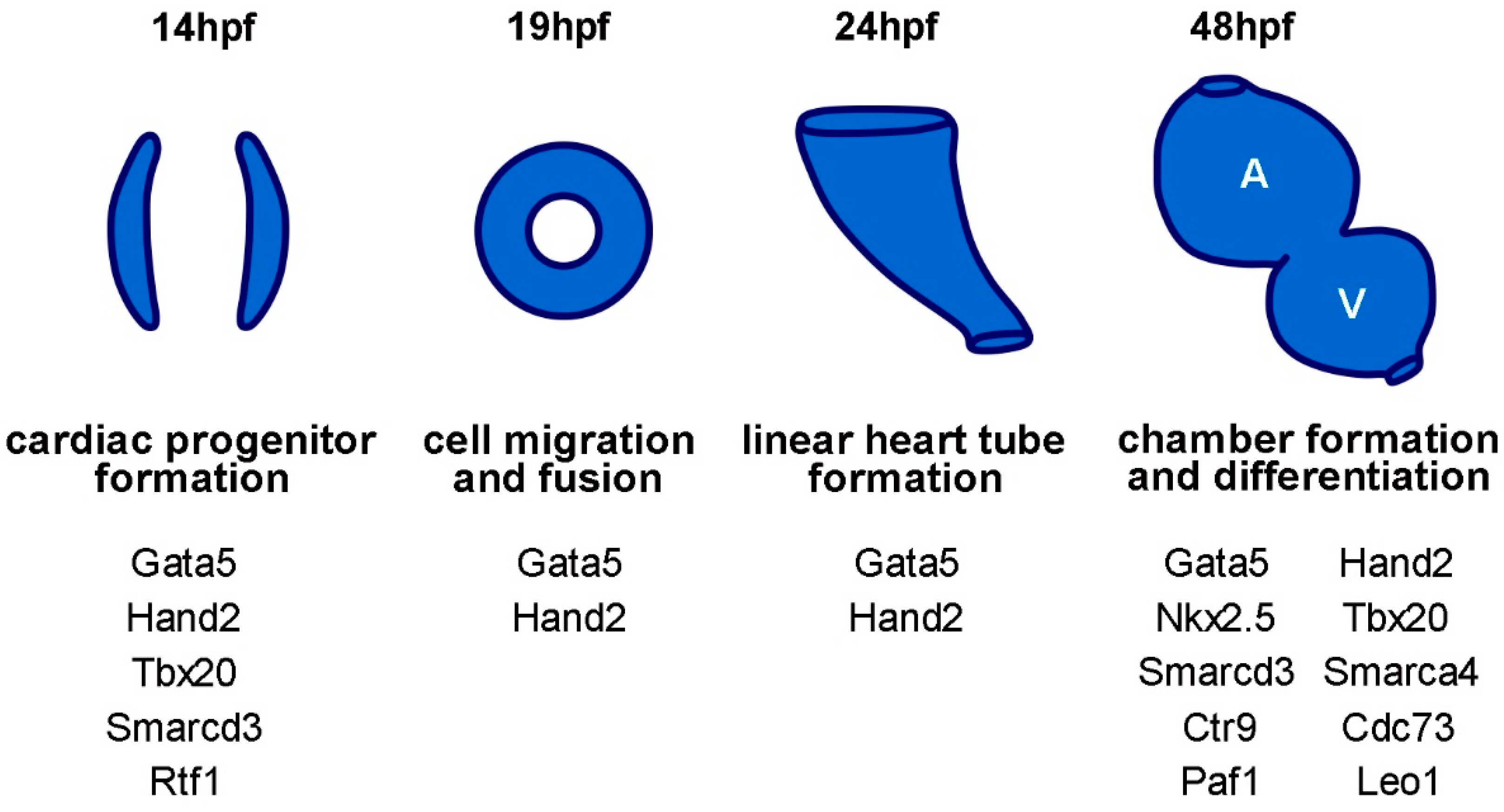
:1. Introduction
2. Cardiac Transcription Factors
2.1. GATA Factors Control Cardiomyocyte Progenitor Migration and Regulate Cardiac Fate Specification by Restricting Wnt Signaling
2.2. Hand2 Regulates Cardiac Differentiation and Morphogenesis
2.3. Nkx2.5 Maintains Cardiac Chamber Identity
2.4. Tbx20 Is Required for Cardiac Progenitor Formation
3. Epigenetic and Transcriptional Regulators
3.1. BAF Chromatin Remodeling Complex Induces a Cardiac Mesoderm Fate
3.2. The PAF1 Complex Is Involved in Cardiac Progenitor Formation and Cardiomyocyte Differentiation
4. Summary
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bruneau, B.G. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb. Perspect. Biol. 2013, 5, a008292. [Google Scholar] [CrossRef] [PubMed]
- Paige, S.L.; Plonowska, K.; Xu, A.; Wu, S.M. Molecular regulation of cardiomyocyte differentiation. Circ. Res. 2015, 116, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Rana, M.S.; Christoffels, V.M.; Moorman, A.F. A molecular and genetic outline of cardiac morphogenesis. Acta Physiol. (Oxford, England) 2013, 207, 588–615. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.P.; Bruneau, B.G. Epigenetics and cardiovascular development. Annu. Rev. Physiol. 2012, 74, 41–68. [Google Scholar] [CrossRef] [PubMed]
- Boland, M.J.; Nazor, K.L.; Loring, J.F. Epigenetic regulation of pluripotency and differentiation. Circ. Res. 2014, 115, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Nuhrenberg, T.; Gilsbach, R.; Preissl, S.; Schnick, T.; Hein, L. Epigenetics in cardiac development, function, and disease. Cell Tissue Res. 2014, 356, 585–600. [Google Scholar] [CrossRef] [PubMed]
- Stainier, D.Y.; Lee, R.K.; Fishman, M.C. Cardiovascular development in the zebrafish. I. Myocardial fate map and heart tube formation. Development 1993, 119, 31–40. [Google Scholar] [PubMed]
- Zhou, Y.; Cashman, T.J.; Nevis, K.R.; Obregon, P.; Carney, S.A.; Liu, Y.; Gu, A.; Mosimann, C.; Sondalle, S.; Peterson, R.E.; et al. Latent TGF-β binding protein 3 identifies a second heart field in zebrafish. Nature 2011, 474, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Reiter, J.F.; Alexander, J.; Rodaway, A.; Yelon, D.; Patient, R.; Holder, N.; Stainier, D.Y. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999, 13, 2983–2995. [Google Scholar] [CrossRef] [PubMed]
- Reiter, J.F.; Kikuchi, Y.; Stainier, D.Y. Multiple roles for Gata5 in zebrafish endoderm formation. Development 2001, 128, 125–135. [Google Scholar] [PubMed]
- Kuo, C.T.; Morrisey, E.E.; Anandappa, R.; Sigrist, K.; Lu, M.M.; Parmacek, M.S.; Soudais, C.; Leiden, J.M. Gata4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997, 11, 1048–1060. [Google Scholar] [CrossRef] [PubMed]
- Molkentin, J.D.; Lin, Q.; Duncan, S.A.; Olson, E.N. Requirement of the transcription factor Gata4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997, 11, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Narita, N.; Bielinska, M.; Wilson, D.B. Wild-type endoderm abrogates the ventral developmental defects associated with GATA-4 deficiency in the mouse. Dev. Biol. 1997, 189, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Holtzinger, A.; Evans, T. Gata5 and Gata6 are functionally redundant in zebrafish for specification of cardiomyocytes. Dev. Biol. 2007, 312, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Moskowitz, I.P.; Wang, J.; Peterson, M.A.; Pu, W.T.; Mackinnon, A.C.; Oxburgh, L.; Chu, G.C.; Sarkar, M.; Berul, C.; Smoot, L.; et al. Transcription factor genes Smad4 and Gata4 cooperatively regulate cardiac valve development. [Corrections]. Proc. Nal. Acad. Sci. USA 2011, 108, 4006–4011. [Google Scholar] [CrossRef] [PubMed]
- Novikov, N.; Evans, T. Tmem88a mediates GATA-dependent specification of cardiomyocyte progenitors by restricting wnt signaling. Development 2013, 140, 3787–3798. [Google Scholar] [CrossRef] [PubMed]
- Palpant, N.J.; Pabon, L.; Rabinowitz, J.S.; Hadland, B.K.; Stoick-Cooper, C.L.; Paige, S.L.; Bernstein, I.D.; Moon, R.T.; Murry, C.E. Transmembrane protein 88: A wnt regulatory protein that specifies cardiomyocyte development. Development 2013, 140, 3799–3808. [Google Scholar] [CrossRef] [PubMed]
- Schoenebeck, J.J.; Keegan, B.R.; Yelon, D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev. Cell 2007, 13, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Palencia-Desai, S.; Rost, M.S.; Schumacher, J.A.; Ton, Q.V.; Craig, M.P.; Baltrunaite, K.; Koenig, A.L.; Wang, J.; Poss, K.D.; Chi, N.C.; et al. Myocardium and BMP signaling are required for endocardial differentiation. Development 2015, 142, 2304–2315. [Google Scholar] [CrossRef] [PubMed]
- Trinh, L.A.; Stainier, D.Y. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev. cell 2004, 6, 371–382. [Google Scholar] [CrossRef]
- Trinh, L.A.; Yelon, D.; Stainier, D.Y. Hand2 regulates epithelial formation during myocardial diferentiation. Curr. Biol.: CB 2005, 15, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Yelon, D.; Ticho, B.; Halpern, M.E.; Ruvinsky, I.; Ho, R.K.; Silver, L.M.; Stainier, D.Y. The BHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development 2000, 127, 2573–2582. [Google Scholar] [PubMed]
- Garavito-Aguilar, Z.V.; Riley, H.E.; Yelon, D. Hand2 ensures an appropriate environment for cardiac fusion by limiting fibronectin function. Development 2010, 137, 3215–3220. [Google Scholar] [CrossRef] [PubMed]
- Azpiazu, N.; Frasch, M. Tinman and bagpipe: Two homeo box genes that determine cell fates in the dorsal mesoderm of drosophila. Genes Dev. 1993, 7, 1325–1340. [Google Scholar] [CrossRef] [PubMed]
- Bodmer, R. The gene tinman is required for specification of the heart and visceral muscles in drosophila. Development 1993, 118, 719–729. [Google Scholar] [PubMed]
- Lyons, I.; Parsons, L.M.; Hartley, L.; Li, R.; Andrews, J.E.; Robb, L.; Harvey, R.P. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2–5. Genes Dev. 1995, 9, 1654–1666. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Wechsler, S.B.; Lee, I.W.; Yamasaki, N.; Lawitts, J.A.; Izumo, S. Complex modular cis-acting elements regulate expression of the cardiac specifying homeobox gene Csx/Nkx2.5. Development 1999, 126, 1439–1450. [Google Scholar] [PubMed]
- Targoff, K.L.; Schell, T.; Yelon, D. Nkx genes regulate heart tube extension and exert differential effects on ventricular and atrial cell number. Dev. Biol. 2008, 322, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.N.; Fishman, M.C. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development 1996, 122, 3809–3816. [Google Scholar] [PubMed]
- Lee, K.H.; Xu, Q.; Breitbart, R.E. A new tinman-related gene, Nkx2.7, anticipates the expression of Nkx2.5 and Nkx2.3 in zebrafish heart and pharyngeal endoderm. Dev. Biol. 1996, 180, 722–731. [Google Scholar] [CrossRef] [PubMed]
- Targoff, K.L.; Colombo, S.; George, V.; Schell, T.; Kim, S.H.; Solnica-Krezel, L.; Yelon, D. Nkx genes are essential for maintenance of ventricular identity. Development 2013, 140, 4203–4213. [Google Scholar] [CrossRef] [PubMed]
- George, V.; Colombo, S.; Targoff, K.L. An early requirement for Nkx2.5 ensures the first and second heart field ventricular identity and cardiac function into adulthood. Dev. Biol. 2015, 400, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Ahn, D.G.; Ruvinsky, I.; Oates, A.C.; Silver, L.M.; Ho, R.K. TBX20, a new vertebrate T-box gene expressed in the cranial motor neurons and developing cardiovascular structures in zebrafish. Mech. Dev. 2000, 95, 253–258. [Google Scholar] [CrossRef]
- Griffin, K.J.; Stoller, J.; Gibson, M.; Chen, S.; Yelon, D.; Stainier, D.Y.; Kimelman, D. A conserved role for H15-related T-box transcription factors in zebrafish and drosophila heart formation. Dev. Biol. 2000, 218, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.D.; Binder, O.; Pagratis, M.; Parr, B.A.; Conlon, F.L. Developmental expression of the Xenopus laevis TBX20 orthologue. Dev. Genes Evol. 2003, 212, 604–607. [Google Scholar] [PubMed]
- Shen, T.; Aneas, I.; Sakabe, N.; Dirschinger, R.J.; Wang, G.; Smemo, S.; Westlund, J.M.; Cheng, H.; Dalton, N.; Gu, Y.; et al. TBX20 regulates a genetic program essential to adult mouse cardiomyocyte function. J. Clin. Investig. 2011, 121, 4640–4654. [Google Scholar] [CrossRef] [PubMed]
- Kraus, F.; Haenig, B.; Kispert, A. Cloning and expression analysis of the mouse T-box gene TBX20. Mech. Dev. 2001, 100, 87–91. [Google Scholar] [CrossRef]
- Chakraborty, S.; Yutzey, K.E. TBX20 regulation of cardiac cell proliferation and lineage specialization during embryonic and fetal development in vivo. Dev. Biol. 2012, 363, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.L.; Zhou, W.; Yang, L.; Bu, L.; Qyang, Y.; Zhang, X.; Li, X.; Rosenfeld, M.G.; Chen, J.; Evans, S. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development 2005, 132, 2475–2487. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.K.; Christoffels, V.M.; Dias, J.M.; Trowe, M.O.; Petry, M.; Schuster-Gossler, K.; Burger, A.; Ericson, J.; Kispert, A. TBX20 is essential for cardiac chamber differentiation and repression of TBX2. Development 2005, 132, 2697–2707. [Google Scholar] [CrossRef] [PubMed]
- Stennard, F.A.; Costa, M.W.; Lai, D.; Biben, C.; Furtado, M.B.; Solloway, M.J.; McCulley, D.J.; Leimena, C.; Preis, J.I.; Dunwoodie, S.L.; et al. Murine T-box transcription factor TBX20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development 2005, 132, 2451–2462. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.K.; Mileikovskaia, M.; Koshiba-Takeuchi, K.; Heidt, A.B.; Mori, A.D.; Arruda, E.P.; Gertsenstein, M.; Georges, R.; Davidson, L.; Mo, R.; et al. TBX20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development 2005, 132, 2463–2474. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Mohapatra, B.; Akasaka, T.; Liu, J.; Ocorr, K.; Towbin, J.A.; Bodmer, R. Transcription factor neuromancer/TBX20 is required for cardiac function in drosophila with implications for human heart disease. Proc. Natl. Acad. Sci. USA 2008, 105, 19833–19838. [Google Scholar] [CrossRef] [PubMed]
- Kirk, E.P.; Sunde, M.; Costa, M.W.; Rankin, S.A.; Wolstein, O.; Castro, M.L.; Butler, T.L.; Hyun, C.; Guo, G.; Otway, R.; et al. Mutations in cardiac t-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am. J. Hum. Genet. 2007, 81, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shen, A.; Li, X.; Jiao, W.; Zhang, X.; Li, Z. T-box transcription factor TBX20 mutations in Chinese patients with congenital heart disease. Eur. J. Med. Genet. 2008, 51, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Hammer, S.; Toenjes, M.; Lange, M.; Fischer, J.J.; Dunkel, I.; Mebus, S.; Grimm, C.H.; Hetzer, R.; Berger, F.; Sperling, S. Characterization of TBX20 in human hearts and its regulation by TFAP2. J. Cell. Biochem. 2008, 104, 1022–1033. [Google Scholar] [CrossRef] [PubMed]
- Posch, M.G.; Gramlich, M.; Sunde, M.; Schmitt, K.R.; Lee, S.H.; Richter, S.; Kersten, A.; Perrot, A.; Panek, A.N.; Al Khatib, I.H.; et al. A gain-of-function TBX20 mutation causes congenital atrial septal defects, patent foramen ovale and cardiac valve defects. J. Med. Genet. 2010, 47, 230–235. [Google Scholar] [CrossRef] [PubMed]
- De Jong, R.N.; Truffault, V.; Diercks, T.; Ab, E.; Daniels, M.A.; Kaptein, R.; Folkers, G.E. Structure and DNA binding of the human Rtf1 Plus3 domain. Structure 2008, 16, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Geng, R.; Zhou, N.; Zheng, G.F.; Zhao, H.; Wang, J.; Zhao, C.M.; Qiu, X.B.; Yang, Y.Q.; Liu, X.Y. TBX20 loss-of-function mutation contributes to double outlet right ventricle. Int. J. Mol. Med. 2015, 35, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Liu, J.; Bodmer, R. Neuromancer TBX20-related genes (H15/midline) promote cell fate specification and morphogenesis of the drosophila heart. Dev. Biol. 2005, 279, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Szeto, D.P.; Griffin, K.J.; Kimelman, D. HrT is required for cardiovascular development in zebrafish. Development 2002, 129, 5093–5101. [Google Scholar] [PubMed]
- Brown, D.D.; Martz, S.N.; Binder, O.; Goetz, S.C.; Price, B.M.; Smith, J.C.; Conlon, F.L. TBX5 and TBX20 act synergistically to control vertebrate heart morphogenesis. Development 2005, 132, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.D.; Kim, E.; Koun, S.; Ham, H.J.; Rhee, M.; Kim, M.J.; Huh, T.L. Proper activity of histone H3 lysine 4 (H3K4) methyltransferase is required for morphogenesis during zebrafish cardiogenesis. Mol. Cells 2015, 38, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Kim, M.J.; Koo, T.H.; Kim, J.D.; Koun, S.; Ham, H.J.; Lee, Y.M.; Rhee, M.; Yeo, S.Y.; Huh, T.L. Histone deacetylase is required for the activation of Wnt/β-catenin signaling crucial for heart valve formation in zebrafish embryos. Biochem. Biophys. Res. Commun. 2012, 423, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Hang, C.T.; Yang, J.; Han, P.; Cheng, H.-L.; Shang, C.; Ashley, E.; Zhou, B.; Chang, C.-P. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 2010, 466, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Schlesinger, J.; Schoenhals, S.; Tonjes, M.; Dunkel, I.; Meierhofer, D.; Cano, E.; Schulz, K.; Berger, M.F.; Haack, T.; et al. Phosphorylation of the chromatin remodeling factor DPF3a induces cardiac hypertrophy through releasing hey repressors from DNA. Nucl. Acids Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Lange, M.; Kaynak, B.; Forster, U.B.; Tonjes, M.; Fischer, J.J.; Grimm, C.; Schlesinger, J.; Just, S.; Dunkel, I.; Krueger, T.; et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the baf chromatin remodeling complex. Genes Dev. 2008, 22, 2370–2384. [Google Scholar] [CrossRef] [PubMed]
- Bajpai, R.; Chen, D.A.; Rada-Iglesias, A.; Zhang, J.; Xiong, Y.; Helms, J.; Chang, C.P.; Zhao, Y.; Swigut, T.; Wysocka, J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 2010, 463, 958–962. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, H.; Fujiki, R.; Yoshimura, K.; Mezaki, Y.; Uematsu, Y.; Matsui, D.; Ogawa, S.; Unno, K.; Okubo, M.; Tokita, A.; et al. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell 2003, 113, 905–917. [Google Scholar] [CrossRef]
- Lickert, H.; Takeuchi, J.K.; Von Both, I.; Walls, J.R.; McAuliffe, F.; Adamson, S.L.; Henkelman, R.M.; Wrana, J.L.; Rossant, J.; Bruneau, B.G. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 2004, 432, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Deshwar, A.R.; Crump, J.G.; Scott, I.C. Smarcd3b and Gata5 promote a cardiac progenitor fate in the zebrafish embryo. Development 2011, 138, 3113–3123. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Fulcoli, F.G.; Ferrentino, R.; Martucciello, S.; Illingworth, E.A.; Baldini, A. Transcriptional control in cardiac progenitors: TBX1 interacts with the BAF chromatin remodeling complex and regulates wnt5a. PLoS Genet. 2012, 8, e1002571. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.K.; Lou, X.; Alexander, J.M.; Sugizaki, H.; Delgado-Olguin, P.; Holloway, A.K.; Mori, A.D.; Wylie, J.N.; Munson, C.; Zhu, Y.; et al. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat. Commun. 2011, 2, 187. [Google Scholar] [CrossRef] [PubMed]
- Gregg, R.G.; Willer, G.B.; Fadool, J.M.; Dowling, J.E.; Link, B.A. Positional cloning of the young mutation identifies an essential role for the brahma chromatin remodeling complex in mediating retinal cell differentiation. Proc. Natl. Acad. Sci. USA 2003, 100, 6535–6540. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.K.; Bruneau, B.G. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 2009, 459, 708–711. [Google Scholar] [CrossRef] [PubMed]
- Tomson, B.N.; Arndt, K.M. The many roles of the conserved eukaryotic PAF1 complex in regulating transcription, histone modifications, and disease states. Biochim. Biophys. Acta (BBA)—Gene Regul. Mech. 2013, 1829, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.; Wollmann, P.; Dahl, M.; Lottspeich, F. A role for Ctr9p and PAF1p in the regulation G1 cyclin expression in yeast. Nucl. Acids Res. 1999, 27, 2126–2134. [Google Scholar] [CrossRef] [PubMed]
- Akanuma, T.; Koshida, S.; Kawamura, A.; Kishimoto, Y.; Takada, S. PAF1 complex homologues are required for notch-regulated transcription during somite segmentation. EMBO Rep. 2007, 8, 858–863. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.T.; Langenbacher, A.; Hsieh, M.; Chen, J.N. The PAF1 complex component Leo1 is essential for cardiac and neural crest development in zebrafish. Dev. Biol. 2010, 341, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Paszkowski-Rogacz, M.; Nitzsche, A.; Slabicki, M.M.; Heninger, A.K.; de Vries, I.; Kittler, R.; Junqueira, M.; Shevchenko, A.; Schulz, H.; et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the PAF1 complex for embryonic stem cell identity. Cell Stem Cell 2009, 4, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Langenbacher, A.D.; Nguyen, C.T.; Cavanaugh, A.M.; Huang, J.; Lu, F.; Chen, J.N. The PAF1 complex differentially regulates cardiomyocyte specification. Dev. Biol. 2011, 353, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Mbogning, J.; Nagy, S.; Page, V.; Schwer, B.; Shuman, S.; Fisher, R.P.; Tanny, J.C. The PAF complex and Prf1/Rtf1 delineate distinct Cdk9-dependent pathways regulating transcription elongation in fission yeast. PLoS Genet. 2013, 9, e1004029. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.F.; Yamamoto, J.; Isobe, T.; Tateno, S.; Murase, Y.; Chen, Y.; Handa, H.; Yamaguchi, Y. Characterization of the human transcription elongation factor Rtf1: Evidence for nonoverlapping functions of Rtf1 and the PAF1 complex. Mol. Cell. Biol. 2015, 35, 3459–3470. [Google Scholar] [CrossRef] [PubMed]
- Rozenblatt-Rosen, O.; Hughes, C.M.; Nannepaga, S.J.; Shanmugam, K.S.; Copeland, T.D.; Guszczynski, T.; Resau, J.H.; Meyerson, M. The parafibromin tumor suppressor protein is part of a human PAF1 complex. Mol. Cell. Biol. 2005, 25, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Mandal, S.S.; Pham, A.D.; Zheng, Y.; Erdjument-Bromage, H.; Batra, S.K.; Tempst, P.; Reinberg, D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005, 19, 1668–1673. [Google Scholar] [CrossRef] [PubMed]
- Adelman, K.; Wei, W.; Ardehali, M.B.; Werner, J.; Zhu, B.; Reinberg, D.; Lis, J.T. Drosophila PAF1 modulates chromatin structure at actively transcribed genes. Mol. Cell. Biol. 2006, 26, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Nordick, K.; Hoffman, M.G.; Betz, J.L.; Jaehning, J.A. Direct interactions between the PAF1 complex and a cleavage and polyadenylation factor are revealed by dissociation of PAF1 from RNA polymerase ii. Eukaryot. Cell 2008, 7, 1158–1167. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.; Sleep, E.; Raya, M.; Marti, M.; Raya, A.; Belmonte, J.C.I. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010, 464, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Holdway, J.E.; Werdich, A.A.; Anderson, R.M.; Fang, Y.; Egnaczyk, G.F.; Evans, T.; MacRae, C.A.; Stainier, D.Y.R.; Poss, K.D. Primary contribution to zebrafish heart regeneration by Gata4+ cardiomyocytes. Nature 2010, 464, 601–605. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Gemberling, M.; Karra, R.; Rosenfeld, G.E.; Evans, T.; Poss, K.D. An injury-responsive Gata4 program shapes the zebrafish cardiac ventricle. Curr. Biol. 2013, 23, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Schindler, Y.L.; Garske, K.M.; Wang, J.; Firulli, B.A.; Firulli, A.B.; Poss, K.D.; Yelon, D. Hand2 elevates cardiomyocyte production during zebrafish heart development and regeneration. Development 2014, 141, 3112–3122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lepilina, A.; Coon, A.N.; Kikuchi, K.; Holdway, J.E.; Roberts, R.W.; Burns, C.G.; Poss, K.D. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 2006, 127, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Holdway, J.E.; Major, R.J.; Blum, N.; Dahn, R.D.; Begemann, G.; Poss, K.D. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 2011, 20, 397–404. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, F.; Langenbacher, A.D.; Chen, J.-N. Transcriptional Regulation of Heart Development in Zebrafish. J. Cardiovasc. Dev. Dis. 2016, 3, 14. https://doi.org/10.3390/jcdd3020014
Lu F, Langenbacher AD, Chen J-N. Transcriptional Regulation of Heart Development in Zebrafish. Journal of Cardiovascular Development and Disease. 2016; 3(2):14. https://doi.org/10.3390/jcdd3020014
Chicago/Turabian StyleLu, Fei, Adam D. Langenbacher, and Jau-Nian Chen. 2016. "Transcriptional Regulation of Heart Development in Zebrafish" Journal of Cardiovascular Development and Disease 3, no. 2: 14. https://doi.org/10.3390/jcdd3020014




