Advances in the Study of Heart Development and Disease Using Zebrafish
Abstract
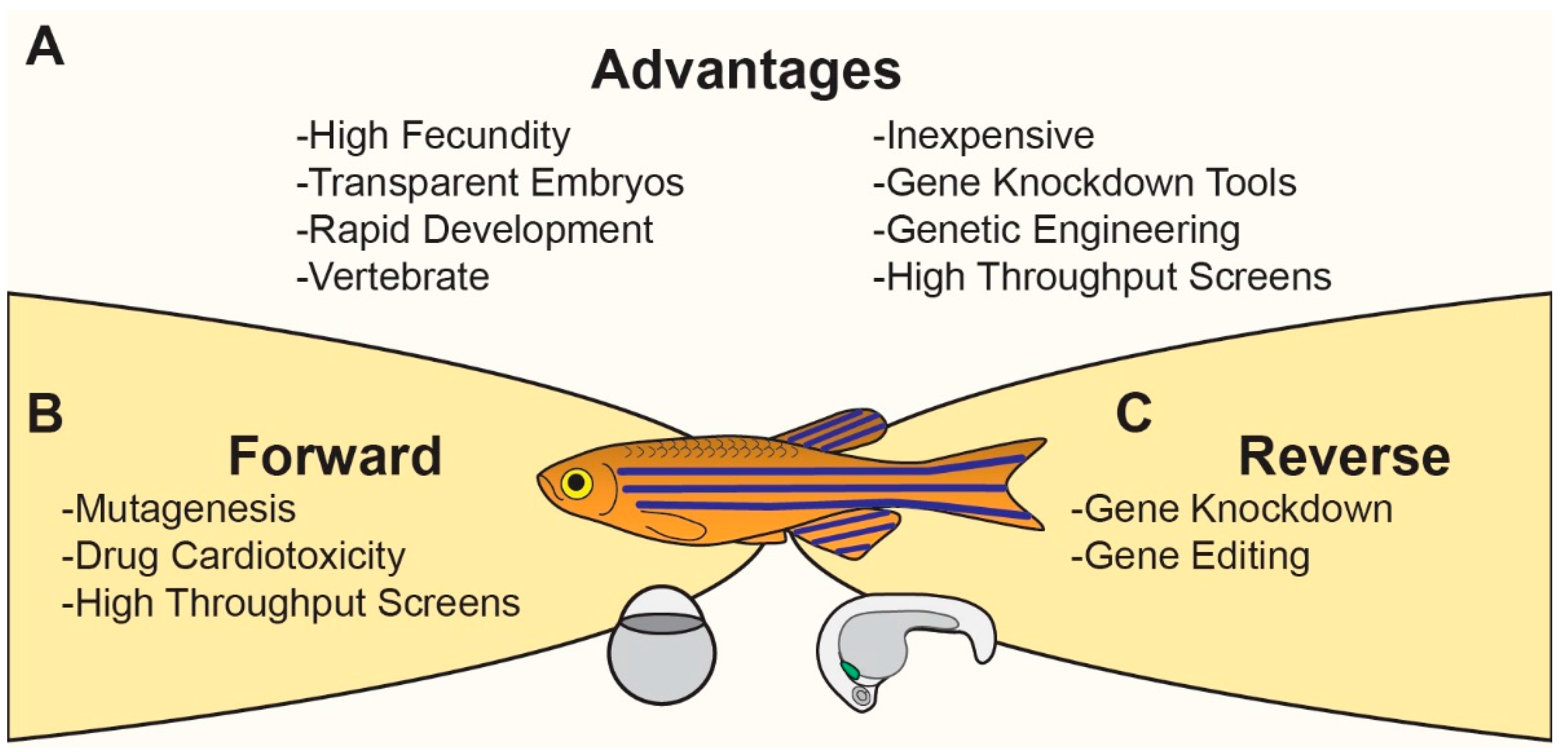
:1. Introduction
Modeling Cardiovascular Development and Disease in Zebrafish
2. Cardiovascular Development in Zebrafish
2.1. Zebrafish Cardiac Morphogenesis
2.2. Cardiac Morphogenesis Phenotypes in Zebrafish
2.2.1. Congenital Heart Disease Modeled in Zebrafish
2.2.2. Genes That Cause CHD in Humans Cause Heart Malformation in Zebrafish
2.2.3. Non-Genetic Factors That Cause CHD in Humans Cause Heart Malformation in Zebrafish
2.2.4. Cardiomyopathy Modeled in Zebrafish
3. Experimental Approaches to Studying CHDs in Zebrafish
3.1. Hypothesis Driven/Reverse Genetics
3.1.1. Transgenesis
- (1)
- Spatial control of gene expression is achieved by engineering transgenes to have cell type specific promoter sequences driving gene expression. By varying the gene expressed, transgenes can label subcellular features, trace cell lineages, or even selectively kill a specific cell type. For example, the promoter for the cardiac myosin light chain 2 (aka, myl7), is expressed exclusively in CMs. Thus, CMs in Tg(cmlc2:GFP) zebrafish express the green fluorescent protein, GFP [111]. Tg(cmlc2:dsRed) zebrafish express red fluorescent protein dsRed [112]. Similarly, Tg(cmlc2:DTA) zebrafish express DTA (diphtheria toxin A) to selectively kill CMs [113]. Likewise, Tg(cmlc2:Cre-ERT2) CMs express the fusion protein of Cre recombinase and ERT2 [114], which is a tamoxifen-inducible portion of the estrogen receptor. In the presence of tamoxifen, Cre-ERT2 translocates to the nuclease where it recognizes and recombines loxP sites. The Cre-Lox system can be used for gene deletion or lineage tracing.
- (2)
- Temporal control of gene expression has been achieved using heat-shock protein promoters. For example, in Tg(Hsp70:sema3aa) zebrafish [115], when the organism is exposed to elevated temperature, Heat Shock Factor binds to the Hsp70 promoter to transiently activate transcription of the downstream gene sema3aa.
- (3)
- Overexpression: The functional consequences of over-activation of the gene of interest can be assessed by designing a transgenic construct where the promoter drives expression of the gene of interest. Expression level can be titrated by altering transgene copy number and promoter strength. This is particularly useful if the gene of interest has dominant activity.
- (4)
- Biosensors: Transgenic reporter lines can be made to express biosensors under the control of signaling pathway-responsive elements, making useful and reliable molecular tools to decipher the activation or inhibition of distinct cell signaling events. These biosensors are additionally, valuable resources for drug screening. For example, in Tg(tp1:EGFP) zebrafish, the Notch response element tp1 is upstream of the fluorescent protein EGFP such that cells with active Notch signaling express EGFP [116].
3.1.2. Morpholinos
- Efficacy:
- (1)
- Dose-dependent reduction in endogenous protein
- (2)
- Dose-dependent reduction in properly spliced transcript (splice-blocking MO only)
- (3)
- Dose-dependent reduction in tagged version of target protein
- Specificity:
- (1)
- MO sequence selection (BLAST)
- (2)
- Comparison to existing mutant homozygous for null allele
- (3)
- Co-injection with in vitro transcribed target RNA
- (4)
- Control morpholino (standard control, 5-base pair mismatch, and p53 MOs)
- (5)
- Multiple MOs targeting the same gene lead to similar, synergistic phenotypes
3.1.3. Genome Editing
- (1)
- Strategic gRNA design—Use computational methods to predict guide RNA specificity to the target site [142].
- (2)
- (3)
- For each desired genome editing process, generate at least 2 independent lines using different gRNAs.
- (4)
- Outcross to wild type to isolate desired mutation from non-linked off-target mutations.
4. Adult Functional Assays
5. Data Driven/Forward Approach
5.1. Mutagenesis
5.1.1. Forward Chemical Genetic Screens
5.1.2. Zebrafish in High Throughput Chemical Screens
5.1.3. Cardiotoxicity Screens
6. Future Directions
6.1. Characterizing Cardiac Phenotypes in Embryos
6.2. Adult Zebrafish Disease Model
6.3. Personalized Medicine
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Moran, A.E.; Roth, G.A.; Narula, J.; Mensah, G.A. 1990–2010 global cardiovascular disease atlas. Global heart 2014, 9, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Vos, T.; Barber, R.M.; Bell, B.; Bertozzi-Villa, A.; Biryukov, S.; Bolliger, I.; Charlson, F.; Davis, A.; Degenhardt, L.; Dicker, D.; et al. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: A systematic analysis for the global burden of disease study 2013. Lancet 2015, 386, 743–800. [Google Scholar]
- Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; Das, S.R.; de Ferranti, S.; Despres, J.P.; Fullerton, H.J.; et al. Heart disease and stroke statistics—2016 update: A report from the american heart association. Circulation 2015. [Google Scholar] [CrossRef] [PubMed]
- Teekakirikul, P.; Kelly, M.A.; Rehm, H.L.; Lakdawala, N.K.; Funke, B.H. Inherited cardiomyopathies: Molecular genetics and clinical genetic testing in the postgenomic era. J. Mol. Diagn. 2013, 15, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Moorman, A.F.; Christoffels, V.M. Cardiac chamber formation: Development, genes, and evolution. Physiol. Rev. 2003, 83, 1223–1267. [Google Scholar] [CrossRef] [PubMed]
- Fahed, A.C.; Gelb, B.D.; Seidman, J.G.; Seidman, C.E. Genetics of congenital heart disease the glass half empty. Circ. Res. 2013, 112, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Asnani, A.; Peterson, R.T. The zebrafish as a tool to identify novel therapies for human cardiovascular disease. Dis. Models Mech. 2014, 7, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Barros, T.P.; Alderton, W.K.; Reynolds, H.M.; Roach, A.G.; Berghmans, S. Zebrafish: An emerging technology for in vivo pharmacological assessment to identify potential safety liabilities in early drug discovery. Br. J. Pharmacol. 2008, 154, 1400–1413. [Google Scholar] [CrossRef] [PubMed]
- Delvecchio, C.; Tiefenbach, J.; Krause, H.M. The zebrafish: A powerful platform for in vivo, hts drug discovery. Assay Drug Dev. Technol. 2011, 9, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Stewart, A.M.; Gerlai, R.; Kalueff, A.V. Developing higher-throughput zebrafish screens for in-vivo cns drug discovery. Front. Behav. Neurosci. 2015, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Yozzo, K.L.; Isales, G.M.; Raftery, T.D.; Volz, D.C. High-content screening assay for identification of chemicals impacting cardiovascular function in zebrafish embryos. Environ. Sci. Technol. 2013, 47, 11302–11310. [Google Scholar] [CrossRef] [PubMed]
- Kitambi, S.S.; Nilsson, E.S.; Sekyrova, P.; Ibarra, C.; Tekeoh, G.N.; Andang, M.; Ernfors, P.; Uhlen, P. Small molecule screening platform for assessment of cardiovascular toxicity on adult zebrafish heart. BMC Physiol. 2012, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Kessler, M.; Rottbauer, W.; Just, S. Recent progress in the use of zebrafish for novel cardiac drug discovery. Expert Opin. Drug Discov. 2015, 10, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Ruzicka, L.; Bradford, Y.M.; Frazer, K.; Howe, D.G.; Paddock, H.; Ramachandran, S.; Singer, A.; Toro, S.; Van Slyke, C.E.; Eagle, A.E.; et al. Zfin, the zebrafish model organism database: Updates and new directions. Genesis 2015, 53, 498–509. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Stainier, D.Y. Zebrafish in the study of early cardiac development. Circ. Res. 2012, 110, 870–874. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The zebrafish book. A Guide for the Laboratory Use of Zebrafish (Danio Rerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- Stainier, D.Y.; Lee, R.K.; Fishman, M.C. Cardiovascular development in the zebrafish. I. Myocardial fate map and heart tube formation. Development 1993, 119, 31–40. [Google Scholar] [PubMed]
- Huang, C.J.; Tu, C.T.; Hsiao, C.D.; Hsieh, F.J.; Tsai, H.J. Germ-line transmission of a myocardium-specific gfp transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 2003, 228, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Jinn, S.W.; Beisl, D.; Mitchell, T.; Chen, J.N.; Stainier, D.Y.R. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 2005, 132, 5199–5209. [Google Scholar] [CrossRef] [PubMed]
- Perner, B.; Englert, C.; Bollig, F. The wilms tumor genes wt1a and wt1b control different steps during formation of the zebrafish pronephros. Dev. Biol. 2007, 309, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Long, Q.M.; Meng, A.M.; Wang, H.; Jessen, J.R.; Farrell, M.J.; Lin, S. Gata-1 expression pattern can be recapitulated in living transgenic zebrafish using gfp reporter gene. Development 1997, 124, 4105–4111. [Google Scholar] [PubMed]
- Bang, A.; Gronkjaer, P.; Malte, H. Individual variation in the rate of oxygen consumption by zebrafish embryos. J. Fish Biol. 2004, 64, 1285–1296. [Google Scholar] [CrossRef]
- Strecker, R.; Seiler, T.B.; Hollert, H.; Braunbeck, T. Oxygen requirements of zebrafish (Danio rerio) embryos in embryo toxicity tests with environmental samples. Comp. Biochem. Phys. C 2011, 153, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.N.; Haffter, P.; Odenthal, J.; Vogelsang, E.; Brand, M.; van Eeden, F.J.; Furutani-Seiki, M.; Granato, M.; Hammerschmidt, M.; Heisenberg, C.P.; et al. Mutations affecting the cardiovascular system and other internal organs in zebrafish. Development 1996, 123, 293–302. [Google Scholar] [PubMed]
- Stainier, D.Y.; Fouquet, B.; Chen, J.N.; Warren, K.S.; Weinstein, B.M.; Meiler, S.E.; Mohideen, M.A.; Neuhauss, S.C.; Solnica-Krezel, L.; Schier, A.F.; et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development 1996, 123, 285–292. [Google Scholar] [PubMed]
- Sehnert, A.J.; Huq, A.; Weinstein, B.M.; Walker, C.; Fishman, M.; Stainier, D.Y. Cardiac troponin t is essential in sarcomere assembly and cardiac contractility. Nat. Genet. 2002, 31, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Samsa, L.A.; Yang, B.; Liu, J. Embryonic cardiac chamber maturation: Trabeculation, conduction, and cardiomyocyte proliferation. Am. J. Med. Genet. C 2013, 163C, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Sedmera, D.; Pexieder, T.; Vuillemin, M.; Thompson, R.P.; Anderson, R.H. Developmental patterning of the myocardium. Anat. Rec. 2000, 258, 319–337. [Google Scholar] [CrossRef]
- Kirby, M.L. Cardiac Development; Oxford University Press: New York, NY, USA, 2007. [Google Scholar]
- Sabaliauskas, N.A.; Foutz, C.A.; Mest, J.R.; Budgeon, L.R.; Sidor, A.T.; Gershenson, J.A.; Joshi, S.B.; Cheng, K.C. High-throughput zebrafish histology. Methods 2006, 39, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Tsao-Wu, G.S.; Weber, C.H.; Budgeon, L.R.; Cheng, K.C. Agarose-embedded tissue arrays for histologic and genetic analysis. BioTechniques 1998, 25, 614–618. [Google Scholar] [PubMed]
- Keegan, B.R.; Meyer, D.; Yelon, D. Organization of cardiac chamber progenitors in the zebrafish blastula. Development 2004, 131, 3081–3091. [Google Scholar] [CrossRef] [PubMed]
- Misfeldt, A.M.; Boyle, S.C.; Tompkins, K.L.; Bautch, V.L.; Labosky, P.A.; Baldwin, H.S. Endocardial cells are a distinct endothelial lineage derived from Flk1+ multipotent cardiovascular progenitors. Dev. Biol. 2009, 333, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Milgrom-Hoffman, M.; Harrelson, Z.; Ferrara, N.; Zelzer, E.; Evans, S.M.; Tzahor, E. The heart endocardium is derived from vascular endothelial progenitors. Development 2011, 138, 4777–4787. [Google Scholar] [CrossRef] [PubMed]
- Bussmann, J.; Bakkers, J.; Schulte-Merker, S. Early endocardial morphogenesis requires Scl/Tal1. Plos Genet. 2007, 3, 1425–1437. [Google Scholar] [CrossRef] [PubMed]
- De la Pompa, J.L.; Timmerman, L.A.; Takimoto, H.; Yoshida, H.; Elia, A.J.; Samper, E.; Potter, J.; Wakeham, A.; Marengere, L.; Langille, B.L.; et al. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature 1998, 392, 182–186. [Google Scholar] [PubMed]
- Trinh, L.A.; Stainier, D.Y.R. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev. Cell 2004, 6, 371–382. [Google Scholar] [CrossRef]
- Yelon, D.; Horne, S.A.; Stainier, D.Y. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol. 1999, 214, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Palencia-Desai, S.; Rost, M.S.; Schumacher, J.A.; Ton, Q.V.; Craig, M.P.; Baltrunaite, K.; Koenig, A.L.; Wang, J.; Poss, K.D.; Chi, N.C.; et al. Myocardium and bmp signaling are required for endocardial differentiation. Development 2015, 142, 2304–2315. [Google Scholar] [CrossRef] [PubMed]
- Holtzman, N.G.; Schoenebeck, J.J.; Tsai, H.J.; Yelon, D. Endocardium is necessary for cardiomyocyte movement during heart tube assembly. Development 2007, 134, 2379–2386. [Google Scholar] [CrossRef] [PubMed]
- Garavito-Aguilar, Z.V.; Riley, H.E.; Yelon, D. Hand2 ensures an appropriate environment for cardiac fusion by limiting fibronectin function. Development 2010, 137, 3215–3220. [Google Scholar] [CrossRef] [PubMed]
- Rohr, S.; Otten, C.; Abdelilah-Seyfried, S. Asymmetric involution of the myocardial field drives heart tube formation in zebrafish. Circ. Res. 2008, 102, E12–E19. [Google Scholar] [CrossRef] [PubMed]
- Lazic, S.; Scott, I.C. Mef2cb regulates late myocardial cell addition from a second heart field-like population of progenitors in zebrafish. Dev. Biol. 2011, 354, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Cashman, T.J.; Nevis, K.R.; Obregon, P.; Carney, S.A.; Liu, Y.; Gu, A.; Mosimann, C.; Sondalle, S.; Peterson, R.E.; et al. Latent TGF-β binding protein 3 identifies a second heart field in zebrafish. Nature 2011, 474, 645–648. [Google Scholar] [CrossRef] [PubMed]
- Hami, D.; Grimes, A.C.; Tsai, H.J.; Kirby, M.L. Zebrafish cardiac development requires a conserved secondary heart field. Development 2011, 138, 2389–2398. [Google Scholar] [CrossRef] [PubMed]
- De Pater, E.; Clijsters, L.; Marques, S.R.; Lin, Y.F.; Garavito-Aguilar, Z.V.; Yelon, D.; Bakkers, J. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development 2009, 136, 1633–1641. [Google Scholar] [CrossRef] [PubMed]
- Baker, K.; Holtzman, N.G.; Burdine, R.D. Direct and indirect roles for nodal signaling in two axis conversions during asymmetric morphogenesis of the zebrafish heart. Proc. Natl. Acad. Sci. USA 2008, 105, 13924–13929. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.N.; vanEeden, F.J.M.; Warren, K.S.; Chin, A.; NussleinVolhard, C.; Haffter, P.; Fishman, M.C. Left-right pattern of cardiac bmp4 may drive asymmetry of the heart in zebrafish. Development 1997, 124, 4373–4382. [Google Scholar] [PubMed]
- Beis, D.; Bartman, T.; Jin, S.W.; Scott, I.C.; D’Amico, L.A.; Ober, E.A.; Verkade, H.; Frantsve, J.; Field, H.A.; Wehman, A.; et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development 2005, 132, 4193–4204. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.T.; Bartman, T. Analysis of heart valve development in larval zebrafish. Dev. Dyn. 2009, 238, 1796–1802. [Google Scholar] [CrossRef] [PubMed]
- Grimes, A.C.; Kirby, M.L. The outflow tract of the heart in fishes: Anatomy, genes and evolution. J. Fish Biol. 2009, 74, 983–1036. [Google Scholar] [CrossRef] [PubMed]
- Hofsteen, P.; Plavicki, J.; Johnson, S.D.; Peterson, R.E.; Heideman, W. Sox9b is required for epicardium formation and plays a role in tcdd-induced heart malformation in zebrafish. Mol. Pharmacol. 2013, 84, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Bressan, M.; Hassel, D.; Huisken, J.; Staudt, D.; Kikuchi, K.; Poss, K.D.; Mikawa, T.; Stainier, D.Y. A dual role for erbb2 signaling in cardiac trabeculation. Development 2010, 137, 3867–3875. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; von Gise, A.; Ma, Q.; Rivera-Feliciano, J.; Pu, W.T. Nkx2-5- and lsl1-expressing cardiac progenitors contribute to proepicardium. Biochem. Biophys. Res. Commun. 2008, 375, 450–453. [Google Scholar] [CrossRef] [PubMed]
- Serluca, F.C. Development of the proepicardial organ in the zebrafish. Dev. Biol. 2008, 315, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Peralta, M.; Steed, E.; Harlepp, S.; Gonzalez-Rosa, J.M.; Monduc, F.; Ariza-Cosano, A.; Cortes, A.; Rayon, T.; Gomez-Skarmeta, J.L.; Zapata, A.; et al. Heartbeat-driven pericardiac fluid forces contribute to epicardium morphogenesis. Curr. Biol. 2013, 23, 1726–1735. [Google Scholar] [CrossRef] [PubMed]
- Plavicki, J.S.; Hofsteen, P.; Yue, M.S.; Lanham, K.A.; Peterson, R.E.; Heideman, W. Multiple modes of proepicardial cell migration require heartbeat. BMC Dev. Biol. 2014, 14. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Gupta, V.; Wang, J.H.; Holdway, J.E.; Wills, A.A.; Fang, Y.; Poss, K.D. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development 2011, 138, 2895–2902. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Rosa, J.M.; Peralta, M.; Mercader, N. Pan-epicardial lineage tracing reveals that epicardium derived cells give rise to myofibroblasts and perivascular cells during zebrafish heart regeneration. Dev. Biol. 2012, 370, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Peralta, M.; Gonzalez-Rosa, J.M.; Marques, I.J.; Mercader, N. The epicardium in the embryonic and adult zebrafish. J. Dev. Biol. 2014, 2, 101–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Icardo, J.M.; Fernandez-Teran, A. Morphologic study of ventricular trabeculation in the embryonic chick heart. Acta Anat. 1987, 130, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Samsa, L.A.; Givens, C.; Tzima, E.; Stainier, D.Y.; Qian, L.; Liu, J. Cardiac contraction activates endocardial notch signaling to modulate chamber maturation in zebrafish. Development 2015, 142, 4080–4091. [Google Scholar] [CrossRef] [PubMed]
- Staudt, D.W.; Liu, J.D.; Thorn, K.S.; Stuurman, N.; Liebling, M.; Stainier, D.Y.R. High-resolution imaging of cardiomyocyte behavior reveals two distinct steps in ventricular trabeculation. Development 2014, 141, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Scherz, P.J.; Huisken, J.; Sahai-Hernandez, P.; Stainier, D.Y.R. High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development 2008, 135, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Timmerman, L.A.; Grego-Bessa, J.; Raya, A.; Bertran, E.; Perez-Pomares, J.M.; Diez, J.; Aranda, S.; Palomo, S.; McCormick, F.; Izpisua-Belmonte, J.C.; et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004, 18, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Singleman, C.; Holtzman, N.G. Analysis of postembryonic heart development and maturation in the zebrafish, Danio rerio. Dev. Dyn. 2012, 241, 1993–2004. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Poss, K.D. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature 2012, 484, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.R.M.; Bussmann, J.; Huang, Y.; Zhao, L.; Osorio, A.; Burns, C.G.; Burns, C.E.; Sucov, H.M.; Siekmann, A.F.; Lien, C.L. Chemokine-guided angiogenesis directs coronary vasculature formation in zebrafish. Dev. Cell 2015, 33, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Gemberling, M.; Karra, R.; Rosenfeld, G.E.; Evans, T.; Poss, K.D. An injury-responsive gata4 program shapes the zebrafish cardiac ventricle. Curr. Biol. 2013, 23, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Miura, G.I.; Yelon, D. A guide to analysis of cardiac phenotypes in the zebrafish embryo. Methods Cell Biol. 2011, 101, 161–180. [Google Scholar] [PubMed]
- Granados-Riveron, J.T.; Brook, J.D. The impact of mechanical forces in heart morphogenesis. Circ: Cardiovasc. Genet. 2012, 5, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, S.E.; Butcher, J.T.; Yalcin, H.C. Mechanical regulation of cardiac development. Front. Physiol. 2014, 5, 318. [Google Scholar] [CrossRef] [PubMed]
- Hove, J.R.; Koster, R.W.; Forouhar, A.S.; Acevedo-Bolton, G.; Fraser, S.E.; Gharib, M. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature 2003, 421, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Sedmera, D.; Pexieder, T.; Rychterova, V.; Hu, N.; Clark, E.B. Remodeling of chick embryonic ventricular myoarchitecture under experimentally changed loading conditions. Anat. Rec. 1999, 254, 238–252. [Google Scholar] [CrossRef]
- Reckova, M.; Rosengarten, C.; deAlmeida, A.; Stanley, C.P.; Wessels, A.; Gourdie, R.G.; Thompson, R.P.; Sedmera, D. Hemodynamics is a key epigenetic factor in development of the cardiac conduction system. Circ. Res. 2003, 93, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Peshkovsky, C.; Totong, R.; Yelon, D. Dependence of cardiac trabeculation on neuregulin signaling and blood flow in zebrafish. Dev. Dyn. 2011, 240, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Steed, E.; Boselli, F.; Vermot, J. Hemodynamics driven cardiac valve morphogenesis. Biochim. Biophys. Acta 2015. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.Y.; Chang, W.T.; Lai, Y.C.; Liau, I. Toward functional screening of cardioactive and cardiotoxic drugs with zebrafish in vivo using pseudodynamic three-dimensional imaging. Anal. Chem. 2014, 86, 2213–2220. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.C.; Monte, A.; Cook, J.M.; Kabir, M.S.; Peterson, K.P. Zebrafish heart failure models for the evaluation of chemical probes and drugs. Assay Drug Dev. Technol. 2013, 11, 561–572. [Google Scholar] [CrossRef] [PubMed]
- Milan, D.J.; Peterson, T.A.; Ruskin, J.N.; Peterson, R.T.; MacRae, C.A. Drugs that induce repolarization abnormalities cause bradycardia in zebrafish. Circulation 2003, 107, 1355–1358. [Google Scholar] [CrossRef] [PubMed]
- Kopf, P.G.; Walker, M.K. Overview of developmental heart defects by dioxins, pcbs, and pesticides. J. Environ. Sci. Health C 2009, 27, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Kartiko, S.; Finnell, R.H. Importance of gene-environment interactions in the etiology of selected birth defects. Clin. Genet. 2009, 75, 409–423. [Google Scholar] [CrossRef] [PubMed]
- Incardona, J.P.; Linbo, T.L.; Scholz, N.L. Cardiac toxicity of 5-ring polycyclic aromatic hydrocarbons is differentially dependent on the aryl hydrocarbon receptor 2 isoform during zebrafish development. Toxicol. Appl. Pharm. 2011, 257, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Hicken, C.E.; Linbo, T.L.; Baldwin, D.H.; Willis, M.L.; Myers, M.S.; Holland, L.; Larsen, M.; Stekoll, M.S.; Rice, S.D.; Collier, T.K.; et al. Sublethal exposure to crude oil during embryonic development alters cardiac morphology and reduces aerobic capacity in adult fish. Proc. Natl Acad Sci USA 2011, 108, 7086–7090. [Google Scholar] [CrossRef] [PubMed]
- Incardona, J.P.; Day, H.L.; Collier, T.K.; Scholz, N.L. Developmental toxicity of 4-ring polycyclic aromatic hydrocarbons in zebrafish is differentially dependent on ah receptor isoforms and hepatic cytochrome p4501a metabolism. Toxicol. Appl. Pharm. 2006, 217, 308–321. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.R.; Clark, B.W.; Garner, L.V.; Di Giulio, R.T. Zebrafish cardiotoxicity: The effects of CYP1A inhibition and AHR2 knockdown following exposure to weak aryl hydrocarbon receptor agonists. Environ. Sci. Pollut. Res. Int. 2015, 22, 8329–8338. [Google Scholar] [CrossRef] [PubMed]
- Garner, L.V.T.; Brown, D.R.; Di Giulio, R.T. Knockdown of ahr1a but not ahr1b exacerbates pah and PCB-126 toxicity in zebrafish (Danio rerio) embryos. Aquat. Toxicol. 2013, 142, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Padilla, S.; Corum, D.; Padnos, B.; Hunter, D.L.; Beam, A.; Houck, K.A.; Sipes, N.; Kleinstreuer, N.; Knudsen, T.; Dix, D.J.; et al. Zebrafish developmental screening of the toxcast phase i chemical library. Reprod. Toxicol. 2012, 33, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Sipes, N.S.; Padilla, S.; Knudsen, T.B. Zebrafish: As an integrative model for twenty-first century toxicity testing. Birth Defects Res. C Embryo Today 2011, 93, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Fahed, A.C.; Roberts, A.E.; Mital, S.; Lakdawala, N.K. Heart failure in congenital heart disease: A confluence of acquired and congenital. Heart Fail. Clin. 2014, 10, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.L.; Meiler, S.E.; Zhong, T.P.; Mohideen, M.; Crossley, D.A.; Burggren, W.W.; Fishman, M.C. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat. Genet. 2002, 30, 205–209. [Google Scholar] [PubMed]
- Berdougo, E.; Coleman, H.; Lee, D.H.; Stainier, D.Y.R.; Yelon, D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development 2003, 130, 6121–6129. [Google Scholar] [CrossRef] [PubMed]
- Alday, A.; Alonso, H.; Gallego, M.; Urrutia, J.; Letamendia, A.; Callol, C.; Casis, O. Ionic channels underlying the ventricular action potential in zebrafish embryo. Pharmacol. Res. 2014, 84, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Turer, A.T.; Malloy, C.R.; Newgard, C.B.; Podgoreanu, M.V. Energetics and metabolism in the failing heart: Important but poorly understood. Curr. Opin. Clin. Nutr. Metab. Care 2010, 13, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.; Basson, C.T. The molecular genetics of congenital heart disease: A review of recent developments. Curr. Opin. Cardiol. 2010, 25, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Sehnert, A.J.; Stainier, D.Y.R. A window to the heart: Can zebrafish mutants help us understand heart disease in humans? Trends Genet. 2002, 18, 491–494. [Google Scholar] [CrossRef]
- Irion, U.; Krauss, J.; Nusslein-Volhard, C. Precise and efficient genome editing in zebrafish using the crispr/cas9 system. Development 2014, 141, 4827–4830. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, P.R.; Campbell, J.M.; Clark, K.J.; Ekker, S.C. The crispr system-keeping zebrafish gene targeting fresh. Zebrafish 2013, 10, 116–118. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, R.N.; Jopling, C.; van Eeden, F.J.M. Zebrafish as a model of cardiac disease. Prog. Mol. Biol. Transl. 2014, 124, 65–91. [Google Scholar]
- Yu, C.; Zhang, Y.G.; Yao, S.H.; Wei, Y.Q. A pcr based protocol for detecting indel mutations induced by talens and crispr/cas9 in zebrafish. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.Y.; Fu, Y.F.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.R.J.; Joung, J.K. Efficient genome editing in zebrafish using a crispr-cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.N.; Davey, C.F.; Whitebirch, A.C.; Miller, A.C.; Moens, C.B. Rapid reverse genetic screening using crispr in zebrafish. Nat. Methods 2015, 12, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Zhu, Z.; Lin, S.; Zhang, B. Reverse genetic approaches in zebrafish. Yi Chuan Xue Bao 2012, 39, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Lawson, N.D.; Wolfe, S.A. Forward and reverse genetic approaches for the analysis of vertebrate development in the zebrafish. Dev. Cell 2011, 21, 48–64. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. Corrigendum: The zebrafish reference genome sequence and its relationship to the human genome. Nature 2014, 505, 248. [Google Scholar] [CrossRef]
- Kimura, Y.; Hisano, Y.; Kawahara, A.; Higashijima, S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by crispr/cas9-mediated genome engineering. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Auer, T.O.; Del Bene, F. Crispr/cas9 and talen-mediated knock-in approaches in zebrafish. Methods 2014, 69, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Huttner, I.G.; Trivedi, G.; Jacoby, A.; Mann, S.A.; Vandenberg, J.I.; Fatkin, D. A transgenic zebrafish model of a human cardiac sodium channel mutation exhibits bradycardia, conduction-system abnormalities and early death. J. Mol. Cell. Cardiol. 2013, 61, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.J.; Urban, M.D.; Skuster, K.J.; Ekker, S.C. Transgenic zebrafish using transposable elements. Methods Cell Biol. 2011, 104, 137–149. [Google Scholar] [PubMed]
- Huang, C.J.; Tu, C.T.; Hsiao, C.D.; Hsieh, F.J.; Tsai, H.J. Germ-line transmission of a myocardium-specific gfp transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 2003, 228, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, S.C.; Lahvic, J.; Francescatto, L.; McLeod, J.J.; Burgess, S.M.; Tombes, R.M. CaMK-II activation is essential for zebrafish inner ear development and acts through δ-notch signaling. Dev. Biol. 2013, 381, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Panakova, D.; Kikuchi, K.; Holdway, J.E.; Gemberling, M.; Burris, J.S.; Singh, S.P.; Dickson, A.L.; Lin, Y.F.; Sabeh, M.K.; et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 2011, 138, 3421–3430. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Holdway, J.E.; Werdich, A.A.; Anderson, R.M.; Fang, Y.; Egnaczyk, G.F.; Evans, T.; Macrae, C.A.; Stainier, D.Y.; Poss, K.D. Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature 2010, 464, 601–605. [Google Scholar] [CrossRef] [PubMed]
- Shoji, W.; Isogai, S.; Sato-Maeda, M.; Obinata, M.; Kuwada, J.Y. Semaphorin3a1 regulates angioblast migration and vascular development in zebrafish embryos. Development 2003, 130, 3227–3236. [Google Scholar] [CrossRef] [PubMed]
- Parsons, M.J.; Pisharath, H.; Yusuff, S.; Moore, J.C.; Siekmann, A.F.; Lawson, N.; Leach, S.D. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech. Dev. 2009, 126, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Nasevicius, A.; Ekker, S.C. Effective targeted gene “knockdown” in zebrafish. Nat. Genet. 2000, 26, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Eisen, J.S.; Smith, J.C. Controlling morpholino experiments: Don’t stop making antisense. Development 2008, 135, 1735–1743. [Google Scholar] [CrossRef] [PubMed]
- Bill, B.R.; Petzold, A.M.; Clark, K.J.; Schimmenti, L.A.; Ekker, S.C. A primer for morpholino use in zebrafish. Zebrafish 2009, 6, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Prasch, A.L.; Carney, S.A.; Heideman, W.; Peterson, R.E. Morpholino knockdown of ahr2 in the zebrafish embryo protects against tcdd developmental toxicity. Toxicol. Sci. 2003, 72, 366. [Google Scholar]
- Van Tiem, L.A.; Di Giulio, R.T. AHR2 knockdown prevents PAH-mediated cardiac toxicity and XRE- and ARE-associated gene induction in zebrafish (Danio rerio). Toxicol. Appl. Pharm. 2011, 254, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Matson, C.W.; Clark, B.W.; Jenny, M.J.; Fleming, C.R.; Hahn, M.E.; Di Giulio, R.T. Development of the morpholino gene knockdown technique in Fundulus heteroclitus: A tool for studying molecular mechanisms in an established environmental model. Aquat. Toxicol. 2008, 87, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Clark, B.W.; Matson, C.W.; Jung, D.; Di Giulio, R.T. AHR2 mediates cardiac teratogenesis of polycyclic aromatic hydrocarbons and PCB-126 in atlantic killifish (fundulus heteroclitus). Aquat. Toxicol. 2010, 99, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Schulte-Merker, S.; Stainier, D.Y. Out with the old, in with the new: Reassessing morpholino knockdowns in light of genome editing technology. Development 2014, 141, 3103–3104. [Google Scholar] [CrossRef] [PubMed]
- Kok, F.O.; Shin, M.; Ni, C.W.; Gupta, A.; Grosse, A.S.; van Impel, A.; Kirchmaier, B.C.; Peterson-Maduro, J.; Kourkoulis, G.; Male, I.; et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell 2015, 32, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.; Kontarakis, Z.; Gerri, C.; Nolte, H.; Holper, S.; Kruger, M.; Stainier, D.Y. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 2015, 524, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Summerton, J.; Weller, D. Morpholino antisense oligomers: Design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997, 7, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; De Robertis, E.M.; Wallingford, J.B.; Niehrs, C. Morpholinos: Antisense and sensibility. Dev. Cell 2015, 35, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, B.B.; Ren, Y.G.; Gu, S.Y.; Xiang, Y.H.; Huang, C.; Du, J.L. Intron targeting-mediated and endogenous gene integrity-maintaining knockin in zebrafish using the crispr/cas9 system. Cell. Res. 2015, 25, 634–637. [Google Scholar] [CrossRef] [PubMed]
- Varshney, G.K.; Pei, W.H.; LaFave, M.C.; Idol, J.; Xu, L.S.; Gallardo, V.; Carrington, B.; Bishop, K.; Jones, M.; Li, M.Y.; et al. High-throughput gene targeting and phenotyping in zebrafish using crispr/cas9. Genome Res. 2015, 25, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Straimer, J.; Lee, M.C.; Lee, A.H.; Zeitler, B.; Williams, A.E.; Pearl, J.R.; Zhang, L.; Rebar, E.J.; Gregory, P.D.; Llinas, M.; et al. Site-specific genome editing in plasmodium falciparum using engineered zinc-finger nucleases. Nat. Methods 2012, 9, 993–998. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Luo, D.; Lei, Y.; Hu, W.; Zhao, H.; Cheng, C.H. A highly effective talen-mediated approach for targeted gene disruption in xenopus tropicalis and zebrafish. Methods 2014, 69, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Ablain, J.; Durand, E.M.; Yang, S.; Zhou, Y.; Zon, L.I. A crispr/cas9 vector system for tissue-specific gene disruption in zebrafish. Dev. Cell 2015, 32, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Hruscha, A.; Schmid, B. Generation of zebrafish models by crispr/cas9 genome editing. Methods Mol. Biol 2015, 1254, 341–350. [Google Scholar] [PubMed]
- Hruscha, A.; Krawitz, P.; Rechenberg, A.; Heinrich, V.; Hecht, J.; Haass, C.; Schmid, B. Efficient crispr/cas9 genome editing with low off-target effects in zebrafish. Development 2013, 140, 4982–4987. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Pandey, M.; Kahler, J.F.; Loshakov, A.; Harris, B.; Dagur, P.K.; Mo, Y.Y.; Simonds, W.F. Improving the specificity and efficacy of crispr/cas9 and grna through target specific DNA reporter. J. Biotechnol. 2014, 189, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Kim, S.; Kim, Y.; Kweon, J.; Kim, H.S.; Bae, S.; Kim, J.S. Analysis of off-target effects of crispr/cas-derived rna-guided endonucleases and nickases. Genome Res. 2014, 24, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Tee, L.Y.; Wang, X.G.; Huang, Q.S.; Yang, S.H. Off-target effects in crispr/cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 2015, 4, e264. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, R.; von Kalle, C.; Schmidt, M. Mapping the precision of genome editing. Nat. Biotechnol. 2015, 33, 150–152. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.I.; Suresh, B.; Kim, H.; Ramakrishna, S. CRISPR/Cas9 system as an innovative genetic engineering tool: Enhancements in sequence specificity and delivery methods. BBA-Rev. Cancer 2015, 1856, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Koo, T.; Lee, J.; Kim, J.S. Measuring and reducing off-target activities of programmable nucleases including crispr-cas9. Mol. Cells 2015, 38, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by rna-guided crispr cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Guilinger, J.P.; Thompson, D.B.; Liu, D.R. Fusion of catalytically inactive cas9 to foki nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014, 32, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Widmer, S.; Moore, F.B.G.; Bagatto, B. The effects of chronic developmental hypoxia on swimming performance in zebrafish. J. Fish. Biol 2006, 69, 1885–1891. [Google Scholar] [CrossRef]
- Sfakianakis, D.G.; Leris, I.; Kentouri, M. Effect of developmental temperature on swimming performance of zebrafish (Danio rerio) juveniles. Environ. Biol. Fish. 2011, 90, 421–427. [Google Scholar] [CrossRef]
- Barrionuevo, W.R.; Burggren, W.W. O2 consumption and heart rate in developing zebrafish (Danio rerio): Influence of temperature and ambient O2. Am. J. Physiol. Regul. Integr. Comp. Physiol. 1999, 276, R505–R513. [Google Scholar]
- Abdallah, S.J.; Thomas, B.S.; Jonz, M.G. Aquatic surface respiration and swimming behaviour in adult and developing zebrafish exposed to hypoxia. J. Exp. Biol. 2015, 218, 1777–1786. [Google Scholar] [CrossRef] [PubMed]
- Ram, R.; Mickelsen, D.M.; Theodoropoulos, C.; Blaxall, B.C. New approaches in small animal echocardiography: Imaging the sounds of silence. Am. J. Physiol. Heart C 2011, 301, H1765–H1780. [Google Scholar] [CrossRef] [PubMed]
- Abduch, M.C.D.; Assad, R.S.; Mathias, W.; Aiello, V.D. The echocardiography in the cardiovascular laboratory: A guide to research with animals. Arq. Bras. Cardiol. 2014, 102, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Hein, S.J.; Lehmann, L.H.; Kossack, M.; Juergensen, L.; Fuchs, D.; Katus, H.A.; Hassel, D. Advanced echocardiography in adult zebrafish reveals delayed recovery of heart function after myocardial cryoinjury. PLoS ONE 2015, 10, e0122665. [Google Scholar] [CrossRef] [PubMed]
- Marcaletti, S.; Thomas, C.; Feige, J.N. Exercise performance tests in mice. Curr. Protocols Mouse Biol. 2011, 1, 141–154. [Google Scholar]
- Budick, S.A.; O’malley, D.M. Locomotor repertoire of the larval zebrafish: Swimming, turning and prey capture. J. Exp. Biol. 2000, 203, 2565–2579. [Google Scholar] [PubMed]
- Drucker, E.G. The use of gait transition speed in comparative studies of fish locomotion. Am. Zool 1996, 36, 555–566. [Google Scholar] [CrossRef]
- Dalziel, A.C.; Schulte, P.M. Correlates of prolonged swimming performance in f2 hybrids of migratory and non-migratory threespine stickleback. J. Exp. Biol. 2012, 215, 3587–3596. [Google Scholar] [CrossRef] [PubMed]
- Brett, J.R. The respiratory metabolism and swimming performance of young sockeye salmon. J. Fish. Res. Board Can. 1964, 21, 1183–1226. [Google Scholar] [CrossRef]
- Jain, K.E.; Hamilton, J.C.; Farrell, A.P. Use of a ramp velocity test to measure critical swimming speed in rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. A 1997, 117, 441–444. [Google Scholar] [CrossRef]
- Claireaux, G.; McKenzie, D.J.; Genge, A.G.; Chatelier, A.; Aubin, J.; Farrell, A.P. Linking swimming performance, cardiac pumping ability and cardiac anatomy in rainbow trout. J. Exp. Biol 2005, 208, 1775–1784. [Google Scholar] [CrossRef] [PubMed]
- Kolok, A.S.; Farrell, A.P. Individual variation in the swimming performance and cardiac-performance of northern squawfish, ptychocheilus-oregonensis. Physiol. Zool. 1994, 67, 706–722. [Google Scholar] [CrossRef]
- Gallaugher, P.E.; Thorarensen, H.; Kiessling, A.; Farrell, A.P. Effects of high intensity exercise training on cardiovascular function, oxygen uptake, internal oxygen transport and osmotic balance in chinook salmon (Oncorhynchus tshawytscha) during critical speed swimming. J. Exp. Biol. 2001, 204, 2861–2872. [Google Scholar] [PubMed]
- Palstra, A.P.; Tudorache, C.; Rovira, M.; Brittijn, S.A.; Burgerhout, E.; van den Thillart, G.E.E.J.M.; Spaink, H.P.; Planas, J.V. Establishing zebrafish as a novel exercise model: Swimming economy, swimming-enhanced growth and muscle growth marker gene expression. PLoS ONE 2010, 5, e14483. [Google Scholar] [CrossRef] [PubMed]
- Plaut, I.; Gordon, M.S. Swimming metabolism of wild-type and cloned zebrafish brachydanio-rerio. J. Exp. Biol. 1994, 194, 209–223. [Google Scholar] [PubMed]
- Pelster, B.; Sanger, A.M.; Siegele, M.; Schwerte, T. Influence of swim training on cardiac activity, tissue capillarization, and mitochondrial density in muscle tissue of zebrafish larvae. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003, 285, R339–R347. [Google Scholar] [CrossRef] [PubMed]
- Lieschke, G.J.; Currie, P.D. Animal models of human disease: Zebrafish swim into view. Nat. Rev. Genet. 2007, 8, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, J.R.; Jobin, C. Think small: Zebrafish as a model system of human pathology. J. Biomed. Biotechnol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Haffter, P.; Granato, M.; Brand, M.; Mullins, M.C.; Hammerschmidt, M.; Kane, D.A.; Odenthal, J.; van Eeden, F.J.; Jiang, Y.J.; Heisenberg, C.P.; et al. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 1996, 123, 1–36. [Google Scholar] [PubMed]
- Knapik, E.W. Enu mutagenesis in zebrafish—from genes to complex diseases. Mamm. Genome 2000, 11, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Draper, B.W.; McCallum, C.M.; Stout, J.L.; Slade, A.J.; Moens, C.B. A high-throughput method for identifying n-ethyl-n-nitrosourea (enu)-induced point mutations in zebrafish. Method Cell. Biol. 2004, 77, 91–112. [Google Scholar]
- Hrabe de Angelis, M.; Balling, R. Large scale enu screens in the mouse: Genetics meets genomics. Mutat. Res. 1998, 400, 25–32. [Google Scholar] [CrossRef]
- Schneeberger, K. Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat. Rev. Genet. 2014, 15, 662–676. [Google Scholar] [CrossRef] [PubMed]
- Obholzer, N.; Swinburne, I.A.; Schwab, E.; Nechiporuk, A.V.; Nicolson, T.; Megason, S.G. Rapid positional cloning of zebrafish mutations by linkage and homozygosity mapping using whole-genome sequencing. Development 2012, 139, 4280–4290. [Google Scholar] [CrossRef] [PubMed]
- Henke, K.; Bowen, M.E.; Harris, M.P. Perspectives for identification of mutations in the zebrafish: Making use of next-generation sequencing technologies for forward genetic approaches. Methods 2013, 62, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Driever, W.; Solnica-Krezel, L.; Schier, A.F.; Neuhauss, S.C.; Malicki, J.; Stemple, D.L.; Stainier, D.Y.; Zwartkruis, F.; Abdelilah, S.; Rangini, Z.; et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 1996, 123, 37–46. [Google Scholar] [PubMed]
- Sams-Dodd, F. Drug discovery: Selecting the optimal approach. Drug Discov. Today 2006, 11, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Zon, L.I.; Peterson, R.T. In vivo drug discovery in the zebrafish. Nature reviews. Drug Discov. 2005, 4, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Letamendia, A.; Quevedo, C.; Ibarbia, I.; Virto, J.M.; Holgado, O.; Diez, M.; Belmonte, J.C.I.; Callol-Massot, C. Development and validation of an automated high-throughput system for zebrafish in vivo screenings. PLoS ONE 2012, 7, e36690. [Google Scholar] [CrossRef] [PubMed]
- Burns, C.G.; Milan, D.J.; Grande, E.J.; Rottbauer, W.; MacRae, C.A.; Fishman, M.C. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat. Chem. Biol. 2005, 1, 263–264. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Doberstein, S.K. Hts technologies in biopharmaceutical discovery. Drug Discov. Today 2006, 11, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Macrae, C.A. Cardiac arrhythmia: In vivo screening in the zebrafish to overcome complexity in drug discovery. Expert Opin. Drug Discov. 2010, 5, 619–632. [Google Scholar] [CrossRef] [PubMed]
- Bowman, T.V.; Zon, L.I. Swimming into the future of drug discovery: In vivo chemical screens in zebrafish. ACS Chem. Biol. 2010, 5, 159–161. [Google Scholar] [CrossRef] [PubMed]
- Spomer, W.; Pfriem, A.; Alshut, R.; Just, S.; Pylatiuk, C. High-throughput screening of zebrafish embryos using automated heart detection and imaging. J. Lab. Autom. 2012, 17, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.T.; Shaw, S.Y.; Peterson, T.A.; Milan, D.J.; Zhong, T.P.; Schreiber, S.L.; MacRae, C.A.; Fishman, M.C. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat. Biotechnol. 2004, 22, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Zhong, T.P.; Rosenberg, M.; Mohideen, M.A.; Weinstein, B.; Fishman, M.C. Gridlock, an HLH gene required for assembly of the aorta in zebrafish. Science 2000, 287, 1820–1824. [Google Scholar] [CrossRef] [PubMed]
- Kopp, R.; Schwerte, T.; Pelster, B. Cardiac performance in the zebrafish breakdance mutant. J. Exp. Biol 2005, 208, 2123–2134. [Google Scholar] [CrossRef] [PubMed]
- Peal, D.S.; Mills, R.W.; Lynch, S.N.; Mosley, J.M.; Lim, E.; Ellinor, P.T.; January, C.T.; Peterson, R.T.; Milan, D.J. Novel chemical suppressors of long QT syndrome identified by an in vivo functional screen. Circulation 2011, 123, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Incardona, J.P.; Carls, M.G.; Day, H.L.; Sloan, C.A.; Bolton, J.L.; Collier, T.K.; Scholz, N.L. Cardiac arrhythmia is the primary response of embryonic pacific herring (Clupea pallasi) exposed to crude oil during weathering. Environ. Sci. Technol. 2009, 43, 201–207. [Google Scholar] [CrossRef] [PubMed]
- MacRae, C.A. Recent advances in in vivo screening for antiarrhythmic drugs. Exp. Opin. Drug Discov. 2013, 8, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Meder, B.; Scholz, E.P.; Hassel, D.; Wolff, C.; Just, S.; Berger, I.M.; Patzel, E.; Karle, C.; Katus, H.A.; Rottbauer, W. Reconstitution of defective protein trafficking rescues long-QT syndrome in zebrafish. Biochem. Biophys. Res. Commun. 2011, 408, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.L.; Getzinger, G.J.; Cooper, E.M.; Clark, B.W.; Garner, L.V.T.; Di Giulio, R.T.; Ferguson, P.L.; Stapleton, H.M. Effect-directed analysis of elizabeth river porewater: Developmental toxicity in zebrafish (Danio rerio). Environ. Toxicol. Chem. 2014, 33, 2767–2774. [Google Scholar] [CrossRef] [PubMed]
- Asnani, A.; Peterson, R.T. The zebrafish as a tool to identify novel therapies for human cardiovascular disease. Dis. Model. Mech. 2014, 7, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Kari, G.; Dicker, A.P.; Rodeck, U.; Koch, W.J.; Force, T. A novel preclinical strategy for identifying cardiotoxic kinase inhibitors and mechanisms of cardiotoxicity. Circ. Res. 2011, 109, 1401–1409. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Guan, J.; Plovie, E.; Seldin, D.C.; Connors, L.H.; Merlini, G.; Falk, R.H.; MacRae, C.A.; Liao, R. Human amyloidogenic light chain proteins result in cardiac dysfunction, cell death, and early mortality in zebrafish. Am. J. Physiol. Heart Circ. Physiol. 2013, 305, H95–H103. [Google Scholar] [CrossRef] [PubMed]
- Milan, D.J.; Peterson, T.A.; Ruskin, J.N.; Peterson, R.T.; MacRae, C.A. Drugs that induce repolarization abnormalities cause bradycardia in zebrafish. Circulation 2003, 107, 1355–1358. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.J.; Hoage, T.; Bai, P.; Ding, Y.H.; Chen, Z.Y.; Zhang, R.L.; Huang, W.; Jahangir, A.; Paw, B.; Li, Y.G.; et al. Cardiac hypertrophy involves both myocyte hypertrophy and hyperplasia in anemic zebrafish. PLoS ONE 2009, 4, e6596. [Google Scholar] [CrossRef] [PubMed]


| Gene | Gene name | Human CHD | Zebrafish | Pubmed ID |
|---|---|---|---|---|
| Transcription Factors | ||||
| GATA4/5/6 | GATA4 transcription factor | Septal defects, valve malformation, Tetrology of Fallot | CM specification | 12845333, 24638895, 23289003, 24841381, 16079152, 10580005, 17950269, 17869240 |
| NKX2.5 | Homeobox containing transcription factor 2-5 | Septal defects, conduction abnormalities | Looping, CM proliferation, CM differentiation | 9651244, 19158954 |
| TBX5 | T-box 5 transcription factor | Septal defects and Tetrology of Fallot in Holt-Oram Syndrome | Bradycardia, looping | 8988164, 12223419 |
| HAND2 | Helix-loop-helix transcription factor | Tetrology of Fallot | Cardiac differentiation | 26676105, 17681136, 10821756 |
| Cell Signaling and Growth Factors | ||||
| NOTCH1 | Notch homolog1 | Valve malformation, outflow tract | Valvulogenesis, conduction tissue specification, trabeculation | 18593716, 16025100, 16481353, 14701881, 26628092 |
| SMAD6 | SMAD family member 6 | Septal defects, valve malformation, coractation of the aorta | CM proliferation | 22275001, 22247485 |
| FLK1 | Vascular endothelial growth factor | Coractation of the aorta, outflow tract defects | Valvulogenesis | 20420808, 16170785 |
| SEMA3 | Semaphorin 3 | Anomalous pulmonary vein connection | Primary heart field size | 23685842, 16860789 |
| Cardiomyocyte Function | ||||
| MYH6 | Alpha myosin heavy chain | Atrial septal defect, left ventricular non-compaction, cardiomyopathy | Atrial contraction | 15735645, 17611253, 14573521 |
| ACTC | Alpha cardiac actin | Atrial septal defect | Endocardial cushion morphogenesis | 17947298, 22751927 |
| TITIN | Titin | Cardiomyopathy | Sarcomere assembly | 22335739, 11788825, 9007227 |
| KCNH2 | Potassium channel, voltage gated eag related subfamily H, member 2 | Long QT syndrome, short QT syndrome, atrial fibrillation | Ventricular asystole, QT intreval | 15828882, 19668779, 17592134, 18250272, 14678746 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brown, D.R.; Samsa, L.A.; Qian, L.; Liu, J. Advances in the Study of Heart Development and Disease Using Zebrafish. J. Cardiovasc. Dev. Dis. 2016, 3, 13. https://doi.org/10.3390/jcdd3020013
Brown DR, Samsa LA, Qian L, Liu J. Advances in the Study of Heart Development and Disease Using Zebrafish. Journal of Cardiovascular Development and Disease. 2016; 3(2):13. https://doi.org/10.3390/jcdd3020013
Chicago/Turabian StyleBrown, Daniel R., Leigh Ann Samsa, Li Qian, and Jiandong Liu. 2016. "Advances in the Study of Heart Development and Disease Using Zebrafish" Journal of Cardiovascular Development and Disease 3, no. 2: 13. https://doi.org/10.3390/jcdd3020013






