Evaluation of Commercial Prototype Bacteriophage Intervention Designed for Reducing O157 and Non-O157 Shiga-Toxigenic Escherichia coli (STEC) on Beef Cattle Hide
Abstract
:1. Introduction
2. Materials and Methods
2.1. STEC Host Range and Spot Titration of Phages
2.2. Efficiency of Plating Assay for Phage Infection of STEC Hosts
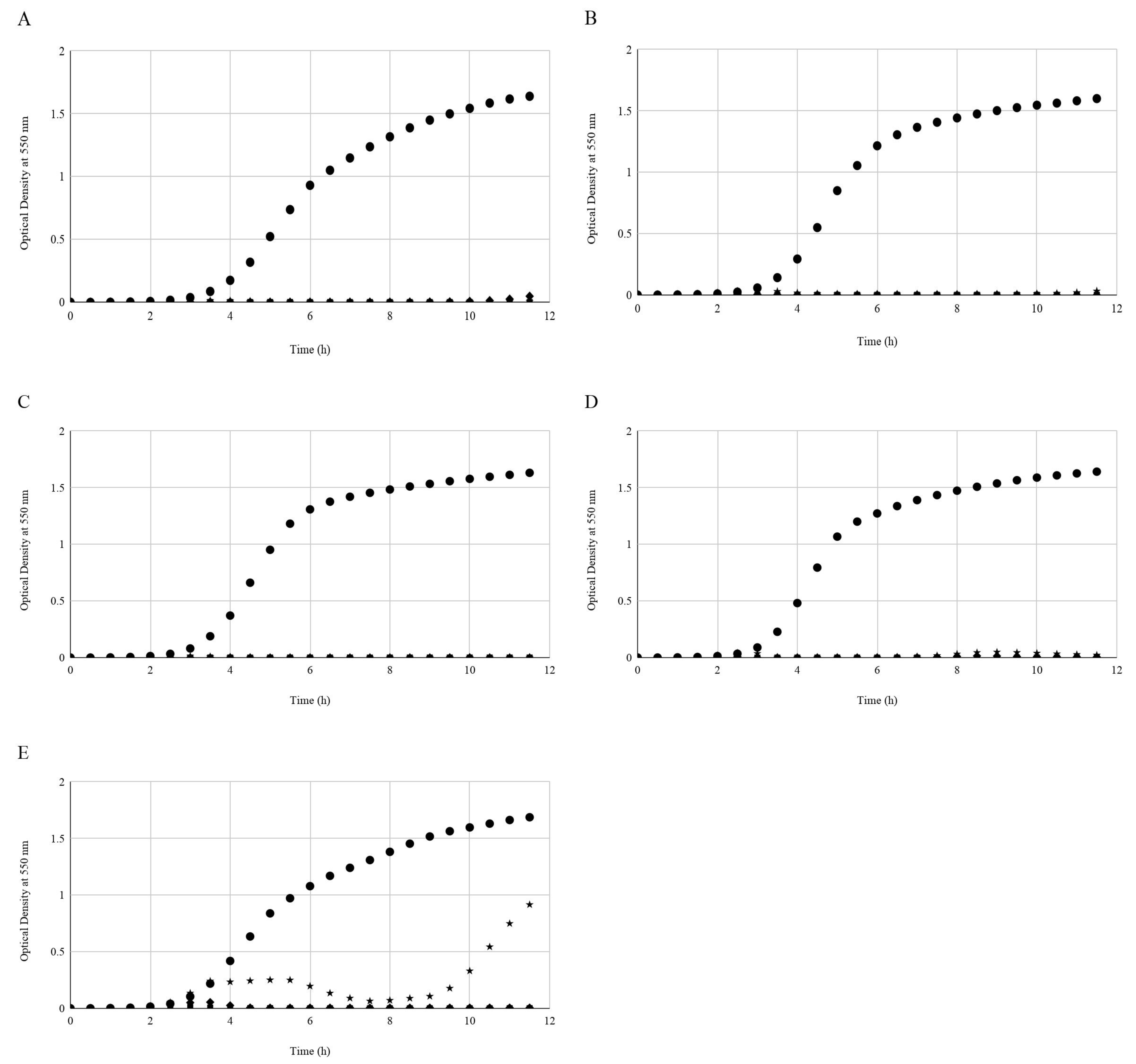
2.3. In Vitro Growth Inhibition of STEC by Phages
2.4. Application of Decontamination Treatments to STEC-Inoculated Cattle Hide Pieces and Determination of Phage: Host Multiplicity of Infection (MOI)
2.5. Statistical Analysis of Data
3. Results
3.1. Efficiency of Plating of Individual and Cocktailed Phages against STEC Isolates
3.2. STEC Growth In Vitro Challenged with Serially Diluted Phages
3.3. STEC Reduction on Cattle Hide Pieces by Differing Antimicrobial Treatments and Estimation of Numbers of Phages Required to Infect Host Cells by the Multiplicity of Infection (MOI) Determination
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Scallan, E.; Hoekstra, R.M.; Mahon, B.E.; Jones, T.F.; Griffin, P.M. An assessment of the human health impact of seven leading foodborne pathogens in the United States using disability adjusted life years. Epidemiol. Infect. 2015, 143, 2795–2804. [Google Scholar] [CrossRef] [PubMed]
- Hancock, D.D.; Besser, T.E.; Rice, D.H.; Ebel, E.D.; Herriott, D.E.; Carpenter, L.V. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the northwestern USA. Prev. Vet. Med. 1998, 35, 11–19. [Google Scholar] [CrossRef]
- Keen, J.E.; Elder, R.O. Isolation of Shiga-toxigenic Escherichia coli O157 from hide surfaces and the oral cavity of finished beef feedlot cattle. J. Am. Vet. Med. Assoc. 2002, 220, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Stanford, K.; Bryan, M.; Peters, J.; González, L.A.; Stephens, T.P.; Schwartzkopf-Genswein, K.S. Effects of long- or short-haul transportation of slaughter heifers and cattle liner microclimate on hide contamination with Escherichia coli O157. J. Food Prot. 2011, 74, 1605–1610. [Google Scholar] [CrossRef] [PubMed]
- Laine, E.S.; Scheftel, J.M.; Boxrud, D.J.; Vought, K.J.; Danila, R.N.; Elfering, K.M.; Smith, K.E. Outbreak of Escherichia coli O157:H7 infections associated with nonintact blade-tenderized frozen steaks sold by door-to-door vendors. J. Food Prot. 2005, 68, 1198–1202. [Google Scholar] [CrossRef] [PubMed]
- Pihkala, N.; Bauer, N.; Eblen, D.; Evans, P.; Johnson, R.; Webb, J.; Williams, C. Risk profile for pathogenic non-O157 Shiga toxin-producing Escherichia coli (non-O157 STEC). Available online: https://www.fsis.usda.gov/shared/PDF/Non_O157_STEC_Risk_Profile_May2012.pdf (accessed on 28 May 2017).
- Luna-Gierke, R.E.; Griffin, P.M.; Gould, L.H.; Herman, K.; Bopp, C.A.; Strockbine, N.; Mody, R.K. Outbreaks of non-O157 Shiga toxin-producing Escherichia coli infection: USA. Epidemiol. Infect. 2014, 142, 2270–2280. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). E. coli (Escherichia coli). Available online: https://www.cdc.gov/ecoli/index.html (accessed on 16 March 2018).
- Arthur, T.M.; Bosilevac, J.M.; Brichta-Harhay, D.M.; Kalchayanand, N.; Shackelford, S.D.; Wheeler, T.L.; Koohmaraie, M. Effects of a minimal hide wash cabinet on the levels and prevalence of Escherichia coli O157:H7 and Salmonella on the hides of beef cattle at slaughter. J. Food Prot. 2007, 70, 1076–1079. [Google Scholar] [CrossRef] [PubMed]
- Dorsa, W.J.; Cutter, C.N.; Siragusa, G.R.; Koohmaraie, M. Microbial decontamination of beef and sheep carcasses by steam, hot water spray washes, and a steam-vacuum sanitizer. J. Food Prot. 1996, 59, 127–135. [Google Scholar] [CrossRef]
- Dorsa, W.J.; Cutter, C.N.; Siragusa, G.R. Effects of steam-vacuuming and hot water spray wash on the microflora of refrigerated beef carcass surface tissue inoculated with Escherichia coli O157:H7, Listeria innocua, and Clostridium sporogenes. J. Food Prot. 1997, 60, 114–119. [Google Scholar] [CrossRef]
- Koohmaraie, M.; Arthur, T.M.; Bosilevac, J.M.; Guerini, M.; Shackelford, S.D.; Wheeler, T.L. Post-harvest interventions to reduce/eliminate pathogens in beef. Meat Sci. 2005, 71, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Castillo, A.; Lucia, L.M.; Goodson, K.J.; Savell, J.W.; Acuff, G.R. Comparison of water wash, trimming, and combined hot water and lactic acid treatments for reducing bacteria of fecal origin on beef carcasses. J. Food Prot. 1998, 61, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Ellebracht, E.A.; Castillo, A.; Lucia, L.M.; Miller, R.K.; Acuff, G.R. Reduction of pathogens using hot water and lactic acid on beef trimmings. J. Food Sci. 1999, 64, 1094–1099. [Google Scholar] [CrossRef]
- Castillo, A.; Lucia, L.M.; Goodson, K.J.; Savell, J.W.; Acuff, G.R. Use of hot water for beef carcass decontamination. J. Food Prot. 1998, 61, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Abuladze, T.; Li, M.; Menetrez, M.Y.; Dean, T.; Senecal, A.; Sulakvelidze, A. Bacteriophages reduce experimental contamination of hard surfaces, tomato, spinach, broccoli, and ground beef by Escherichia coli O157:H7. Appl. Environ. Microbiol. 2008, 74, 6230–6238. [Google Scholar] [CrossRef] [PubMed]
- Bigwood, T.; Hudson, J.A.; Billington, C. Influence of host and bacteriophage concentrations on the inactivation of food-borne pathogenic bacteria by two phages. FEMS Microbiol. Lett. 2009, 291, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Anany, H.; Chen, W.; Pelton, R.; Griffiths, M.W. Biocontrol of Listeria monocytogenes and Escherichia coli O157:H7 in meat by using phages immobilized on modified cellulose membranes. Appl. Environ. Microbiol. 2011, 77, 6379–6387. [Google Scholar] [CrossRef] [PubMed]
- Bigwood, T.; Hudson, J.A.; Billington, C.; Carey-Smith, G.V.; Heinemann, J.A. Phage inactivation of foodborne pathogens on cooked and raw meat. Food Microbiol. 2008, 25, 400–406. [Google Scholar] [CrossRef] [PubMed]
- USDA-FSIS Directive 7120.1, Rev. 45: Safe and Suitable Ingredients Used in the Production of Meat, Poultry, and Egg Products. Available online: https://www.fsis.usda.gov/wps/wcm/connect/bab10e09-aefa-483b-8be8-809a1f051d4c/7120.1.pdf?MOD=AJPERES (accessed on 13 February 2018).
- Rozema, E.A.; Stephens, T.P.; Bach, S.J.; Okine, E.K.; Johnson, R.P.; Stanford, K.; McAllister, T.A. Oral and rectal administration of bacteriophages for control of Escherichia coli O157:H7 in feedlot cattle. J. Food Prot. 2009, 72, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Goodridge, L.D.; Bisha, B. Phage-based biocontrol strategies to reduce foodborne pathogens in foods. Bacteriophage 2011, 1, 130–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waddell, T.; Mazzocco, A.; Johnson, R.P.; Pacan, J.; Campbell, S.; Perets, A.; MacKinnon, J.; Holtslander, B.; Pope, B.; Gyles, C. Control of Escherichia coli O157:H7 infection of calves by bacteriophages. In 4th International Symposium and Workshop on Shiga Toxin (Verocytotoxin)-Producing Escherichia coli Infections, Kyoto, Japan; European Centre for Disease Prevention and Control (ECDC): Stockholm, Sweden, 2000. [Google Scholar]
- Coffey, B.; Rivas, L.; Duffy, G.; Coffey, A.; Ross, R.P.; McAuliffe, O. Assessment of Escherichia coli O157:H7-specific bacteriophages e11/2 and e4/1c in model broth and hide environments. Int. J. Food Microbiol. 2011, 147, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Arthur, T.M.; Kalchayanand, N.; Agga, G.E.; Wheeler, T.L.; Koohmaraie, M. Evaluation of bacteriophage application to cattle in lairage at beef processing plants to reduce Escherichia coli O157:H7 prevalence on hides and carcasses. Foodborne Pathog. Dis. 2017, 14, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H. Bacteriophages; Interscience Publishers: New York, NY, USA, 1959. [Google Scholar]
- Khan Mirzaei, M.; Nilsson, A.S. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wahab, L.; Gill, J.J. Development and validation of a microtiter plate-based assay for determination of bacteriophage host range and virulence. Viruses 2018, 10, 189. [Google Scholar] [CrossRef] [PubMed]
- Gudro, I.; Valeika, V.; Sirvaitytė, J. Short term preservation of hide using vacuum: Influence on properties of hide and of processed leather. PLoS ONE 2014, 9, e112783. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, K.R.; Tolen, T.N.; Hudson, J.C.; Castillo, A.; Griffin, D.; Taylor, T.M. Effectiveness of a commercial lactic acid bacteria intervention applied to inhibit Shiga toxin-producing Escherichia coli on refrigerated vacuum-aged beef. Int. J. Food Sci. 2017, 2017, 8070515. [Google Scholar] [CrossRef] [PubMed]
- Tuttle, J.; Gomez, T.; Doyle, M.P.; Wells, J.G.; Zhao, T.; Tauxe, R.V.; Griffin, P.M. Lessons from a large outbreak of Escherichia coli O157:H7 infections: Insights into the infectious dose and method of widespread contamination of hamburger patties. Epidemiol. Infect. 1999, 122, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Griffin, P.M.; Tauxe, R.V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 1991, 13, 60–98. [Google Scholar] [CrossRef] [PubMed]
- Sofos, J.N.; Kochevar, S.L.; Reagan, J.O.; Smith, G.C. Extent of beef carcass contamination with Escherichia coli and probabilities of passing U.S. regulatory criteria. J. Food Prot. 1999, 62, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Sofos, J.N.; Kochevar, S.L.; Bellinger, G.R.; Buege, D.R.; Hancock, D.D.; Ingham, S.C.; Morgan, J.B.; Reagan, J.O.; Smith, G.C. Sources and extent of microbiological contamination of beef carcasses in seven United States slaughtering plants. J. Food Prot. 1999, 62, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Arthur, T.M.; Bosilevac, J.M.; Nou, X.; Shackelford, S.D.; Wheeler, T.L.; Kent, M.P.; Jaroni, D.; Pauling, B.; Allen, D.M.; Koohmaraie, M. Escherichia coli O157 prevalence and enumeration of aerobic bacteria, Enterobacteriaceae, and Escherichia coli O157 at various steps in commercial beef processing plants. J. Food Prot. 2004, 67, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Jadeja, R.; Hung, Y.-C. Efficacy of near neutral and alkaline pH electrolyzed oxidizing waters to control Escherichia coli O157:H7 and Salmonella Typhimurium DT 104 from beef hides. Food Control 2014, 41, 17–20. [Google Scholar] [CrossRef]
- Elramady, M.G.; Aly, S.S.; Rossitto, P.V.; Crook, J.A.; Cullor, J.S. Synergistic effects of lactic acid and sodium dodecyl sulfate to decontaminate Escherichia coli O157:H7 on cattle hide sections. Foodborne Pathog. Dis. 2013, 10, 661–663. [Google Scholar] [CrossRef] [PubMed]
| STEC Isolate 1 | Sero-ID | Phage Cocktail Titer (log10 PFU/mL) 2 | Phage Efficiency of Plating (EOP) | ||||
|---|---|---|---|---|---|---|---|
| Phage A | Phage B | Phage C | Phage D | Phage Cocktail 4 | |||
| USDA-FSIS 380-94 | O157:H7 | 8.6 ± 0.1 | - | 0.19 3 | 1.0 | + | 1.0 |
| CDC 96-3258 | O45:H2 | 8.3 ± 0.1 | + | 0.08 | - | + | 0.5 |
| CDC 90-3128 | O103:H2 | 8.4 ± 0.5 | + | 1.0 | 0.42 | 0.25 | 1.0 |
| JB1-95 | O111:H- | 9.1 ± 0.1 | 1.0 | - | - | 0.29 | 3.3 |
| CDC 97-3068 | O121:H19 | 6.7 ± 0.1 | - | - | - | 0.08 | 0.01 |
| STEC Isolate 1. | Sero-ID | Phages | Water | Control | Phage:Host MOI 3 |
|---|---|---|---|---|---|
| USDA-FSIS 380-94 | O157:H7 | 5.6 ± 0.2B 2 | 6.1 ± 0.2A | 6.1 ± 0.3A | 11.2 |
| CDC 96-3258 | O45:H2 | 5.8 ± 0.1AB | 5.9 ± 0.1AB | 5.8 ± 0.2AB | 47.3 |
| CDC 90-3128 | O103:H2 | 5.2 ± 0.4C | 6.0 ± 0.2A | 5.9 ± 0.2AB | 15.6 |
| JB1-95 | O111:H- | 5.9 ± 0.1A | 5.9 ± 0.2AB | 6.0 ± 0.2A | 41.7 |
| CDC 97-3068 | O121:H19 | 4.7 ± 0.3D | 5.1 ± 0.1C | 5.1 ± 0.2C | 2.2 |
| Pooled Standard Error = 0.07 | |||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tolen, T.N.; Xie, Y.; Hairgrove, T.B.; Gill, J.J.; Taylor, T.M. Evaluation of Commercial Prototype Bacteriophage Intervention Designed for Reducing O157 and Non-O157 Shiga-Toxigenic Escherichia coli (STEC) on Beef Cattle Hide. Foods 2018, 7, 114. https://doi.org/10.3390/foods7070114
Tolen TN, Xie Y, Hairgrove TB, Gill JJ, Taylor TM. Evaluation of Commercial Prototype Bacteriophage Intervention Designed for Reducing O157 and Non-O157 Shiga-Toxigenic Escherichia coli (STEC) on Beef Cattle Hide. Foods. 2018; 7(7):114. https://doi.org/10.3390/foods7070114
Chicago/Turabian StyleTolen, Tamra N., Yicheng Xie, Thomas B. Hairgrove, Jason J. Gill, and T. Matthew Taylor. 2018. "Evaluation of Commercial Prototype Bacteriophage Intervention Designed for Reducing O157 and Non-O157 Shiga-Toxigenic Escherichia coli (STEC) on Beef Cattle Hide" Foods 7, no. 7: 114. https://doi.org/10.3390/foods7070114





