1. Introduction
Models for mixtures of distributions—first discussed by Newcomb [
1] and Pearson [
2]—are currently very popular in clustering. Wolfe [
3,
4] and Day [
5] proposed a multivariate normal mixture model in cluster analysis. The most important problems in clustering are choosing the number of components and identifying the structure of the covariance matrix, based on modeling with multivariate normal distributions for each component that forms the data set. Oliveira-Brochado and Martins [
6] examined information criteria used in the determination of the number of components in the mixture model. Despite the many criteria used in the determination of the number of components, these criteria cannot always give accurate results. In particular, information criteria on real data sets with a known number of clusters give different results. In this study, commonly used methods for the determination of the number of clusters—Akaike Information Criterion (AIC), corrected Akaike Information Criterion (AIC
c), Bayesian Information Criterion (BIC), Classification Likelihood Criterion (CLC), Approximate Weight of Evidence Criterion (AWE), Normalized Entropy Criterion (NEC), Kullback Information Criterion (KIC), corrected Kullback Information Criterion (KIC
c), and an approximation of Kullback Information Criterion (AKIC
c) are compared according to the effectiveness of the information criteria, determined by the number of components, and determined by the success in the selection of appropriate covariance matrices and classification accuracy (CA).
2. Clustering Based on Multivariate Finite Mixture Distributions
Mixture cluster analysis based on the mixture of multivariate distributions assumes that the data to be clustered are from several subgroups or clusters, with distinct multivariate distributions. In mixture cluster analysis, each cluster is mathematically represented by a parametric distribution, such as multivariate normal distribution. The entire data set is modeled by a mixture of these distributions.
Assume that there are
observations with
-dimensions, such that an observed random sample is expressed as
. The probability density function of finite mixture distribution models is given by [
7],
where
are probability density functions of the components and
are the mixing proportions or weights. Here,
and
. The parameter vector
contains all of the parameters of the mixture models. Here
denotes unknown parameters of the probability density function of the
ith components (subgroup or cluster) in the mixture models. In Equation (1), the number of components or clusters is represented by
.
The mixture likelihood approach can be used for estimation of the parameters in the mixture models. This approach assumes that the probability function can be the sum of weighted component densities. If the mixture likelihood approach is used for clustering, the clustering problem becomes a problem of estimating the parameters of a mixture distribution model. The maximum-likelihood function is given as follows [
8],
The most widely used approach for parameter estimation is the Expectation-Maximization (EM) algorithm [
9].
In the EM framework, the data
are considered incomplete because their associated component label vectors
are not observed. The component label variables
are consequently introduced, where
is defined to be one or zero, according to whether
did or did not arise from the
ith component of the mixture model (
;
). The completed data vector is represented as follows
where
is the unobservable vector of component-indicator variables. The log-likelihood function for the completed data is shown as
3. EM Algorithm
The EM algorithm is applied to this problem by treating the
as missing data. In this part, the E and M steps of the EM algorithm are described for the mixture distribution models [
7].
E step: Log-likelihood function of the complete data, since
is linear in terms of its label values in the E step; given
observed value, the instant conditional expected values of the categorical variables of
are calculated. Here,
is a random variable corresponding to
. For parameter vector
, the initial value
is assigned. In the first loop of the EM algorithm, while
is given for the E step, the conditional expected value of
is calculated with the initial value of
.
In the
(k + 1
)th loop of the E step of the EM algorithm, the expression
must be represented. Here,
is a value of vector
which is obtained from the
kth step of EM. Since the
Eth step of the
(k + 1
)th loop of the EM algorithm, where
and
the formula below is calculated.
Here, the expression of
is the membership probability of pattern
in segment
(posterior probability). While
is given using the expression in Equation (7), the conditional probability in Equation (5) can be calculated as follows
M step: In the (k + 1)th loop of the EM algorithm, the estimated value of , defined in parameter space , that makes the function maximum is calculated. In the finite mixture probability distribution model, the current estimate of is done independently from the updated vector of the unknown parameters in component density .
If
’s are observed, the maximum likelihood estimation of
for completed data can be found as
If the logarithm takes in completed data in the Eth step of the EM algorithm,
values are used instead of the
expression. Similarly, when the current estimate
of
is calculated,
is used instead of the
expression in Equation (9), as shown below:
In the (
k + 1)th iteration of the
Mth step of the EM algorithm, the current value
of
is defined as
The E and M steps are repeated until the convergence criterion in the EM algorithm is satisfied. As a convenient stopping rule for convergence, if the difference of is quite small or stable, the algorithm is terminated.
4. The Mixture of Multivariate Normal Distribution
The mixture density function of the multivariate normal distribution is given by [
7];
where
is a multivariate normal distribution function, such that
Here, the mean vector is
, and the covariance matrix is
,
, and
. In this case, all unknown parameters of the model are shown as
. Here,
occurs from the mean compound vectors
and the compound covariance matrix
of the parameters of the compound probability density function in the mixture distribution model. Posterior probability is given as
Maximum likelihood estimators of updated mixture proportions
, and mean vector
of the (
k + 1)th iteration of the
Mth step is calculated, respectively, by
Current estimates of the covariance matrix (
) of the component probability density are calculated via the following formula
6. Application and Results
In this section, the performances of the information criteria used for the determination of the number of clusters are compared. Moreover, the efficiency of the different types of covariance matrices are investigated in the model based on clustering. The comparison of the information criteria is performed using two different settings. First, commonly used real data sets are used. Second, synthetic data sets are generated by using the properties of these real data sets, and they are used for comparison.
The appropriate number of clusters is determined as the value which gives the minimum information criteria. According to
Table 2, the number of clusters of the Liver Disorders data set is correctly determined via AWE, BIC, KIC
c, and NEC. In
Table 3, AIC
c and KIC could accurately determine the number of clusters of the Iris data set. The number of clusters of the Wine data set is correctly determined via AIC and KIC in
Table 4.
According to
Table 5, the number of clusters of the Ruspini [
23] data set is correctly determined via AIC
c, AKIC
c, BIC, and KIC
c. In
Table 6, the number of clusters of the Vehicle Silhouettes data set is correctly determined by AIC, BIC, CLC, and KIC.
According to
Table 7, the number of clusters for the Landsat Satellite data set is correctly determined via AIC and KIC. In
Table 8, the number of clusters for the Image Segmentation data set is correctly determined by all information criteria.
In
Table 2,
Table 3,
Table 4,
Table 5,
Table 6,
Table 7 and
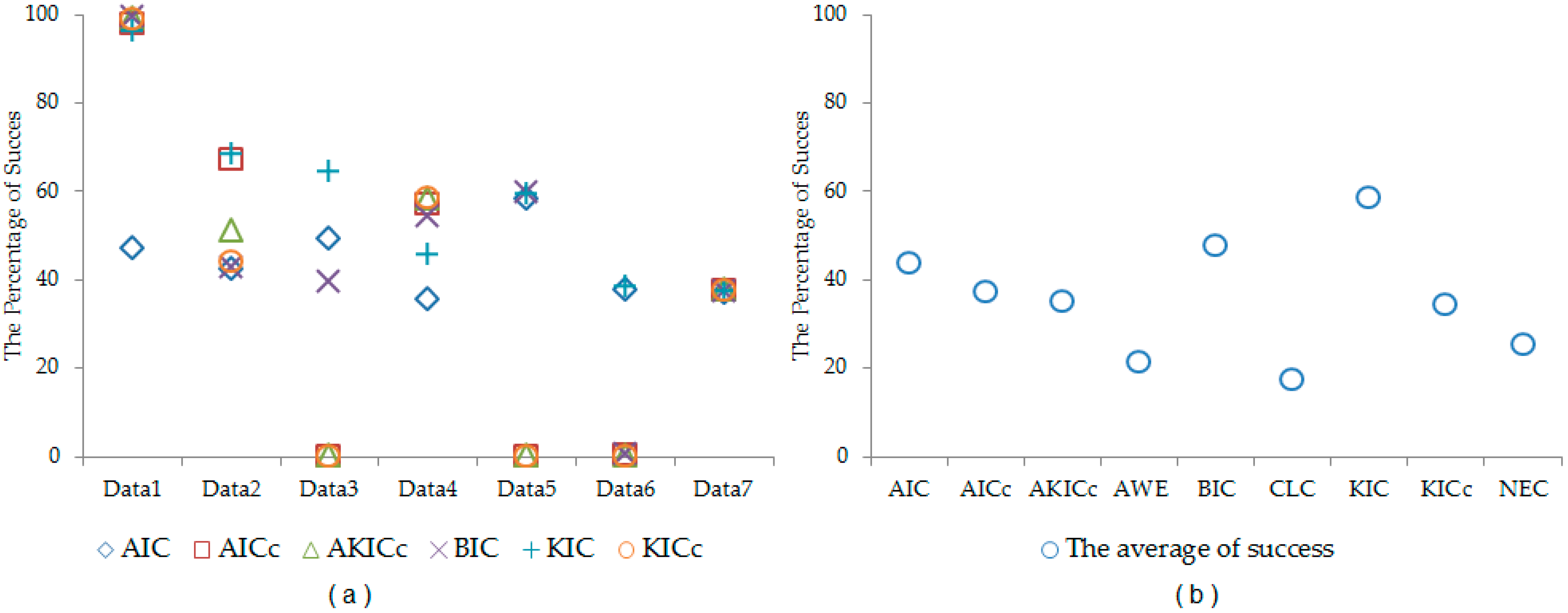
Table 8, the performance of each information criterion varies in each data set. In order to make general conclusions, a simulation study is provided. By using the properties of each real data set, synthetic data sets are generated. In this simulation, we generated 1000 data sets according to each real data set. The synthetic data sets are generated from Liver, Iris, Wine, Ruspini, Vehicle, Landsat, and Image data sets. The cluster number determination accuracy is computed for each information criterion. The results are given in
Table 9 and
Figure 1. According to simulation results, better results are obtained by using KIC.
The efficiency of different types of covariance structures in mixture clustering based on a mixture of multivariate normal distributions is investigated.
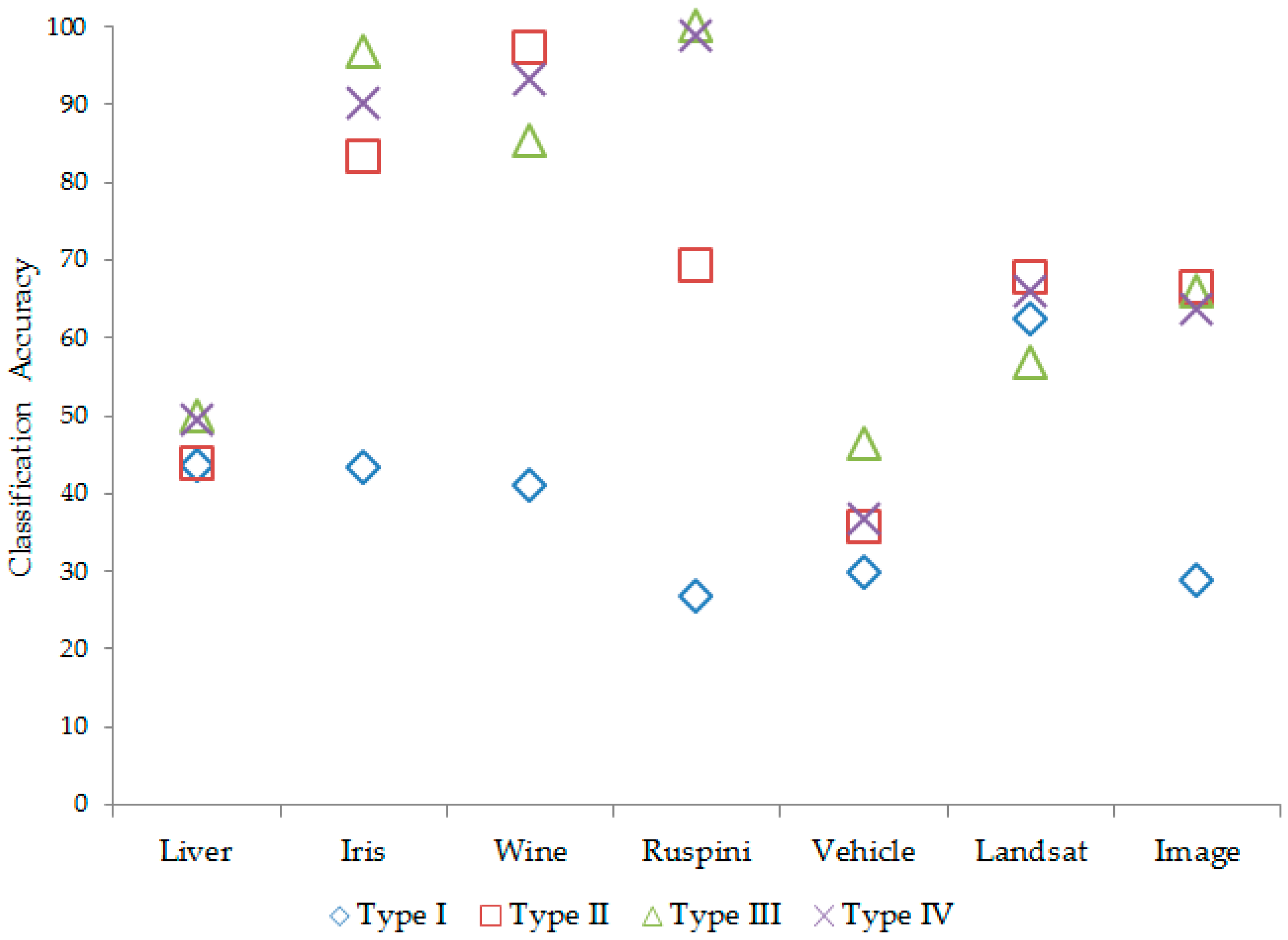
According to the number of clusters regarding each data set, classification accuracy and information criteria are computed for each covariance structure. The results are given in
Table 10.
According to
Table 10, the Type III
covariance matrix of each subgroup has generally performed better in the results, both in terms of the correct classification and the minimum information criteria value. The classification accuracy in mixture clustering based on a mixture of multivariate normal distributions according to covariance types is given in
Figure 2.