Applying Fourier Transform Mid Infrared Spectroscopy to Detect the Adulteration of Salmo salar with Oncorhynchus mykiss
Abstract
:1. Introduction
2. Material and Methods
2.1. Sampling
2.2. Microbial Analysis
2.3. Determination of Moisture Content
2.4. Determination of Free Fat Content/Soxhlet Extraction
2.5. Fourier Transform Infrared Measurement
2.6. Mathematical Treatment
2.6.1. Principal Component Analysis
2.6.2. Partial Least Squares Regression
3. Results and Discussion
3.1. Microbial Analysis
3.2. Determination of Moisture Content
3.3. Determination of Free Fat Content/Soxhlet Extraction
3.4. Fourier Transform Infrared Measured Spectra
3.4.1. Preliminary Analysis of the Spectral Dataset
3.4.2. Partial Least Squares Regression Models for Prediction Based on Spectral Dataset
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Castejón, D. NMR-detection of methylamine compounds in Atlantic salmon (Salmo salar) subjected to E-beam irradiation. Food Control 2016, 60 (Suppl. C), 455–460. [Google Scholar] [CrossRef]
- Haq, M. Modifications of Atlantic salmon by-product oil for obtaining different ω-3 polyunsaturated fatty acids concentrates: An approach to comparative analysis. J. Food Drug Anal. 2017. [Google Scholar] [CrossRef] [PubMed]
- Cai, L. Effects of different freezing treatments on physicochemical responses and microbial characteristics of Japanese sea bass (Lateolabrax japonicas) fillets during refrigerated storage. LWT Food Sci. Technol. 2014, 59, 122–129. [Google Scholar] [CrossRef]
- Fidalgo, L.G. Microbial and physicochemical evolution during hyperbaric storage at room temperature of fresh Atlantic salmon (Salmo salar). Innov. Food Sci. Emerg. Technol. 2018, 45, 264–272. [Google Scholar] [CrossRef]
- Lundebye, A.-K. Lower levels of Persistent Organic Pollutants, metals and the marine omega 3-fatty acid DHA in farmed compared to wild Atlantic salmon (Salmo salar). Environ. Res. 2017, 155 (Suppl. C), 49–59. [Google Scholar] [CrossRef] [PubMed]
- Choubert, G.; Baccaunaud, M. Colour changes of fillets of rainbow trout (Oncorhynchus mykiss W.) fed astaxanthin or canthaxanthin during storage under controlled or modified atmosphere. LWT Food Sci. Technol. 2006, 39, 1203–1213. [Google Scholar] [CrossRef]
- Rady, A.; Adedeji, A. Assessing different processed meats for adulterants using visible-near-infrared spectroscopy. Meat Sci. 2018, 136 (Suppl. C), 59–67. [Google Scholar] [CrossRef] [PubMed]
- Nunes, K.M. Detection and characterisation of frauds in bovine meat in natura by non-meat ingredient additions using data fusion of chemical parameters and ATR-FTIR spectroscopy. Food Chem. 2016, 205 (Suppl. C), 14–22. [Google Scholar] [CrossRef] [PubMed]
- Spink, J.; Moyer, D.C.; Whelan, P. The role of the public private partnership in Food Fraud prevention—Includes implementing the strategy. Curr. Opin. Food Sci. 2016, 10 (Suppl. C), 68–75. [Google Scholar] [CrossRef]
- Boyacı, İ.H. A novel method for discrimination of beef and horsemeat using Raman spectroscopy. Food Chem. 2014, 148 (Suppl. C), 37–41. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.-H. Integration of classifiers analysis and hyperspectral imaging for rapid discrimination of fresh from cold-stored and frozen-thawed fish fillets. J. Food Eng. 2015, 161 (Suppl. C), 33–39. [Google Scholar] [CrossRef]
- Ottavian, M. Foodstuff authentication from spectral data: Toward a species-independent discrimination between fresh and frozen–thawed fish samples. J. Food Eng. 2013, 119, 765–775. [Google Scholar] [CrossRef]
- Standal, I.B.; Axelson, D.E.; Aursand, M. 13C NMR as a tool for authentication of different gadoid fish species with emphasis on phospholipid profiles. Food Chem. 2010, 121, 608–615. [Google Scholar] [CrossRef]
- Al-Kahtani, H.A.; Ismail, E.A.; Asif Ahmed, M. Pork detection in binary meat mixtures and some commercial food products using conventional and real-time PCR techniques. Food Chem. 2017, 219 (Suppl. C), 54–60. [Google Scholar] [CrossRef] [PubMed]
- Kamruzzaman, M.; Makino, Y.; Oshita, S. Non-invasive analytical technology for the detection of contamination, adulteration, and authenticity of meat, poultry, and fish: A review. Anal. Chim. Acta 2015, 853 (Suppl. C), 19–29. [Google Scholar] [CrossRef] [PubMed]
- Alamprese, C. Detection of minced beef adulteration with turkey meat by UV–vis, NIR and MIR spectroscopy. LWT Food Sci. Technol. 2013, 53, 225–232. [Google Scholar] [CrossRef]
- Lohumi, S. A review of vibrational spectroscopic techniques for the detection of food authenticity and adulteration. Trends Food Sci. Technol. 2015, 46, 85–98. [Google Scholar] [CrossRef]
- Beasley, M.M. Comparison of transmission FTIR, ATR, and DRIFT spectra: Implications for assessment of bone bioapatite diagenesis. J. Archaeol. Sci. 2014, 46, 16–22. [Google Scholar] [CrossRef]
- Roggo, Y. A review of near infrared spectroscopy and chemometrics in pharmaceutical technologies. J. Pharm. Biomed. Anal. 2007, 44, 683–700. [Google Scholar] [CrossRef] [PubMed]
- International Organization for Standardization. Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Microorganisms—Colony-Count Technique at 30 °C; ISO 4833:2003; International Organization for Standardization: Genova, Switzerland, 2003; pp. 1–9. [Google Scholar]
- NP 2307:1987 = Microbiologie alimentaire: Directives Générales Pour le Dénombrement de Micro-Organismes Psychrotrophes/Instituto Português da Qualidade; elab. CT 61.-Lisboa: Instituto Português da Qualidade. 1988. Available online: http://biblioteca.esa.ipcb.pt/NormasPortuguesas.pdf (accessed on 1 January 2017).
- International Organization for Standardization. Meat and Meat Products—Determination of Free Fat Content; ISO 1444:1996; International Organization for Standardization: Genova, Switzerland, 1996. [Google Scholar]
- Savitzky, A.; Golay, M.J. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Statist. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Liang, Y.-Z.; Kvalheim, O.M. Robust methods for multivariate analysis—A tutorial review. Chemom. Intell. Lab. Syst. 1996, 32, 1–10. [Google Scholar] [CrossRef]
- Wentzell, P.D.; Montoto, L.V. Comparison of principal components regression and partial least squares regression through generic simulations of complex mixtures. Chemom. Intel. Lab. Syst. 2003, 65, 257–279. [Google Scholar] [CrossRef]
- Miller, J.N.; Miller, J.C. Statistics and Chemometrics for Analytical Chemistry; Pearson Education: London, UK, 2005. [Google Scholar]
- De Luca, M. Multivariate calibration techniques applied to derivative spectroscopy data for the analysis of pharmaceutical mixtures. Chemom. Intell. Lab. Syst. 2009, 96, 14–21. [Google Scholar] [CrossRef]
- Divya, O.; Mishra, A.K. Combining synchronous fluorescence spectroscopy with multivariate methods for the analysis of petrol–kerosene mixtures. Talanta 2007, 72, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Corgozinho, C.N.; Pasa, V.M.; Barbeira, P.J. Determination of residual oil in diesel oil by spectrofluorimetric and chemometric analysis. Talanta 2008, 76, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Tironi, V.A.; Tomás, M.C.; Añón, M.C. Quality loss during the frozen storage of sea salmon (Pseudopercis semifasciata). Effect of rosemary (Rosmarinus officinalis L.) extract. LWT Food Sci. Technol. 2010, 43, 263–272. [Google Scholar] [CrossRef]
- Rohman, A.; Erwanto, Y.; Man, Y.B.C. Analysis of pork adulteration in beef meatball using Fourier transform infrared (FTIR) spectroscopy. Meat Sci. 2011, 88, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Carton, I.; Goicoechea, E.N.; Uriarte, P.S. Characterization of cod liver oil by spectroscopic techniques. New approaches for the determination of compositional parameters, acyl groups, and cholesterol from 1H nuclear magnetic resonance and Fourier transform infrared spectral data. J. Agric. Food Chem. 2008, 56, 9072–9079. [Google Scholar] [CrossRef] [PubMed]
- Guillén, M.D.; Ruiz, A.; Cabo, N. Study of the oxidative degradation of farmed salmon lipids by means of Fourier transform infrared spectroscopy. Influence of salting. J. Sci. Food Agric. 2004, 84, 1528–1534. [Google Scholar] [CrossRef]
- Picard, R.R.; Cook, R.D. Cross-validation of regression models. J. Am. Stat. Assoc. 1984, 79, 575–583. [Google Scholar] [CrossRef]
- Huishan, L. Application Fourier transform near infrared spectrometer in rapid estimation of soluble solids content of intact citrus fruits. In 2005 ASAE Annual Meeting; American Society of Agricultural and Biological Engineers: St. Joseph, MI, USA, 2005. [Google Scholar]


| Micro-Organisms | Fish Species | Time (h) | |||
|---|---|---|---|---|---|
| 0 | 72 | 168 | 240 | ||
| TVC | SS | 3.44 ± 0.46 | 4.67 ± 0.10 | 6.82 ± 0.23 | 7.75 ± 0.22 |
| OM | 3.89 ± 0.61 | 6.26 ± 1.12 | 7.98 ± 0.25 | 8.81 ± 0.21 | |
| TP | SS | 3.19 ± 0.52 | 4.61 ± 0.03 | 6.16 ± 0.06 | 7.47 ± 0.25 |
| OM | 3.89 ± 0.61 | 5.39 ± 0.31 | 8.00 ± 0.25 | 8.86 ± 0.21 | |
| Mixture (% w/w of OM/SS) | Fat Content (% w/w) |
|---|---|
| 0 | 11.75 ± 0.78 |
| 10 | 11.62 ± 0.71 |
| 20 | 11.71 ± 1.11 |
| 30 | 12.6 ± 1.34 |
| 40 | 12.75 ± 0.81 |
| 50 | 12.62 ± 0.67 |
| 60 | 13.31 ± 0.71 |
| 70 | 13.15 ± 1.19 |
| 80 | 13.78 ± 0.66 |
| 90 | 14.21 ± 0.57 |
| 100 | 13.65 ± 1.35 |
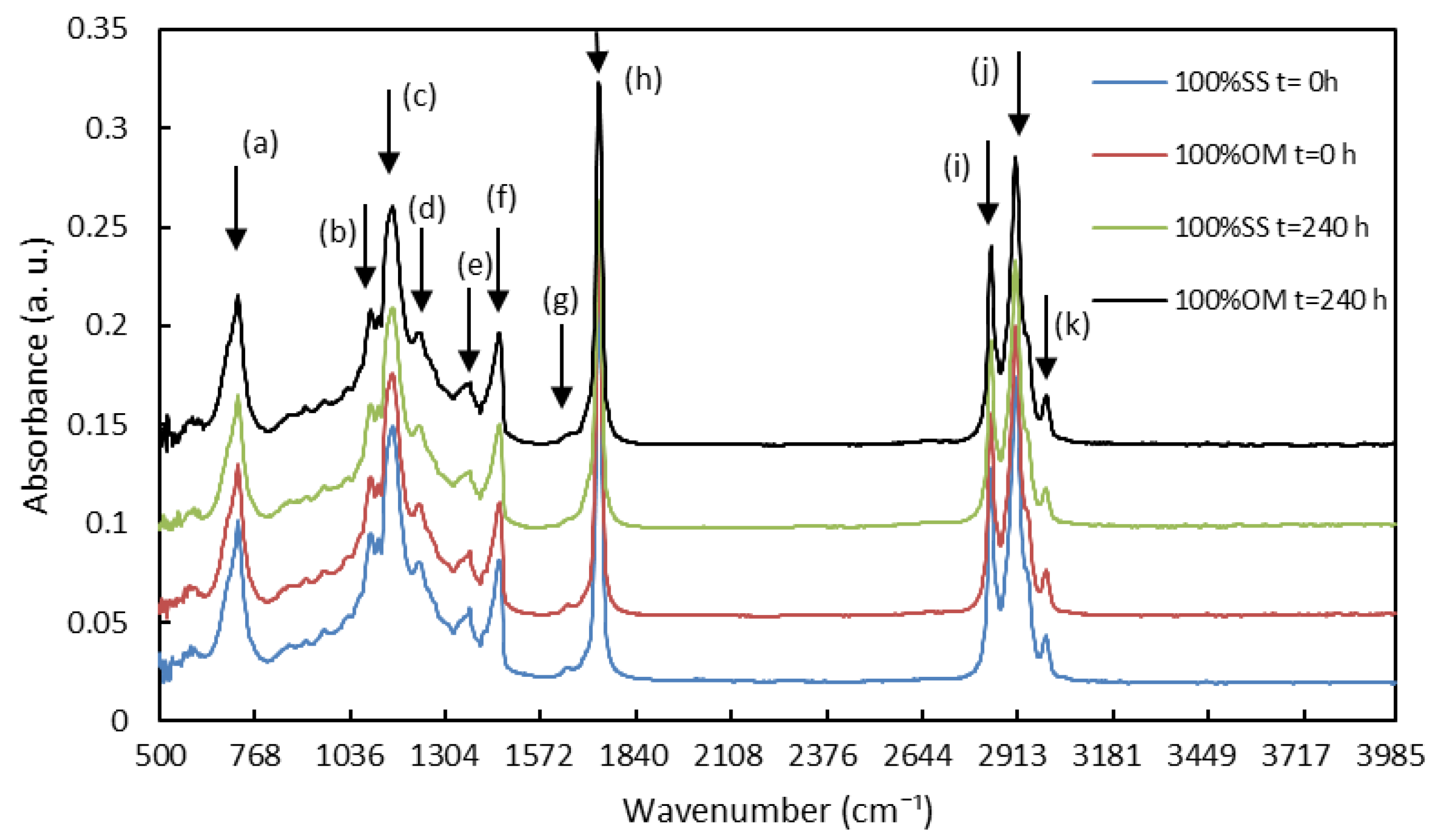
| Assignment | Wavenumber (cm−1) | Functional Group Responsible for IR Absorption |
|---|---|---|
| (a) | 721 | cis-disubstituted olefins (–CH2–, –HC=CH– (cis)) |
| (b) | 1097 | ester of the –C–O group |
| (c) | –C–O, CH2 groups and are correlated with saturated acyl groups | |
| (d) | –C–O, CH2 groups and are correlated with saturated acyl groups | |
| (e) | 1370 | CH3 group |
| (f) | 1464 | CH2 and CH3 |
| (g) | 1655 | unsaturated acyl group (–C=C–) |
| (h) | C=O group of triglycerides | |
| (i) | 2850 to 2925 | symmetrical and asymmetric methylene (CH2) |
| (j) | 2850 to 2925 | symmetrical and asymmetric methylene (CH2) |
| (k) | 3009 | cis olefinic CH double bonds (=C–H) |
| Number of Factors | R2 | RMSE (% w/w of OM/SS) | ||||
|---|---|---|---|---|---|---|
| Calibration | Validation | Prediction | Calibration | Validation | Prediction | |
| 4 | 0.988 | 0.991 | 0.992 | 5.6 | 6.7 | 8.7 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sousa, N.; Moreira, M.J.; Saraiva, C.; De Almeida, J.M.M.M. Applying Fourier Transform Mid Infrared Spectroscopy to Detect the Adulteration of Salmo salar with Oncorhynchus mykiss. Foods 2018, 7, 55. https://doi.org/10.3390/foods7040055
Sousa N, Moreira MJ, Saraiva C, De Almeida JMMM. Applying Fourier Transform Mid Infrared Spectroscopy to Detect the Adulteration of Salmo salar with Oncorhynchus mykiss. Foods. 2018; 7(4):55. https://doi.org/10.3390/foods7040055
Chicago/Turabian StyleSousa, Nuno, Maria João Moreira, Cristina Saraiva, and José M. M. M. De Almeida. 2018. "Applying Fourier Transform Mid Infrared Spectroscopy to Detect the Adulteration of Salmo salar with Oncorhynchus mykiss" Foods 7, no. 4: 55. https://doi.org/10.3390/foods7040055






