Filarial Nematode Infection in Ixodes scapularis Ticks Collected from Southern Connecticut
Abstract
:1. Introduction
2. Experimental Section
2.1. Tick Collection and DNA Preparation
2.2. PCR Protocols
2.3. Sequencing and Sequence Analyses
2.4. Nucleotide Sequence Accession Numbers
2.5. In Situ Hybridization
3. Results and Discussion
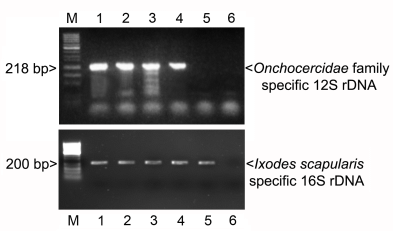
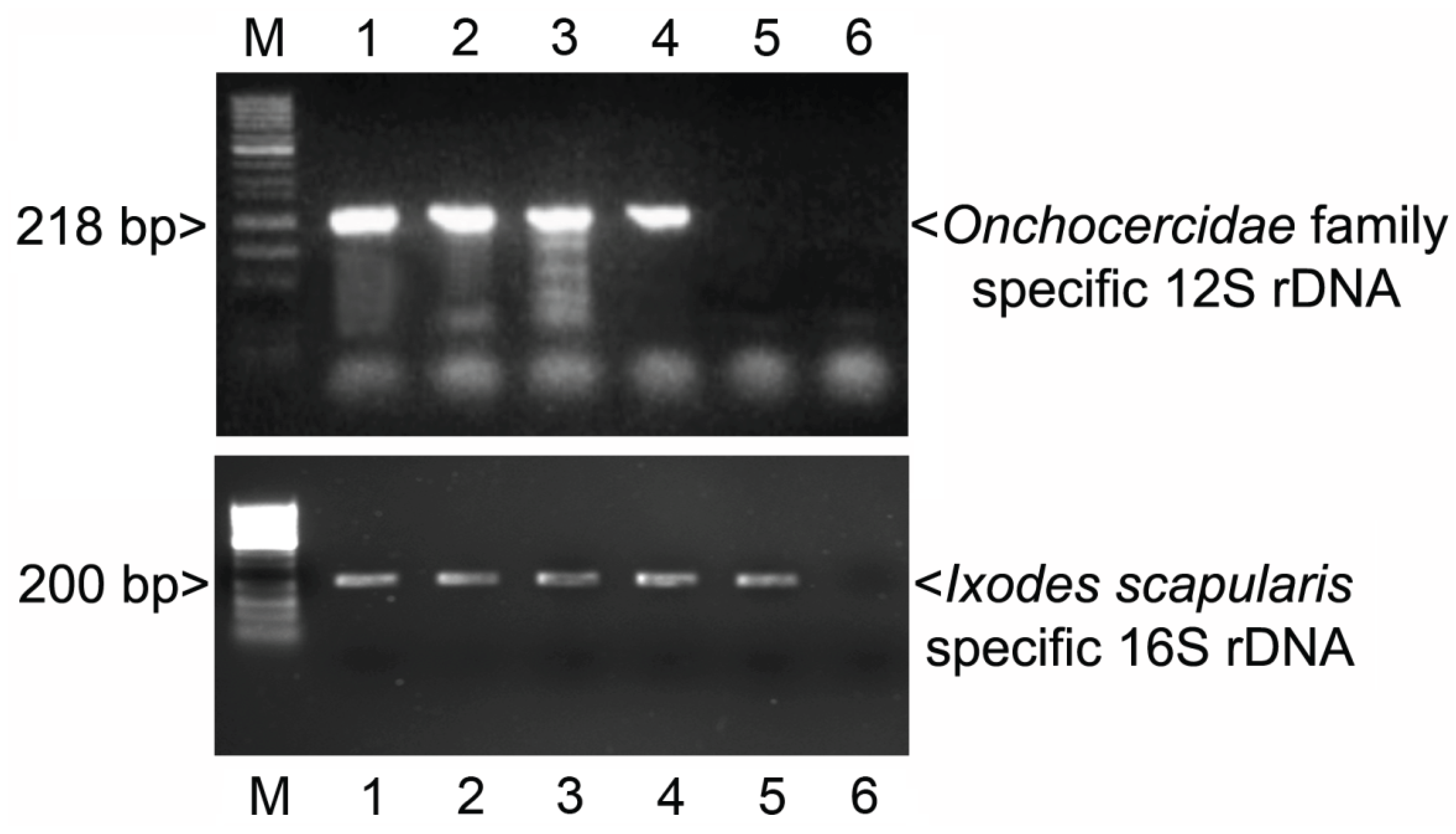
3.1. PCR
3.2. In Situ Confirmation

3.3. Additional PCR and Sequencing Analyses

3.4. Rate of Infection
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Piesman, J.; Hicks, T.C.; Sinsky, R.J.; Obiri, G. Simultaneous transmission of Borrelia burgdorferi and Babesia microti by individual nymphal Ixodes dammini ticks. J. Clin. Microbiol. 1987, 25, 2012–2013. [Google Scholar]
- Levin, M.L.; Fish, D. Acquisition of coinfection and simultaneous transmission of Borrelia burgdorferi and Ehrlichia phagocytophila by Ixodes scapularis ticks. Infect. Immun. 2000, 68, 2183–2186. [Google Scholar] [CrossRef]
- Courtney, J.W.; Dryden, R.L.; Montgomery, J.; Schneider, BS.; Smith, G.; Massung, R.F. Molecular characterization of Anaplasma phagocytophilum and Borrelia burgdorferi in Ixodes scapularis ticks from Pennsylvania. J. Clin. Microbiol. 2003, 41, 1569–1573. [Google Scholar] [CrossRef]
- Adelson, M.E.; Rao, R.V.; Tilton, R.C.; Cabets, K.; Eskow, E.; Fein, L.; Occi, J.L.; Mordechai, E. Prevalence of Borrelia burgdorferi, Bartonella spp., Babesia microti, and Anaplasma phagocytophila in Ixodes scapularis Ticks collected in northern New Jersey. J. Clin. Microbiol. 2004, 42, 2799–2801. [Google Scholar] [CrossRef]
- Holman, M.S.; Caporale, D.A.; Goldberg, J.; Lacombe, E.; Lubelczyk, C.; Rand, P.W.; Smith, R.P. Anaplasma phagocytophilum, Babesia microti, and Borrelia burgdorferi in Ixodes scapularis, southern coastal Maine. Emerg. Infect. Dis. 2004, 10, 744–746. [Google Scholar] [CrossRef]
- Swanson, S.J.; Neitzel, D.; Reed, K.D.; Belongia, E.A. Coinfections acquired from Ixodes ticks. Clin. Microbiol. 2006, 19, 708–727. [Google Scholar] [CrossRef]
- Tokarz, R.; Jain, K.; Bennett, A.; Briese, T.; Lipkin, W.I. Assessment of polymicrobial infections in ticks in New York State. Vector Borne Zoonotic Dis. 2010, 10, 217–221. [Google Scholar] [CrossRef]
- Kjemtrup, AM.; Conrad, P.A. Human babesiosis: An emerging tick-borne disease. Int. J. Parasitol. 2000, 30, 1323–1337. [Google Scholar] [CrossRef]
- Beaver, P.C.; Burgdorfer, W. A microfilaria of exceptional size from the Ixodid tick, Ixodes dammini from Shelter Island New York. J. Parasitol. 1984, 70, 963–966. [Google Scholar] [CrossRef]
- Londono, M.I. Behaviour and characteristics of the filarial Dipetalonema viteae in the tick Ornithodoros tartakowskyi. Antioquia Medica 1973, 23, 515–516. [Google Scholar]
- Olmeda-García, A.S.; Rodríguez-Rodríguez, J.A. Stage specific development of filarial nematodes in vector ticks. J. Helminthol. 1994, 68, 231–235. [Google Scholar] [CrossRef]
- Zhang, X.; Norris, D.E.; Rasgon, J.L. Distribution and molecular characterization of Wolbachia endosymbionts and filarial nematodes in Maryland populations of the lone star tick (Amblyomma americanum). FEMS Microbiol. Ecol. 2011, 77, 50–56. [Google Scholar] [CrossRef]
- Nuchprayoon, S.; Junpee, A.; Poovorawn, Y.; Scott, A. Detection and differentiation of filarial parasites by universal primer and polymerase chain reaction–restriction fragment length polymorphism analysis. Am. J. Trop. Med. Hy. 2005, 73, 895–900. [Google Scholar]
- Tamura, K.; Peterson, D.; Nei, M. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Casiraghi, M.; Anderson, T.J.C.; Bandi, C.; Bazzocchi, C.; Genchi, C. A Phylogenetic analysis of filarial nematode: Comparison with the phylogeny of Wolbachia endosymbionts. Parasitology 2001, 122, 93–103. [Google Scholar] [CrossRef]
- Otranto, D.; Brianti, E.; Dantas-Torres, F.; Weigl, S.; Latrofa, M.S.; Gaglio, G.; Cauquil, L.; Giannetto, S.; Bain, O. Morphological and molecular data on the dermal microfilariae of a species of Cercopithi filaria from a dog in Sicily. Vet. Parasitol. 2011, 182, 221–229. [Google Scholar] [CrossRef]
- Li, B.W.; Rush, A.C.; Tan, J.; Weil, G.J. Quantitative analysis of gender-regulated transcripts in the filarial Nematode Brugia malayi by real-time RT-PCR. Mol. Biochem. Parasitol. 2004, 137, 329–337. [Google Scholar] [CrossRef]
- Hojas, R.M.; Post, R.J. Regional genetic variation in the major sperm protein genes of Onchocerca volvulus and Mansonella ozzardi (Nematoda: Filarioidea). Int. J. Parasitol. 2000, 30, 1459–1465. [Google Scholar] [CrossRef]
- Brianti, B.; Gagglio, G.; Napoli, E.; Giannetto, S.; Dantas-Torres, P.; Bain, O.; Otranto, D. New insights into the ecology and biology of Acanthocheilonema reconditum causing canine subcutaneous filariosis. Parasitology 2012, 139, 530–536. [Google Scholar] [CrossRef]
- Eisenbeiss, W.F.; Apfel, H.; Meyer, T.F. Recovery, distribution, and development of Acanthocheilonema viteae third- and early fourth-stage larvae in adult jirds. J. Parasitol. 1991, 77, 580–586. [Google Scholar] [CrossRef]
- Beaver, P.C.; Meyer, E.A.; Jarroll, E.L. Dipetalonema from the eye of a man in Oregon, USA. A case report. Am. J. Trop. Med. Hyg. 1980, 29, 367–372. [Google Scholar]
- Huynh, T.; Thean, J.; Maini, R. Dipetalonema reconditum in the human eye. Br. J. Ophtalmol. 2001, 85. [Google Scholar] [CrossRef]
- Czajka, C.; Becker, N.; Poppert, S.; Jöst, H.; Schmidt-Chanasit, J.; Krüge, A. Molecular detection of Setaria tundra (Nematoda: Filarioidea) and an unidentified filarial species in mosquitoes in Germany. Parasit. Vectors 2012, 5. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Namrata, P.; Miller, J.M.; Shilpa, M.; Reddy, P.R.; Bandoski, C.; Rossi, M.J.; Sapi, E. Filarial Nematode Infection in Ixodes scapularis Ticks Collected from Southern Connecticut. Vet. Sci. 2014, 1, 5-15. https://doi.org/10.3390/vetsci1010005
Namrata P, Miller JM, Shilpa M, Reddy PR, Bandoski C, Rossi MJ, Sapi E. Filarial Nematode Infection in Ixodes scapularis Ticks Collected from Southern Connecticut. Veterinary Sciences. 2014; 1(1):5-15. https://doi.org/10.3390/vetsci1010005
Chicago/Turabian StyleNamrata, Pabbati, Jamie M. Miller, Madari Shilpa, Patlolla Raghavender Reddy, Cheryl Bandoski, Michael J. Rossi, and Eva Sapi. 2014. "Filarial Nematode Infection in Ixodes scapularis Ticks Collected from Southern Connecticut" Veterinary Sciences 1, no. 1: 5-15. https://doi.org/10.3390/vetsci1010005