New Development of Biomarkers for Gastrointestinal Cancers: From Neoplastic Cells to Tumor Microenvironment
Abstract
:1. Introduction
2. Current Clinical Application of Biomarkers in Gastrointestinal Malignancies
2.1. Tumor Markers
2.2. Targets of Matched Therapies
2.3. Mismatch Repair Genes
3. New Development of Biomarkers
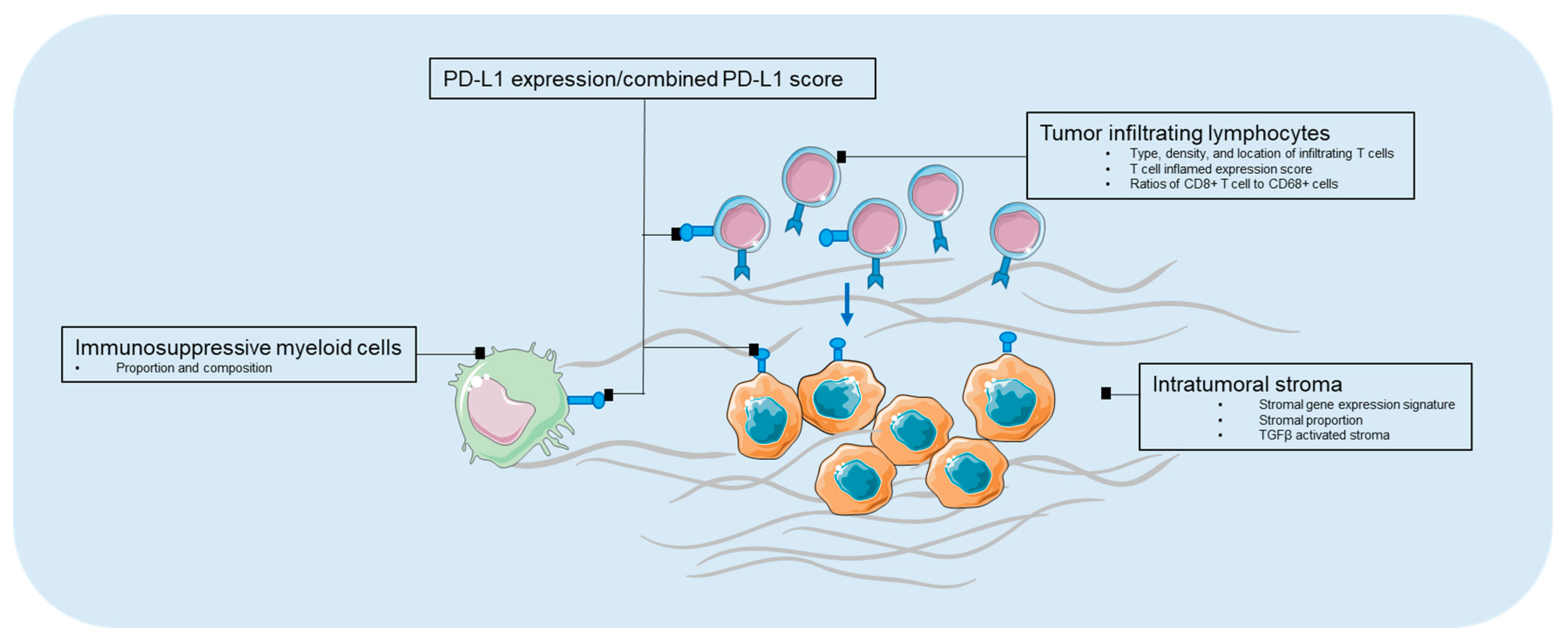
3.1. Biomarkers in Tumor Microenvironment
3.1.1. PD-L1 Expression
3.1.2. Tumor Infiltrating Lymphocyte
3.1.3. Immunosuppressive Myeloid Cells
3.1.4. Intratumoral Stroma
3.2. Biomarkers in Tumor Genomics
3.2.1. Targeted Gene Panels
3.2.2. Mutational Burden
3.3. Biomarkers in Liquid Biopsies
3.3.1. ctDNA
3.3.2. CTC
3.3.3. Exosomes
4. Prospective
Acknowledgments
Conflicts of Interest
References
- Institute National Cancer Biomarkers. Available online: https://www.cancer.gov/publications/dictionaries/cancer-terms/def/biomarker (accessed on 27 July 2018).
- Badreddine, R.; Wang, K.K. Biomarkers in gastrointestinal cancers. Am. J. Gastroenterol. 2008, 103, 2106–2110. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, E.; Metaxa-Mariatou, V.; Tsaousis, G.; Tsoulos, N.; Tsirigoti, A.; Efstathiadou, C.; Apessos, A.; Agiannitopoulos, K.; Pepe, G.; Bourkoula, E.; et al. Molecular predictive markers in tumors of the gastrointestinal tract. World J. Gastrointest. Oncol. 2016, 8, 772–785. [Google Scholar] [CrossRef] [PubMed]
- Deschoolmeester, V.; Lardon, F.; Pauwels, P.; Peeters, M. Biomarkers in Gastrointestinal Cancer: Focus on Colon, Pancreas and Gastric Cancer; InTech: London, UK, 2012. [Google Scholar]
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (toga): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Bang, Y.J.; Feng-Yi, F.; Xu, J.M.; Lee, K.W.; Jiao, S.C.; Chong, J.L.; Lopez-Sanchez, R.I.; Price, T.; Gladkov, O.; et al. HER2 screening data from toga: Targeting HER2 in gastric and gastroesophageal junction cancer. Gastric Cancer 2015, 18, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Okines, A.; Cunningham, D.; Chau, I. Targeting the human EGFR family in esophagogastric cancer. Nat. Rev. Clin. Oncol. 2011, 8, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Yu, I.S.; Cheung, W.Y. A contemporary review of the treatment landscape and the role of predictive and prognostic biomarkers in pancreatic adenocarcinoma. Can. J. Gastroenterol. Hepatol. 2018, 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Murala, S.; Alli, V.; Kreisel, D.; Gelman, A.E.; Krupnick, A.S. Current status of immunotherapy for the treatment of lung cancer. J. Thorac. Dis. 2010, 2, 237–244. [Google Scholar] [PubMed]
- Topalian, S.L.; Drake, C.G.; Pardoll, D.M. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 2015, 27, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Durham, J.N.; Smith, K.N.; Wang, H.; Bartlett, B.R.; Aulakh, L.K.; Lu, S.; Kemberling, H.; Wilt, C.; Luber, B.S.; et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017, 357, 409–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, T.A.; Wolchok, J.D.; Snyder, A. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2015, 373, 1984. [Google Scholar] [CrossRef] [PubMed]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Foppen, M.H.G.; Goldinger, S.M.; et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015, 350, 207–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hugo, W.; Zaretsky, J.M.; Sun, L.; Song, C.; Moreno, B.H.; Hu-Lieskovan, S.; Berent-Maoz, B.; Pang, J.; Chmielowski, B.; Cherry, G.; et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016, 165, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.-L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti–pd-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Ma, Y.; Raoult, D.; Kroemer, G.; Gajewski, T.F. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science 2018, 359, 1366–1370. [Google Scholar] [CrossRef] [PubMed]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M.; et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Dubin, K.; Callahan, M.K.; Ren, B.; Khanin, R.; Viale, A.; Ling, L.; No, D.; Gobourne, A.; Littmann, E.; Huttenhower, C. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 2016, 7, 10391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazama, S.; Tamada, K.; Yamaguchi, Y.; Kawakami, Y.; Nagano, H. Current status of immunotherapy against gastrointestinal cancers and its biomarkers: Perspective for precision immunotherapy. Ann. Gastroenterol. Surg. 2018, 2, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Tsujikawa, T.; Kumar, S.; Borkar, R.N.; Azimi, V.; Thibault, G.; Chang, Y.H.; Balter, A.; Kawashima, R.; Choe, G.; Sauer, D.; et al. Quantitative multiplex immunohistochemistry reveals myeloid-inflamed tumor-immune complexity associated with poor prognosis. Cell Rep. 2017, 19, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Gorris, M.A.J.; Halilovic, A.; Rabold, K.; van Duffelen, A.; Wickramasinghe, I.N.; Verweij, D.; Wortel, I.M.N.; Textor, J.C.; de Vries, I.J.M.; Figdor, C.G. Eight-color multiplex immunohistochemistry for simultaneous detection of multiple immune checkpoint molecules within the tumor microenvironment. J. Immunol. 2018, 200, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, K.; Wakabayashi-Nakao, K.; Ohshima, K.; Sakura, N.; Yamaguchi, K.; Mochizuki, T. Novel protein isoforms of carcinoembryonic antigen are secreted from pancreatic, gastric and colorectal cancer cells. BMC Res. Notes 2013, 6, 381. [Google Scholar] [CrossRef] [PubMed]
- Kuppusamy, P.; Govindan, N.; Yusoff, M.M.; Ichwan, S.J.A. Proteins are potent biomarkers to detect colon cancer progression. Saudi J. Biol. Sci. 2017, 24, 1212–1221. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, R.H. Carcinoembryonic antigen. Ann. Intern. Med. 1986, 104, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, J.P.; Koehl, C.; Adloff, M. Carcinoembryonic antigen (cea) in diagosis and prognosis of colorectal carcinoma. Dis. Colon Rectum 1980, 23, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, M.J.; Mitchell, E.P. Carcinoembryonic antigen in the staging and follow-up of patients with colorectal cancer. Cancer Investig. 2005, 23, 338–351. [Google Scholar] [CrossRef]
- Meyerhardt, J.A.; Mangu, P.B.; Flynn, P.J.; Korde, L.; Loprinzi, C.L.; Minsky, B.D.; Petrelli, N.J.; Ryan, K.; Schrag, D.H.; Wong, S.L.; et al. Follow-up care, surveillance protocol, and secondary prevention measures for survivors of colorectal cancer: American society of clinical oncology clinical practice guideline endorsement. J. Clin. Oncol. 2013, 31, 4465–4470. [Google Scholar] [CrossRef] [PubMed]
- Pleskow, D.K.; Berger, H.J.; Gyves, J.; Allen, E.; McLean, A.; Podolsky, D.K. Evaluation of a serologic marker, ca19-9, in the diagnosis of pancreatic cancer. Ann. Intern. Med. 1989, 110, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Ćwik, G.; Wallner, G.; Skoczylas, T.; Ciechański, A.; Zinkiewicz, K. Cancer antigens 19-9 and 125 in the differential diagnosis of pancreatic mass lesions. Arch. Surg. 2006, 141, 968–973. [Google Scholar] [CrossRef] [PubMed]
- Paganuzzi, M.; Onetto, M.; Marroni, P.; Barone, D.; Conio, M.; Aste, H.; Pugliese, V. Ca 19-9 and Ca 50 in benign and malignant pancreatic and biliary diseases. Cancer 1988, 61, 2100–2108. [Google Scholar] [CrossRef]
- Maithel, S.K.; Maloney, S.; Winston, C.; Gönen, M.; D’Angelica, M.I.; DeMatteo, R.P.; Jarnagin, W.R.; Brennan, M.F.; Allen, P.J. Preoperative ca 19-9 and the yield of staging laparoscopy in patients with radiographically resectable pancreatic adenocarcinoma. Ann. Surg. Oncol. 2008, 15, 3512–3520. [Google Scholar] [CrossRef] [PubMed]
- Bartley, A.N.; Washington, M.K.; Colasacco, C.; Ventura, C.B.; Ismaila, N.; Benson, A.B., 3rd; Carrato, A.; Gulley, M.L.; Jain, D.; Kakar, S.; et al. HER2 testing and clinical decision making in gastroesophageal adenocarcinoma: Guideline from the college of american pathologists, american society for clinical pathology, and the american society of clinical oncology. J. Clin. Oncol. 2017, 35, 446–464. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-B.; Rhim, J.S.; Park, S.-C.; Kimm, S.-W.; Kraus, M.H. Amplification, overexpression, and rearrangement of the erbB-2 protooncogene in primary human stomach carcinomas. Cancer Res. 1989, 49, 6605–6609. [Google Scholar] [PubMed]
- Yonemura, Y.; Ninomiya, I.; Ohoyama, S.; Kimura, H.; Yamaguchi, A.; Fushida, S.; Kosaka, T.; Miwa, K.; Miyazaki, I.; Endou, Y.; et al. Expression of c-erbB-2 oncoprotein in gastric carcinoma. Immunoreactivity for c-erbB-2 protein is an independent indicator of poor short-term prognosis in patients with gastric carcinoma. Cancer 1991, 67, 2914–2918. [Google Scholar] [CrossRef]
- Yonemura, Y.; Yamaguchi, A.; Fushida, S.; Kimura, H.; Ohoyama, S.; Miyazaki, I.; Endou, Y.; Tanaka, M.; Sasaki, T. Evaluation of immunoreactivity for erbB-2 protein as a marker of poor short term prognosis in gastric cancer. Cancer Res. 1991, 51, 1034–1038. [Google Scholar] [PubMed]
- Yoon, H.H.; Shi, Q.; Sukov, W.R.; Wiktor, A.E.; Khan, M.; Sattler, C.A.; Grothey, A.; Wu, T.T.; Diasio, R.B.; Jenkins, R.B.; et al. Association of HER2/ERBB2 expression and gene amplification with pathologic features and prognosis in esophageal adenocarcinomas. Clin. Cancer Res. 2012, 18, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Bandla, S.; Godfrey, T.E.; Tan, D.; Luketich, J.D.; Pennathur, A.; Qiu, X.; Hicks, D.G.; Peters, J.H.; Zhou, Z. HER2 amplification, overexpression and score criteria in esophageal adenocarcinoma. Mod. Pathol. 2011, 24, 899–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Renzo, M.F.; Olivero, M.; Giacomini, A.; Porte, H.; Chastre, E.; Mirossay, L.; Nordlinger, B.; Bretti, S.; Bottardi, S.; Giordano, S.; et al. Overexpression and amplification of the MET/HGF receptor gene during the progression of colorectal cancer. Clin. Cancer Res. 1995, 1, 147–154. [Google Scholar] [PubMed]
- Bradley, C.A.; Salto-Tellez, M.; Laurent-Puig, P.; Bardelli, A.; Rolfo, C.; Tabernero, J.; Khawaja, H.A.; Lawler, M.; Johnston, P.G.; Van Schaeybroeck, S.; et al. Targeting c-MET in gastrointestinal tumours: Rationale, opportunities and challenges. Nat. Rev. Clin. Oncol. 2018, 15, 150. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, M.; Sawada, H.; Yamada, Y.; Watanabe, A.; Tatsumi, M.; Yamashita, J.; Matsuda, M.; Sakaguchi, T.; Hirao, T.; Nakano, H. The prognostic significance of amplification and overexpression of c-MET and c-ERB b-2 in human gastric carcinomas. Cancer 1999, 85, 1894–1902. [Google Scholar] [CrossRef]
- Yu, S.; Yu, Y.; Zhao, N.; Cui, J.; Li, W.; Liu, T. C-MET as a prognostic marker in gastric cancer: A systematic review and meta-analysis. PLoS ONE 2013, 8, e79137. [Google Scholar] [CrossRef] [PubMed]
- Ueki, T.; Fujimoto, J.; Suzuki, T.; Yamamoto, H.; Okamoto, E. Expression of hepatocyte growth factor and its receptor, the c-met proto-oncogene, in hepatocellular carcinoma. Hepatology 1997, 25, 619–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zucali, P.; Santoro, A.; Rodriguez-Lope, C.; Simonelli, M.; Camacho, L.; Senzer, N.; Bolondi, L.; Lamar, M.; Abbadessa, G.; Schwartz, B. Final results from ARQ 197-114: A phase ib safety trial evaluating ARQ 197 in cirrhotic patients (PTS) with hepatocellular carcinoma (HCC). J. Clin. Oncol. 2010, 28, 4137. [Google Scholar] [CrossRef]
- Rimassa, L.; Porta, C.; Borbath, I.; Daniele, B.; Salvagni, S.; Van Laethem, J.L.; Van Vlieberghe, H.; Trojan, J.; Kolligs, F.T.; Weiss, A. Tivantinib (ARQ 197) versus placebo in patients (PTS) with hepatocellular carcinoma (HCC) who failed one systemic therapy: Results of a randomized controlled phase II trial (RCT). Am. Soc. Clin. Oncol. 2012. [Google Scholar] [CrossRef]
- Goyal, L.; Muzumdar, M.D.; Zhu, A.X. Targeting the HGF/C-met pathway in hepatocellular carcinoma. Clin. Cancer Res. 2013, 19, 2310–2318. [Google Scholar] [CrossRef] [PubMed]
- Weekes, C.D.; Clark, J.W.; Zhu, A.X. Tivantinib for advanced hepatocellular carcinoma: Is met still a viable target? Lancet Oncol. 2018, 19, 591–592. [Google Scholar] [CrossRef]
- Rimassa, L.; Assenat, E.; Peck-Radosavljevic, M.; Pracht, M.; Zagonel, V.; Mathurin, P.; Rota Caremoli, E.; Porta, C.; Daniele, B.; Bolondi, L.; et al. Tivantinib for second-line treatment of met-high, advanced hepatocellular carcinoma (metiv-HCC): A final analysis of a phase 3, randomised, placebo-controlled study. Lancet Oncol. 2018, 19, 682–693. [Google Scholar] [CrossRef]
- Souglakos, J.; Philips, J.; Wang, R.; Marwah, S.; Silver, M.; Tzardi, M.; Silver, J.; Ogino, S.; Hooshmand, S.; Kwak, E.; et al. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br. J. Cancer 2009, 101, 465–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreyev, H.J.; Norman, A.R.; Cunningham, D.; Oates, J.R.; Clarke, P.A. Kirsten ras mutations in patients with colorectal cancer: The multicenter “rascal” study. J. Natl. Cancer Inst. 1998, 90, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.H.; Tougeron, D.; Shi, Q.; Alberts, S.R.; Mahoney, M.R.; Nelson, G.D.; Nair, S.G.; Thibodeau, S.N.; Goldberg, R.M.; Sargent, D.J.; et al. KRAS codon 12 and 13 mutations in relation to disease-free survival in braf–wild-type stage III colon cancers from an adjuvant chemotherapy trial (n0147 alliance). Clin. Cancer Res. 2014, 20, 3033–3043. [Google Scholar] [CrossRef] [PubMed]
- Misale, S.; Yaeger, R.; Hobor, S.; Scala, E.; Janakiraman, M.; Liska, D.; Valtorta, E.; Schiavo, R.; Buscarino, M.; Siravegna, G.; et al. Emergence of kras mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012, 486, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Tejpar, S.; Stintzing, S.; Ciardiello, F.; Tabernero, J.; Van Cutsem, E.; Beier, F.; Esser, R.; Lenz, H.J.; Heinemann, V. Prognostic and predictive relevance of primary tumor location in patients with ras wild-type metastatic colorectal cancer: Retrospective analyses of the crystal and fire-3 trials. JAMA Oncol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, N.; Nicolaides, N.C.; Wei, Y.F.; Ruben, S.M.; Carter, K.C.; Rosen, C.A.; Haseltine, W.A.; Fleischmann, R.D.; Fraser, C.M.; Adams, M.D.; et al. Mutation of a mutl homolog in hereditary colon cancer. Science 1994, 263, 1625–1629. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.C.; Rustgi, A.K. DNA mismatch repair and cancer. Gastroenterology 1995, 109, 1685–1699. [Google Scholar] [CrossRef]
- Farris, A.B., III; Demicco, E.G.; Le, L.P.; Finberg, K.E.; Miller, J.; Mandal, R.; Fukuoka, J.; Cohen, C.; Gaissert, H.A.; Zukerberg, L.R. Clinicopathologic and molecular profiles of microsatellite unstable barrett esophagus-associated adenocarcinoma. Am. J. Surg. Pathol. 2011, 35, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Smyth, E.C.; Wotherspoon, A.; Peckitt, C.; Gonzalez, D.; Hulkki-Wilson, S.; Eltahir, Z.; Fassan, M.; Rugge, M.; Valeri, N.; Okines, A.; et al. Mismatch repair deficiency, microsatellite instability, and survival: An exploratory analysis of the medical research council adjuvant gastric infusional chemotherapy (magic) trial. JAMA Oncol. 2017, 3, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Nakata, B.; Wang, Y.Q.; Yashiro, M.; Nishioka, N.; Tanaka, H.; Ohira, M.; Ishikawa, T.; Nishino, H.; Hirakawa, K. Prognostic value of microsatellite instability in resectable pancreatic cancer. Clin. Cancer Res. 2002, 8, 2536–2540. [Google Scholar] [PubMed]
- De la Chapelle, A. Microsatellite instability. N. Engl. J. Med. 2003, 349, 209–210. [Google Scholar] [CrossRef] [PubMed]
- Ribic, C.M.; Sargent, D.J.; Moore, M.J.; Thibodeau, S.N.; French, A.J.; Goldberg, R.M.; Hamilton, S.R.; Laurent-Puig, P.; Gryfe, R.; Shepherd, L.E.; et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N. Engl. J. Med. 2003, 349, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Sargent, D.J.; Marsoni, S.; Monges, G.; Thibodeau, S.N.; Labianca, R.; Hamilton, S.R.; French, A.J.; Kabat, B.; Foster, N.R.; Torri, V.; et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J. Clin. Oncol. 2010, 28, 3219–3226. [Google Scholar] [CrossRef] [PubMed]
- Viale, G.; Trapani, D.; Curigliano, G. Mismatch repair deficiency as a predictive biomarker for immunotherapy efficacy. BioMed Res. Int. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- FDA Approves First Cancer Treatment for Any Solid Tumor with a Specific Genetic Feature. Available online: https://www.fda.gov/newsevents/newsroom/pressannouncements/ucm560167.htm (accessed on 27 July 2018).
- Diaz, L.A.; Le, D.T.; Yoshino, T.; Andre, T.; Bendell, J.C.; Koshiji, M.; Zhang, Y.; Kang, S.P.; Lam, B.; Jaeger, D. Phase 3, open-label, randomized study of first-line pembrolizumab (pembro) vs. investigator-choice chemotherapy for mismatch repair-deficient (dmmr) or microsatellite instability-high (MSI-H) metastatic colorectal carcinoma (MCRC): Keynote-177. J. Clin. Oncol. 2017, 35. [Google Scholar] [CrossRef]
- Frank, A.S.; Qian, S.; Andrew, B.N.; Mody, K.; Levasseur, A.; Dueck, A.C.; Asha, R.D.; Christopher, H.L.; Deirdre, J.C.; Federico, I.R.; et al. Randomized trial of folfox alone or combined with atezolizumab as adjuvant therapy for patients with stage III colon cancer and deficient DNA mismatch repair or microsatellite instability (atomic, alliance a021502). J. Clin. Oncol. 2017, 35. [Google Scholar] [CrossRef]
- Dong, H.; Strome, S.E.; Salomao, D.R.; Tamura, H.; Hirano, F.; Flies, D.B.; Roche, P.C.; Lu, J.; Zhu, G.; Tamada, K.; et al. Tumor-associated B7-H1 promotes T-cell apoptosis: A potential mechanism of immune evasion. Nat. Med. 2002, 8, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.P.; Kurzrock, R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol. Cancer Ther. 2015, 14, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Zhu, Y.; Jiang, J.; Zhao, J.; Zhang, X.G.; Xu, N. Immunohistochemical localization of programmed death-1 ligand-1 (PD-l1) in gastric carcinoma and its clinical significance. Acta Histochem. 2006, 108, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Powles, T.; Eder, J.P.; Fine, G.D.; Braiteh, F.S.; Loriot, Y.; Cruz, C.; Bellmunt, J.; Burris, H.A.; Petrylak, D.P.; Teng, S.-L. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature 2014, 515, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Soria, J.-C.; Kowanetz, M.; Fine, G.D.; Hamid, O.; Gordon, M.S.; Sosman, J.A.; McDermott, D.F.; Powderly, J.D.; Gettinger, S.N.; et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014, 515, 563–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L. PD-L1 expression in pancreatic cancer. JNCI J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef] [PubMed]
- Lutz, E.R.; Wu, A.A.; Bigelow, E.; Sharma, R.; Mo, G.; Soares, K.; Solt, S.; Dorman, A.; Wamwea, A.; Yager, A.; et al. Immunotherapy converts nonimmunogenic pancreatic tumors into immunogenic foci of immune regulation. Cancer Immunol. Res. 2014, 2, 616–631. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Baas, P.; Kim, D.W.; Felip, E.; Perez-Gracia, J.L.; Han, J.Y.; Molina, J.; Kim, J.H.; Arvis, C.D.; Ahn, M.J.; et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (keynote-010): A randomised controlled trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef]
- Carbone, D.P.; Reck, M.; Paz-Ares, L.; Creelan, B.; Horn, L.; Steins, M.; Felip, E.; van den Heuvel, M.M.; Ciuleanu, T.-E.; Badin, F.; et al. First-line nivolumab in stage iv or recurrent non–small-cell lung cancer. N. Engl. J. Med. 2017, 376, 2415–2426. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, L.; Rodríguez-Abreu, D.; Gadgeel, S.; Esteban, E.; Felip, E.; De Angelis, F.; Domine, M.; Clingan, P.; Hochmair, M.J.; Powell, S.F.; et al. Pembrolizumab plus chemotherapy in metastatic non–small-cell lung cancer. N. Engl. J. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Eggermont, A.M.M.; Blank, C.U.; Mandala, M.; Long, G.V.; Atkinson, V.; Dalle, S.; Haydon, A.; Lichinitser, M.; Khattak, A.; Carlino, M.S.; et al. Adjuvant pembrolizumab versus placebo in resected stage iii melanoma. N. Engl. J. Med. 2018, 378, 1789–1801. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, M.D.; Ciuleanu, T.-E.; Pluzanski, A.; Lee, J.S.; Otterson, G.A.; Audigier-Valette, C.; Minenza, E.; Linardou, H.; Burgers, S.; Salman, P.; et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N. Engl. J. Med. 2018, 378, 2094–2104. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, C.S.; Doi, T.; Jang, R.W.; Muro, K.; Satoh, T.; Machado, M.; Sun, W.; Jalal, S.I.; Shah, M.A.; Metges, J.P.; et al. Safety and efficacy of pembrolizumab monotherapy in patients with previously treated advanced gastric and gastroesophageal junction cancer: Phase 2 clinical keynote-059 trial. JAMA Oncol. 2018, 4, e180013. [Google Scholar] [CrossRef] [PubMed]
- FDA Grants Accelerated Approval to Pembrolizumab for Advanced Gastric Cancer. Available online: https://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm577093.htm (accessed on 27 July 2018).
- Mahalingam, D. The coming of age: Immunotherapy in gastrointestinal malignancies. J. Gastrointest. Oncol. 2018, 9, 140–142. [Google Scholar] [CrossRef] [PubMed]
- El-Khoueiry, A.B.; Sangro, B.; Yau, T.; Crocenzi, T.S.; Kudo, M.; Hsu, C.; Kim, T.-Y.; Choo, S.-P.; Trojan, J.; Welling, T.H.; et al. Nivolumab in patients with advanced hepatocellular carcinoma (checkmate 040): An open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 2017, 389, 2492–2502. [Google Scholar] [CrossRef]
- Zhang, J.; Wolfgang, C.L.; Zheng, L. Precision immuno-oncology: Prospects of individualized immunotherapy for pancreatic cancer. Cancers (Basel) 2018, 10, 39. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, Z.; Tian, H.; Li, Y.; Li, M.; Zhao, W.; Zhang, C.; Wang, T.; Liu, J.; Zhang, A.; et al. Circulating MIC-1/GDF15 is a complementary screening biomarker with cea and correlates with liver metastasis and poor survival in colorectal cancer. Oncotarget 2017, 8, 24892–24901. [Google Scholar] [CrossRef] [PubMed]
- Tsao, M.; Kerr, K.; Yatabe, Y.; Hirsch, F.R. PL 03.03 blueprint 2: PD-L1 immunohistochemistry comparability study in real-life, clinical samples. J. Thorac. Oncol. 2017, 12, S1606. [Google Scholar] [CrossRef]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pagès, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Conejo-Garcia, J.R.; Katsaros, D.; Gimotty, P.A.; Massobrio, M.; Regnani, G.; Makrigiannakis, A.; Gray, H.; Schlienger, K.; Liebman, M.N.; et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 2003, 348, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Pages, F.; Sautes-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Amsen, D.; van Gisbergen, K.P.J.M.; Hombrink, P.; van Lier, R.A.W. Tissue-resident memory T cells at the center of immunity to solid tumors. Nat. Immunol. 2018, 19, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Iseki, Y.; Shibutani, M.; Maeda, K.; Nagahara, H.; Fukuoka, T.; Matsutani, S.; Kashiwagi, S.; Tanaka, H.; Hirakawa, K.; Ohira, M. A new method for evaluating tumor-infiltrating lymphocytes (TILs) in colorectal cancer using hematoxylin and eosin (he)-stained tumor sections. PLoS ONE 2018, 13, e0192744. [Google Scholar] [CrossRef] [PubMed]
- Seo, A.N.; Kang, B.W.; Kwon, O.K.; Park, K.B.; Lee, S.S.; Chung, H.Y.; Yu, W.; Bae, H.I.; Jeon, S.W.; Kang, H. Intratumoural PD-L1 expression is associated with worse survival of patients with epstein–barr virus-associated gastric cancer. Br. J. Cancer 2017, 117, 1753. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; He, J.-C.; Yang, Y.; Wang, J.-M.; Qian, Y.-W.; Yang, T.; Ji, L. The prognostic value of tumor-infiltrating lymphocytes in hepatocellular carcinoma: A systematic review and meta-analysis. Sci. Rep. 2017, 7, 7525. [Google Scholar] [CrossRef] [PubMed]
- Gataa, I.; Mezquita, L.; Auclin, E.; Le Moulec, S.; Alemany, P.; Kossai, M.; Massé, J.; Caramella, C.; Remon Masip, J.; Lahmar, J.; et al. 112PPathological evaluation of tumor infiltrating lymphocytes and the benefit of nivolumab in advanced non-small cell lung cancer (NSCLC). Ann. Oncol. 2017, 28. [Google Scholar] [CrossRef] [Green Version]
- Wallden, B.; Pekker, I.; Popa, S.; Dowidar, N.; Sullivan, A.; Hood, T.; Danaher, P.; Mashadi-Hossein, A.; Lunceford, J.K.; Marton, M.J.; et al. Development and analytical performance of a molecular diagnostic for anti-PD1 response on the ncounter dx analysis system. J. Clin. Oncol. 2016, 34. [Google Scholar]
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.R.; Albright, A.; Cheng, J.D.; Kang, S.P.; Shankaran, V.; et al. Ifn-γ–related mrna profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017, 127, 2930–2940. [Google Scholar] [CrossRef] [PubMed]
- Stromnes, I.M.; Hulbert, A.; Pierce, R.H.; Greenberg, P.D.; Hingorani, S.R. T-cell localization, activation, and clonal expansion in human pancreatic ductal adenocarcinoma. Cancer Immunol. Res. 2017, 5, 978–991. [Google Scholar] [CrossRef] [PubMed]
- DeNardo, D.G.; Brennan, D.J.; Rexhepaj, E.; Ruffell, B.; Shiao, S.L.; Madden, S.F.; Gallagher, W.M.; Wadhwani, N.; Keil, S.D.; Junaid, S.A.; et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011, 1, 54–67. [Google Scholar] [CrossRef] [PubMed]
- Ruffell, B.; Chang-Strachan, D.; Chan, V.; Rosenbusch, A.; Ho, C.M.; Pryer, N.; Daniel, D.; Hwang, E.S.; Rugo, H.S.; Coussens, L.M. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell 2014, 26, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Ugel, S.; De Sanctis, F.; Mandruzzato, S.; Bronte, V. Tumor-induced myeloid deviation: When myeloid-derived suppressor cells meet tumor-associated macrophages. J. Clin. Investig. 2015, 125, 3365–3376. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chang, E.W.; Wong, S.C.; Ong, S.M.; Chong, D.Q.; Ling, K.L. Increased myeloid-derived suppressor cells in gastric cancer correlate with cancer stage and plasma s100a8/a9 proinflammatory proteins. J. Immunol. 2013, 190, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Porembka, M.R.; Mitchem, J.B.; Belt, B.A.; Hsieh, C.-S.; Lee, H.-M.; Herndon, J.; Gillanders, W.E.; Linehan, D.C.; Goedegebuure, P. Pancreatic adenocarcinoma induces bone marrow mobilization of myeloid-derived suppressor cells which promote primary tumor growth. Cancer Immunol. Immunother. 2012, 61, 1373–1385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabitass, R.F.; Annels, N.E.; Stocken, D.D.; Pandha, H.A.; Middleton, G.W. Elevated myeloid-derived suppressor cells in pancreatic, esophageal and gastric cancer are an independent prognostic factor and are associated with significant elevation of the TH2 cytokine interleukin-13. Cancer Immunol. Immunother. 2011, 60, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- Iwata, T.; Kondo, Y.; Kimura, O.; Morosawa, T.; Fujisaka, Y.; Umetsu, T.; Kogure, T.; Inoue, J.; Nakagome, Y.; Shimosegawa, T. PD-L1+mdscs are increased in HCC patients and induced by soluble factor in the tumor microenvironment. Sci. Rep. 2016, 6, 39296. [Google Scholar] [CrossRef] [PubMed]
- Michor, F.; Iwasa, Y.; Lengauer, C.; Nowak, M.A. Dynamics of colorectal cancer. Semin. Cancer Biol. 2005, 15, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Richards, C.H.; McMillan, D.C.; Horgan, P.G.; Roxburgh, C.S.D. The relationship between tumour stroma percentage, the tumour microenvironment and survival in patients with primary operable colorectal cancer. Ann. Oncol. 2014, 25, 644–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.; Ham, I.-H.; Son, S.Y.; Han, S.-U.; Kim, Y.-B.; Hur, H. Intratumor stromal proportion predicts aggressive phenotype of gastric signet ring cell carcinomas. Gastric Cancer 2017, 20, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Huijbers, A.; Tollenaar, R.A.E.M.; Pelt, G.W.; Zeestraten, E.C.M.; Dutton, S.; McConkey, C.C.; Domingo, E.; Smit, V.T.H.B.M.; Midgley, R.; Warren, B.F.; et al. The proportion of tumor-stroma as a strong prognosticator for stage II and III colon cancer patients: Validation in the victor trial. Ann. Oncol. 2013, 24, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Ma, W.; Wang, J.; Yu, L.; Zhang, X.; Wang, Z.; Tan, B.; Wang, N.; Bai, B.; Yang, S.; et al. Tumor-stroma ratio is an independent predictor for survival in esophageal squamous cell carcinoma. J. Thorac. Oncol. 2012, 7, 1457–1461. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Grabsch, H.; Ivanova, T.; Tan, I.B.; Murray, J.; Ooi, C.H.; Wright, A.I.; West, N.P.; Hutchins, G.G.A.; Wu, J.; et al. Comprehensive genomic meta-analysis identifies intra-tumoural stroma as a predictor of survival in patients with gastric cancer. Gut 2013, 62, 1100–1111. [Google Scholar] [CrossRef] [PubMed]
- Tauriello, D.V.F.; Palomo-Ponce, S.; Stork, D.; Berenguer-Llergo, A.; Badia-Ramentol, J.; Iglesias, M.; Sevillano, M.; Ibiza, S.; Cañellas, A.; Hernando-Momblona, X.; et al. TGFB drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018, 554, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowery, M.A.; Jordan, E.J.; Basturk, O.; Ptashkin, R.N.; Zehir, A.; Berger, M.F.; Leach, T.; Herbst, B.; Askan, G.; Maynard, H.; et al. Real-time genomic profiling of pancreatic ductal adenocarcinoma: Potential actionability and correlation with clinical phenotype. Clin. Cancer Res. 2017, 23, 6094–6100. [Google Scholar] [CrossRef] [PubMed]
- Forde, P.M.; Chaft, J.E.; Smith, K.N.; Anagnostou, V.; Cottrell, T.R.; Hellmann, M.D.; Zahurak, M.; Yang, S.C.; Jones, D.R.; Broderick, S.; et al. Neoadjuvant PD-1 blockade in resectable lung cancer. N. Engl. J. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.E.; Xiu, J.; Weinberg, B.A.; El-Deiry, W.S.; Weiner, L.M.; Gatalica, Z.; Liu, Z.; El Ghazaly, H.; Xiao, N.; Hwang, J.J.; et al. Characterization of tumor mutation burden (TMB) in gastrointestinal (GI) cancers. J. Clin. Oncol. 2017, 35, 530. [Google Scholar] [CrossRef]
- Llosa, N.J.; Cruise, M.; Tam, A.; Wick, E.C.; Hechenbleikner, E.M.; Taube, J.M.; Blosser, L.; Fan, H.; Wang, H.; Luber, B.; et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015, 5, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Forbes, M.E.; Bitting, R.L.; O’Neill, S.S.; Chou, P.C.; Topaloglu, U.; Miller, L.D.; Hawkins, G.A.; Grant, S.C.; DeYoung, B.R.; et al. Incorporating blood-based liquid biopsy information into cancer staging: Time for a tnmb system? Ann. Oncol. 2018, 29, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.A., Jr.; Bardelli, A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014, 32, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Thiele, J.-A.; Bethel, K.; Králíčková, M.; Kuhn, P. Circulating tumor cells: Fluid surrogates of solid tumors. Annu. Rev.Pathol. 2017, 12, 419–447. [Google Scholar] [CrossRef] [PubMed]
- Reinert, T.; Scholer, L.V.; Thomsen, R.; Tobiasen, H.; Vang, S.; Nordentoft, I.; Lamy, P.; Kannerup, A.S.; Mortensen, F.V.; Stribolt, K.; et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016, 65, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Chan, C.W.; Chan, K.C.; Cheng, S.H.; Wong, J.; Wong, V.W.; Wong, G.L.; Chan, S.L.; Mok, T.S.; Chan, H.L.; et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl. Acad. Sci. USA 2015, 112, E1317–E1325. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Mussolin, B.; Buscarino, M.; Corti, G.; Cassingena, A.; Crisafulli, G.; Ponzetti, A.; Cremolini, C.; Amatu, A.; Lauricella, C.; et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 2015, 21, 795–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabel, L.; Riva, F.; Servois, V.; Livartowski, A.; Daniel, C.; Rampanou, A.; Lantz, O.; Romano, E.; Milder, M.; Buecher, B.; et al. Circulating tumor DNA changes for early monitoring of anti-pd1 immunotherapy: A proof-of-concept study. Ann. Oncol. 2017, 28, 1996–2001. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Zang, W.; Ge, Y.; Weygant, N.; Yu, P.; Li, L.; Rao, G.; Jiang, Z.; Yan, R.; He, L.; et al. RAS/BRAF circulating tumor DNA mutations as a predictor of response to first-line chemotherapy in metastatic colorectal cancer patients. Can. J. Gastroenterol. Hepatol. 2018, 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Balic, M.; Dandachi, N.; Hofmann, G.; Samonigg, H.; Loibner, H.; Obwaller, A.; van der Kooi, A.; Tibbe, A.G.; Doyle, G.V.; Terstappen, L.W.; et al. Comparison of two methods for enumerating circulating tumor cells in carcinoma patients. Cytom. B Clin. Cytom. 2005, 68, 25–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiroya, T.; Yuko, K. Circulating tumor cells in gastrointestinal cancer. J. Hepato-Biliary-Pancreat. Sci. 2010, 17, 577–582. [Google Scholar]
- Li, Y.; Gong, J.; Zhang, Q.; Lu, Z.; Gao, J.; Li, Y.; Cao, Y.; Shen, L. Dynamic monitoring of circulating tumour cells to evaluate therapeutic efficacy in advanced gastric cancer. Br. J. Cancer 2016, 114, 138–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sotelo, M.J.; Sastre, J.; Maestro, M.L.; Veganzones, S.; Viéitez, J.M.; Alonso, V.; Grávalos, C.; Escudero, P.; Vera, R.; Aranda, E.; et al. Role of circulating tumor cells as prognostic marker in resected stage iii colorectal cancer. Ann. Oncol. 2015, 26, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Sullivan, R.J.; Kalinich, M.; Kwan, T.T.; Giobbie-Hurder, A.; Pan, S.; LiCausi, J.A.; Milner, J.D.; Nieman, L.T.; Wittner, B.S.; et al. Molecular signatures of circulating melanoma cells for monitoring early response to immune checkpoint therapy. Proc. Natl. Acad. Sci. USA 2018, 115, 2467–2472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating tumor DNA analysis in patients with cancer: American society of clinical oncology and college of american pathologists joint review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Lee, M.; Jeffrey, S.S. Circulating tumor cells and circulating tumor DNA: Challenges and opportunities on the path to clinical utility. Clin. Cancer Res. 2015, 21, 4786–4800. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Théry, C. Communication by extracellular vesicles: Where we are and where we need to go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshioka, Y.; Kosaka, N.; Konishi, Y.; Ohta, H.; Okamoto, H.; Sonoda, H.; Nonaka, R.; Yamamoto, H.; Ishii, H.; Mori, M.; et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using exoscreen. Nat. Commun. 2014, 5, 3591. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating tumor DNA as a liquid biopsy for cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Kidess-Sigal, E.; Liu, H.E.; Triboulet, M.M.; Che, J.; Ramani, V.C.; Visser, B.C.; Poultsides, G.A.; Longacre, T.A.; Marziali, A.; Vysotskaia, V.; et al. Enumeration and targeted analysis of kras, BRAF and PIK3CA mutations in ctcs captured by a label-free platform: Comparison to ctdna and tissue in metastatic colorectal cancer. Oncotarget 2016, 7, 85349–85364. [Google Scholar] [CrossRef] [PubMed]
- Hardingham, J.E.; Grover, P.; Winter, M.; Hewett, P.J.; Price, T.J.; Thierry, B. Detection and clinical significance of circulating tumor cells in colorectal cancer—20 years of progress. Mol. Med. 2015, 21, S25–S31. [Google Scholar] [CrossRef] [PubMed]
- Lastraioli, E.; Iorio, J.; Arcangeli, A. Ion channel expression as promising cancer biomarker. Biochim. Biophys. Acta BBA 2015, 1848, 2685–2702. [Google Scholar] [CrossRef] [PubMed]
- Nagaraju, G.P.; Basha, R.; Rajitha, B.; Alese, O.B.; Alam, A.; Pattnaik, S.; El-Rayes, B. Aquaporins: Their role in gastrointestinal malignancies. Cancer Lett. 2016, 373, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Pelagalli, A.; Squillacioti, C.; Mirabella, N.; Meli, R. Aquaporins in Health and Disease: An Overview Focusing on the Gut of Different Species. Int. J. Mol. Sci. 2016, 17, 1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Molecule | Tumor Type | Implication |
|---|---|---|
| Tumor markers | ||
| CEA | Colorectal, gastric, and pancreatic cancers | Indicating residual disease, progressive, or recurrent disease |
| Measuring treatment response | ||
| CA19-9 | Pancreatic cancer | Indicating residual disease, progressive, or recurrent disease |
| Measuring treatment response | ||
| Targets of matched therapies | ||
| HER2 | Gastric or esophagogastric-junction cancers | Selecting for targeted therapy |
| KRAS | Colorectal, gastric, and pancreatic cancers | Predicting for treatment unresponsiveness |
| Mismatch repair Genes | ||
| MMR | Solid tumors | Predicting for treatment responsiveness |
| Biomarkers in tumor microenvironment | ||
| PD-L1 expression | Gastric cancer | Enriching patient population responding to anti-PD-1/PD-L1 therapies |
| Molecule | Genomic Alterations | Pathways Involved | Cancer types | Treatment |
|---|---|---|---|---|
| HER2 | Amplification/ overexpression | Activation of the MAPK and the PI3K/AKT axis | Gastric or esophagogastric-junction cancers | Monoclonal antibodies (e.g., cetuximab and trastuzumab) |
| c-MET | Amplification/ overexpression | Activation of GRB2-SOS–RAS–MAPK, the PI3K/AKT axis, and STAT3 pathway | Colorectal cancer, gastric cancer, pancreatic cancers and hepatocellular carcinoma | Monoclonal antibodies (e.g., rilotumumab, ficlatuzumab, and TAK-701); Tyrosine kinase inhibitors (e.g., tivantinib, cabozantinib, and crizotinib) |
| KRAS | Activating mutation within catalytic RAS domain | RAS–RAF–MEK | Colorectal cancer | Downstream pathway inhibitors (e.g., MEK inhibitors selumetinib and trametinib) |
| Molecule | Tumor Type | Implication |
|---|---|---|
| Biomarkers in tumor microenvironment | ||
| PD-L1 expression | Other cancer types, except gastric cancer | Enriching patient population responding to anti-PD-1/PD-L1 therapies |
| Tumor infiltrating lymphocyte | Colon and gastric cancers | Indicating good prognosis |
| Immunosuppressive myeloid cells | Pancreatic, hepatocellular, and gastric cancers | Indicating poor prognosis |
| Intratumoral stroma | Gastric, pancreatic, esophageal, and colon cancers | Indicating poor prognosis |
| Biomarkers in tumor genomics | ||
| Targeted gene panels | Pan-cancer | Selecting patients for targeted therapies |
| Mutational burden | Pan-cancer | Enriching patient population responding to anti-PD-1/PD-L1 therapies |
| Biomarkers in liquid biopsies | ||
| ctDNA/CTC/Exosomes | Pan-cancer | Indicating residual disease, progressive, or recurrent disease |
| Measuring treatment response | ||
| Approaches | Advantages | Disadvantages | References |
|---|---|---|---|
| ctDNA | Higher sensitivity; quick renew/short half-life; maintain tumor-specific genomic aberrations | Not suitable for functional assay, noises from normal cell-free DNA, challenges in methods’ standardization | [134,138,139] |
| CTC | Allow morphological/molecular/functional study; potentials for therapeutic targets | Low specificity, particularly in early stage setting; challenges in methods’ standardization limited capture techniques | [121,127,128,134,140] |
| Exosomes | Higher sensitivity; higher serum concentration; diverse EV contents; Potential for therapeutic targets | Isolation and purification of exosomes; specific exosome marker to identify subset of EVs; not suitable for functional assay; challenges in methods’ standardization | [135,136,137] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Quadri, S.; Wolfgang, C.L.; Zheng, L. New Development of Biomarkers for Gastrointestinal Cancers: From Neoplastic Cells to Tumor Microenvironment. Biomedicines 2018, 6, 87. https://doi.org/10.3390/biomedicines6030087
Zhang J, Quadri S, Wolfgang CL, Zheng L. New Development of Biomarkers for Gastrointestinal Cancers: From Neoplastic Cells to Tumor Microenvironment. Biomedicines. 2018; 6(3):87. https://doi.org/10.3390/biomedicines6030087
Chicago/Turabian StyleZhang, Jiajia, Shafat Quadri, Christopher L. Wolfgang, and Lei Zheng. 2018. "New Development of Biomarkers for Gastrointestinal Cancers: From Neoplastic Cells to Tumor Microenvironment" Biomedicines 6, no. 3: 87. https://doi.org/10.3390/biomedicines6030087





