Quantitative Proteomic Analysis of the Response to Zinc, Magnesium, and Calcium Deficiency in Specific Cell Types of Arabidopsis Roots
Abstract
:1. Introduction
2. Experimental Section
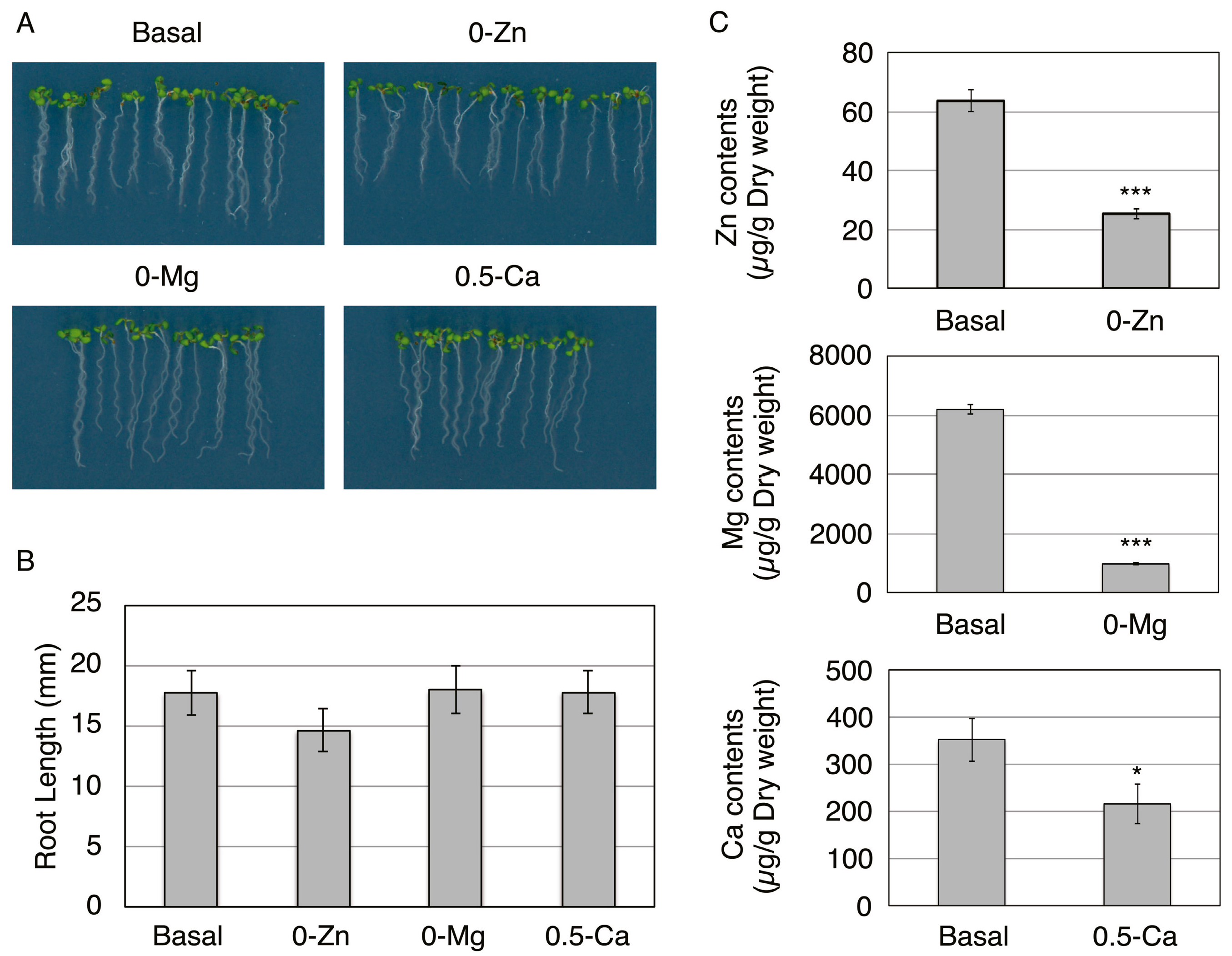
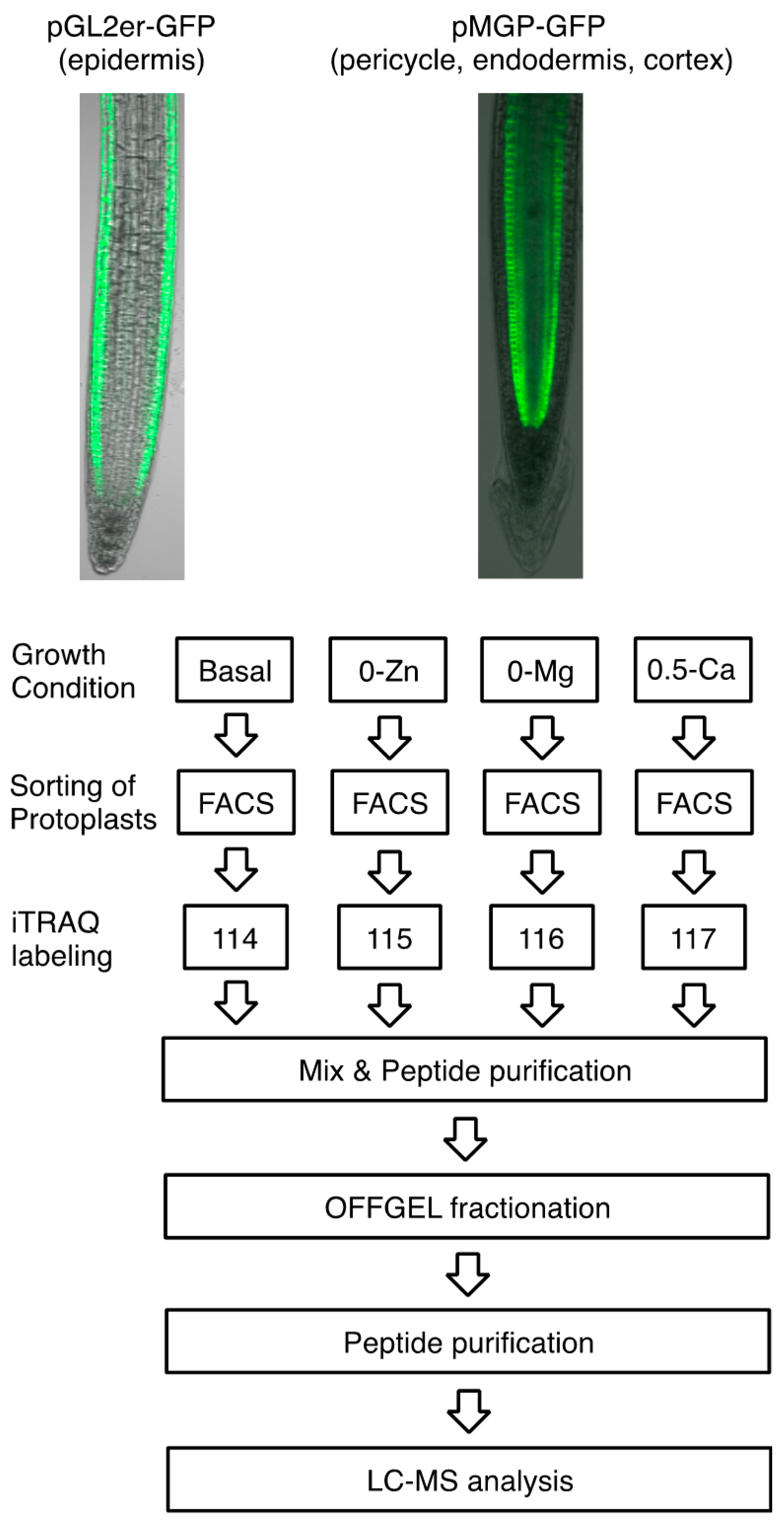
2.1. Plant Growth Condition and Preparation of Protoplasts
2.2. Composition of Basal, 0-Zn, 0-Mg, or 0.5-Ca Medium
2.3. Determination of Elemental Contents
2.4. iTRAQ-OFFGEL Fractionation Analysis
2.5. LC-MS/MS Analysis
2.6. Quantitative Real-Time RT-PCR Analysis
3. Results and Discussion
3.1. Responsive Proteins in the Epidermis or Pericycle, Endodermis, and Cortex to Zn, Mg, and Ca Deficiency

3.2. Overview of Proteins Strongly Downregulated by Zn, Mg, or Ca Deficiency in Epidermal Cells
| AGI Code | Protein | 0-Zn/basal (115/114) | 0-Mg/basal (116/114) | 0.5-Ca/basal (117/114) |
|---|---|---|---|---|
| AT1G48920 | nucleolin like 1 | 1.63 ± 0.24 | 2.03 ± 0.28 | 1.71 ± 0.14 |
| AT4G21960 | Peroxidase superfamily protein | 1.12 ± 0.18 | 2.29 ± 0.47 | 1.73 ± 0.37 |
| AT5G62190 | DEAD box RNA helicase (PRH75) | 1.79 ± 0.27 | 2.23 ± 0.38 | 1.81 ± 0.16 |
| AGI Code | Protein | 0-Zn/basal (115/114) | 0-Mg/basal (116/114) | 0.5-Ca/basal (117/114) |
|---|---|---|---|---|
| AT1G01820 | peroxin 11c | 0.40 ± 0.57 | 0.68 ± 0.65 | 0.38 ± 0.54 |
| AT1G07810 | ER-type Ca2+-ATPase 1 | 0.43 ± 0.74 | 0.45 ± 0.77 | 0.58 ± 1.01 |
| AT1G08830 | copper/zinc superoxide dismutase 1 | 0.46 ± 0.11 | 0.90 ± 0.23 | 1.17 ± 0.23 |
| AT1G12240 | Glycosyl hydrolases family 32 protein | 0.43 ± 0.61 | 0.30 ± 0.42 | 0.57 ± 0.81 |
| AT1G14000 | VH1-interacting kinase | 0.53 ± 0.47 | 0.41 ± 0.71 | N.D. |
| AT1G33490 | unknown protein | 0.62 ± 0.87 | 0.59 ± 0.84 | 0.42 ± 0.60 |
| AT1G35160 | GF14 protein phi chain | 0.33 ± 0.45 | 0.36 ± 0.39 | 0.49 ± 0.69 |
| AT1G35580 | cytosolic invertase 1 | 0.36 ± 0.51 | 0.53 ± 0.75 | 0.49 ± 0.69 |
| AT1G47420 | succinate dehydrogenase 5 | 0.50 ± 0.71 | 0.47 ± 0.66 | 0.51 ± 0.72 |
| AT1G49670 | ARP protein (REF) | 0.37 ± 0.52 | 0.63 ± 0.89 | 0.23 ± 0.32 |
| AT1G71410 | ARM repeat superfamily protein | 0.39 ± 0.67 | 0.38 ± 0.65 | 0.49 ± 0.85 |
| AT1G75330 | ornithine carbamoyltransferase | 0.50 ± 0.70 | 0.61 ± 0.86 | 0.59 ± 0.83 |
| AT1G76550 | Phosphofructokinase family protein | 0.49 ± 0.70 | 0.49 ± 0.70 | 0.46 ± 0.65 |
| AT1G77590 | long chain acyl-CoA synthetase 9 | 0.41 ± 0.58 | N.D. | N.D. |
| AT1G78800 | UDP-Glycosyltransferase superfamily protein | 0.82 ± 0.74 | 0.34 ± 0.58 | 1.00 ± 0.89 |
| AT1G80600 | HOPW1-1-interacting 1 | 0.36 ± 0.62 | 0.83 ± 0.72 | 0.69 ± 0.64 |
| AT2G04305 | Magnesium transporter CorA-like family protein | N.D. | 0.32 ± 0.46 | N.D. |
| AT2G17130 | isocitrate dehydrogenase subunit 2 | 0.47 ± 0.41 | 0.44 ± 0.63 | 0.63 ± 0.89 |
| AT2G28190 | copper/zinc superoxide dismutase 2 | N.D. | 0.40 ± 0.69 | 0.35 ± 0.60 |
| AT2G28520 | vacuolar proton ATPase A1 | 0.47 ± 0.66 | 0.44 ± 0.63 | 0.50 ± 0.71 |
| AT2G42260 | uv-b-insensitive 4 | 0.45 ± 0.64 | 0.54 ± 0.76 | 0.56 ± 0.79 |
| AT2G42500 | protein phosphatase 2A-3 | 0.41 ± 0.58 | 0.42 ± 0.60 | 0.63 ± 0.89 |
| AT2G42810 | protein phosphatase 5.2 | 0.24 ± 0.42 | 0.56 ± 0.97 | 0.69 ± 1.19 |
| AT2G45540 | WD-40 repeat family protein / beige-related | 0.42 ± 0.59 | 0.59 ± 0.83 | 0.60 ± 0.54 |
| AT3G05280 | Integral membrane Yip1 family protein | 0.65 ± 0.23 | 0.82 ± 0.08 | 0.40 ± 0.57 |
| AT3G13235 | ubiquitin family protein | 0.33 ± 0.57 | 0.92 ± 0.95 | 0.82 ± 0.77 |
| AT3G15090 | GroES-like zinc-binding alcohol dehydrogenase family protein | 0.46 ± 0.65 | 0.66 ± 0.93 | 0.54 ± 0.76 |
| AT3G15980 | Coatomer; beta’ subunit | 0.42 ± 0.60 | 1.17 ± 0.21 | 0.96 ± 0.00 |
| AT3G23180 | HR-like lesion-inducing protein-related | 0.42 ± 0.59 | 0.54 ± 0.76 | 0.47 ± 0.67 |
| AT3G28740 | Cytochrome P450 superfamily protein | 0.79 ± 0.79 | 0.45 ± 0.42 | 0.61 ± 0.57 |
| AT3G44340 | clone eighty-four | 0.36 ± 0.50 | 0.40 ± 0.57 | 0.33 ± 0.47 |
| AT3G48560 | chlorsulfuron/imidazolinone resistant 1 | 0.41 ± 0.71 | 0.61 ± 1.05 | 0.54 ± 0.94 |
| AT3G50590 | Transducin/WD40 repeat-like superfamily protein | 0.40 ± 0.57 | 0.51 ± 0.73 | 0.38 ± 0.54 |
| AT3G52140 | tetratricopeptide repeat (TPR)-containing protein | 0.17 ± 0.29 | 0.71 ± 0.64 | 0.67 ± 0.63 |
| AT4G05530 | indole-3-butyric acid response 1 | 0.78 ± 0.75 | 0.75 ± 0.72 | 0.46 ± 0.80 |
| AT4G14950 | SNARE associated Golgi protein family | 0.65 ± 0.57 | 0.51 ± 0.44 | 0.37 ± 0.63 |
| AT4G16630 | DEA(D/H)-box RNA helicase family protein | 0.38 ± 0.65 | 0.27 ± 0.47 | 0.27 ± 0.47 |
| AT5G14600 | S-adenosyl-l-methionine-dependent methyltransferases superfamily protein | 0.40 ± 0.56 | 0.84 ± 0.20 | N.D. |
| AT5G54370 | Late embryogenesis abundant (LEA) protein-related | 0.38 ± 0.66 | 0.36 ± 0.62 | 0.31 ± 0.54 |
| AT5G54640 | Histone superfamily protein | 0.47 ± 0.66 | 0.62 ± 0.88 | 0.60 ± 0.85 |
| AT5G65810 | unknown protein | 0.60 ± 1.04 | 0.37 ± 0.65 | 0.27 ± 0.48 |
3.3. Transcriptional Response of Members of the 14-3-3 Protein Family in Epidermal or Pericycle, Endodermis, and Cortex Cells to Zn, Mg, or Ca Deficiency
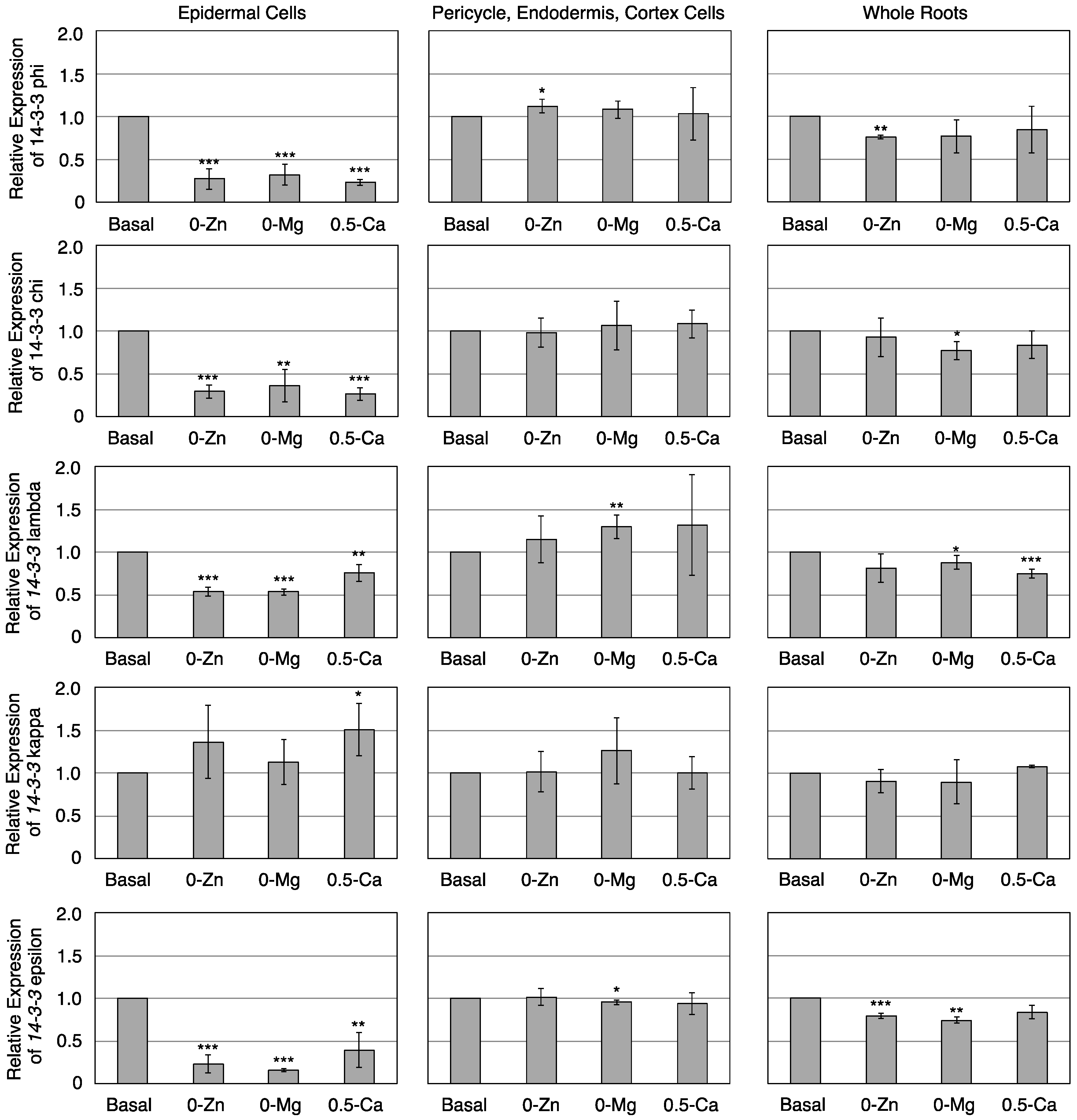
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
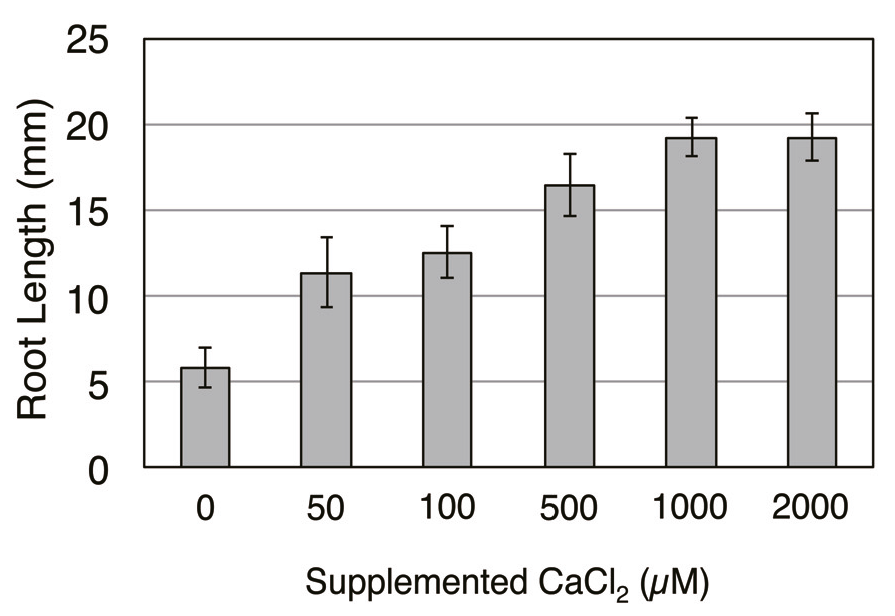
Appendix
References
- Baker, M.A.; Hetherington, L.; Reeves, G.M.; Aitken, R.J. The mouse sperm proteome characterizedvia IPG strip prefractionation and LC-MS/MS identification. Proteomics 2008, 8, 1720–1730. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kou, Z.; Jing, Z.; Zhang, Y.; Guo, X.; Dong, M.; Wilmut, I.; Gao, S. Proteome of mouse oocytes at different developmental stages. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 17639–17644. [Google Scholar] [CrossRef] [PubMed]
- Wasbrough, E.R.; Dorus, S.; Hester, S.; Howard-Murkin, J.; Lilley, K.; Wilkin, E.; Polpitiya, A.; Petritis, K.; Karr, T.L. The Drosophila melanogaster sperm proteome-II (DmSP-II). J. Proteomics 2010, 73, 2171–2185. [Google Scholar] [CrossRef] [PubMed]
- Chik, J.K.; Schriemer, D.C.; Childs, S.J.; McGhee, J.D. Proteome of the Caenorhabditis elegans Oocyte. J. Proteome Res. 2011, 10, 2300–2305. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, M.J.; Siatkowski, M.; Paudel, Y.; Balbach, S.T.; Baeumer, N.; Crosetto, N.; Drexler, H.C.; Fuellen, G.; Boiani, M. Proteomic analysis of mouse oocytes reveals 28 candidate factors of the “Reprogrammome”. J. Proteome Res. 2011, 1, 2140–2153. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, W.; Stanley, B.A.; Assmann, S.M. Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 2008, 20, 3210–3226. [Google Scholar] [CrossRef] [PubMed]
- Grobei, M.A.; Qeli, E.; Brunner, E.; Rehrauer, H.; Zhang, R.; Roschitzki, B.; Basler, K.; Ahrens, C.H.; Grossniklaus, U. Deterministic protein inference for shotgun proteomics data provides new insights into Arabidopsis pollen development and function. Genome Res. 2009, 19, 1786–1800. [Google Scholar] [CrossRef] [PubMed]
- Petricka, J.J.; Schauer, M.A.; Megraw, M.; Breakfield, N.W.; Thompson, J.W.; Georgiev, S.; Soderblom, E.J.; Ohler, U.; Moseley, M.A.; Grossniklaus, U.; et al. The protein expression landscape of the Arabidopsis root. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 6811–6818. [Google Scholar] [CrossRef] [PubMed]
- Fukao, Y.; Yoshida, M.; Kurata, R.; Kobayashi, M.; Nakanishi, M.; Fujiwara, M.; Nakajima, K.; Ferjani, A. Peptide separation methodologies for in-depth proteomics in Arabidopsis. Plant Cell Physiol. 2013, 54, 808–815. [Google Scholar] [CrossRef] [PubMed]
- Dunkley, T.P.J.; Dupree, P.; Watson, R.B.; Lille, K.S. The use of isotope-coded affinity tags (ICAT) to study organelle proteomes in Arabidopsis thaliana. Biochem. Soc. Trans. 2004, 110, 520–523. [Google Scholar] [CrossRef] [PubMed]
- Rudella, A.; Friso, G.; Alonso, J.M.; Ecker, J.R.; van Wijk, K.J. Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell 2006, 18, 1704–1721. [Google Scholar] [CrossRef] [PubMed]
- Fukao, Y.; Ferjani, A.; Tomioka, R.; Nagasaki, N.; Kurata, R.; Nishimori, Y.; Fujiwara, M.; Maeshima, M. iTRAQ analysis reveals mechanisms of growth defects due to excess zinc in Arabidopsis. Plant Physiol. 2011, 155, 1893–1907. [Google Scholar] [CrossRef] [PubMed]
- Gruhler, A.; Olsen, J.V.; Mohammed, S.; Mortensen, P.; Færgeman, N.J.; Mann, M.; Jensen, O.N. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 2005, 4, 310–327. [Google Scholar] [CrossRef] [PubMed]
- Schütz, W.; Hausmann, N.; Krug, K.; Hampp, R.; Macek, B. Extending SILAC to proteomics of plant cell lines. Plant Cell 2011, 23, 1701–1705. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, D.; ten Have, S.; Hodge, K.; Tillemans, V.; Lamond, A.I.; Brown, J.W.S. Plant SILAC: Stable-isotope labelling with amino acids of Arabidopsis seedlings for quantitative proteomics. PLoS ONE 2013, 8, e72207. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Kato, M.; Tomioka, R.; Kurata, R.; Fukao, Y.; Aoyama, T.; Maeshima, M. Characteristics of a root hair-less line of Arabidopsis thaliana under physiological stresses. J. Exp. Bot. 2014, 65, 1497–1512. [Google Scholar] [CrossRef] [PubMed]
- Jorrín, J.V.; Maldonado, A.M.; Castillejo, M.A. Plant proteome analysis: A 2006 update. Proteomics 2007, 7, 2947–2962. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.K.; Sarkar, A.; Righetti, P.G.; Pedreschi, R.; Carpentier, S.; Wang, T.; Barkla, B.J.; Kohli, A.; Ndimba, B.K.; Bykova, N.V.; et al. A decade of plant proteomics and mass spectrometry: Translation of technical advancements to food security and safety issues. Mass. Spec. Rev. 2013, 32, 335–365. [Google Scholar] [CrossRef] [PubMed]
- Zargar, S.; Kurata, R.; Inaba, S.; Fukao, Y. Unraveling the iron deficiency responsive proteome in Arabidopsis shoot by iTRAQ-OFFGEL approach. Plant Signal. Behav. 2013, 9, e26892. [Google Scholar] [CrossRef]
- Waki, T.; Miyashima, S.; Nakanishi, M.; Ikeda, Y.; Hashimoto, T.; Nakajima, K. A GAL4-based targeted activation tagging system in Arabidopsis thaliana. Plant J. 2012, 73, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Miyashima, S.; Hashimoto, T.; Nakajima, K. ARGONAUTE1 acts in Arabidopsis root radial pattern formation independently of the SHR/SCR pathway. Plant Cell Physiol. 2009, 50, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Hörth, P.; Miller, C.A.; Preckel, T.; Wenz, C. Efficient fractionation and improved protein identification by peptide OFFGEL electrophoresis. Mol. Cell. Proteomics 2006, 5, 1968–1974. [Google Scholar] [CrossRef] [PubMed]
- Barberon, M.; Geldner, N. Radial transport of nutrients: the plant root as a polarized epithelium. Plant Physiol. 2014, 166, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Robbins, N.E.; Trontin, C.; Duan, L.; Dinneny, J.R. Beyond the barrier: communication in the root through the endodermis. Plant Physiol. 2014, 166, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Hosmani, P.S.; Kamiya, T.; Danku, J.; Naseer, S.; Geldner, N.; Guerinot, M.L.; Salt, D.E. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 14498–14503. [Google Scholar] [CrossRef] [PubMed]
- Baxter, I.; Hosmani, P.S.; Rus, A.; Lahner, B.; Borevitz, J.O.; Muthukumar, B.; Mickelbart, M.V.; Schreiber, L.; Franke, R.B.; Salt, D.E. Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis. PLoS Genet. 2009, 5, e1000492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volkening, K.; Leystra-Lantz, C.; Yang, W.; Jaffee, H.; Strong, M.J. Tar DNA binding protein of 43 kDa (TDP-43); 14-3-3 proteins and copper/zinc superoxide dismutase (SOD1) interact to modulate NFL mRNA stability. Implications for altered RNA processing in amyotrophic lateral sclerosis (ALS). Brain Res. 2009, 1305, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Garvin, D.F.; Kochian, L.V. Rapid induction of regulatory and transporter genes in response to phosphorus; potassium; and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol. 2002, 130, 1361–1370. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.G.; Pyo, Y.J.; Cho, J.W.; Cho, M.H. Comparative proteome analysis of differentially expressed proteins induced by K+ deficiency in Arabidopsis thaliana. Proteomics 2004, 4, 3549–3559. [Google Scholar] [CrossRef] [PubMed]
- DeLille, J.M.; Sehnke, P.C.; Ferl, R.J. The Arabidopsis 14-3-3 family of signaling regulators. Plant Physiol. 2001, 126, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, D.; Morris, E.R.; Walker, J.C. 14-3-3 and FHA Domains Mediate Phosphoprotein Interactions. Annu. Rev. Plant Biol. 2009, 60, 67–91. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.; JHuber, J.L.; Liao, P.C.; Gage, D.A.; Huber, S.C. The inhibitor protein of phosphorylated nitrate reductase from spinach (Spinacia oleracea) leaves is a 14-3-3 protein. FEBS Lett. 1996, 387, 127–131. [Google Scholar] [CrossRef]
- Palmgren, M.G. Powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 817–847. [Google Scholar] [CrossRef] [PubMed]
- Duby, G.; Boutry, M. The plant plasma membrane proton pump ATPase: A highly regulated P-type ATPase with multiple physiological roles. Eur. J. Physiol. 2008, 457, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Sottocornola, B.; Visconti, S.; Orsi, S.; Gazzarrini, S.; Giacometti, S.; Olivari, C.; Camoni, L.; Aducci, P.; Marra, M.; Abenavoli, A.; et al. The potassium channel KAT1 is activated by plant and animal 14-3-3 proteins. J. Biol. Chem. 2006, 281, 35735–35741. [Google Scholar] [CrossRef] [PubMed]
- Latz, A.; Becker, D.; Hekman, M.; Müller, T.; Beyhl, D.; Marten, I.; Eing, C.; Fischer, A.; Dunkel, M.; Bertl, A.; et al. TPK1; a Ca2+-regulated Arabidopsis vacuole two-pore K+ channel is activated by 14-3-3 proteins. Plant J. 2007, 52, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Voelker, C.; Gomez-Porras, J.L.; Becker, D.; Hamamoto, S.; Uozumi, N.; Gambale, F.; Mueller-Roeber, B.; Czempinski, K.; Dreyer, I. Roles of tandem-pore K+ channels in plants—a puzzle still to be solved. Plant Biol. 2010, 12, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Chang, I.F.; Curran, A.; Woolsey, R.; Quilici, D.; Cushman, J.C.; Mittler, R.; Harmon, A.; Harper, J.F. Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 2009, 9, 2967–2985. [Google Scholar] [CrossRef] [PubMed]
- Arabidopsis Interactome Mapping Consortium. Evidence for network evolution in an Arabidopsis interactome map. Science 2011, 333, 601–607. [Google Scholar]
- Jaspert, N.; Throm, C.; Oecking, C. Arabidopsis 14-3-3 proteins: fascinating and less fascinating aspects. Front. Plant Sci. 2011, 7, 2947–2962. [Google Scholar] [CrossRef] [PubMed]
- Shin, R.; Jez, J.M.; Basra, A.; Zhang, B.; Schachtman, D.P. 14-3-3 Proteins fine-tune plant nutrient metabolism. FEBS Lett. 2011, 585, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.L.; Chen, W.W.; Chen, L.Q.; Qin, C.; Jin, C.W.; Shi, Y.Z.; Zheng, S.J. The 14-3-3 protein GENERAL REGULATORY FACTOR11 (GRF11) acts downstream of nitric oxide to regulate iron acquisition in Arabidopsis thaliana. New Phytol. 2012, 197, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Cao, A.; Jain, A.; Baldwin, J.C.; Raghothama, K.G. Phosphate differentially regulates 14-3-3 family members and GRF9 plays a role in Pi-starvation induced responses. Planta 2007, 226, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Huber, S.C. Polycations globally enhance binding of 14-3-3ω to target proteins in spinach leaves. Plant Cell Physiol. 2006, 47, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Manak, M.S.; Ferl, R.J. Divalent cation effects on interactions between multiple Arabidopsis 14-3-3 isoforms and phosphopeptide targets. Biochemistry 2007, 46, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, K.; Shasha, D.E.; Wang, J.Y.; Jung, J.W.; Lambert, G.M.; Galbraith, D.W.; Benfey, P.N. A gene expression map of the Arabidopsis root. Science 2003, 302, 1956–1960. [Google Scholar] [CrossRef] [PubMed]
- Dinneny, J.R.; Long, T.A.; Wang, J.Y.; Jung, J.W.; Mace, D.; Pointer, S.; Barron, C.; Brady, S.M.; Schiefelbein, J.; Benfey, P.N. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 2008, 320, 942–945. [Google Scholar] [CrossRef] [PubMed]
- Gifford, M.L.; Dean, A.; Gutierrez, R.A.; Coruzzi, G.M.; Birnbaum, K.D. Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.K.; Girke, T.; Pasala, S.; Xie, M.; Reddy, G.V. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl. Acad. Sci. U.S.A. 2009, 105, 4941–4946. [Google Scholar] [CrossRef] [PubMed]
- Grønlund, J.T.; Eyres, A.; Kumar, S.; Buchanan-Wollaston, V.; Gifford, M.L. Cell specific analysis of Arabidopsis leaves using fluorescence activated cell sorting. JoVE 2012, e4214. [Google Scholar] [CrossRef] [PubMed]
- Petersson, S.V.; Johansson, A.I.; Kowalczyk, M.; Makoveychuk, A.; Wang, J.Y.; Moritz, T.; Grebe, M.; Benfey, P.N.; Sandberg, G.; Ljung, K. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 2009, 21, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Rogers, E.D.; Jackson, T.; Moussaieff, A.; Aharoni, A.; Benfey, P.N. Cell type-specific transcriptional profiling: implications for metabolite profiling. Plant J. 2012, 70, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Woll, K.; Borsuk, L.A.; Stransky, H.; Nettleton, D.; Schnable, P.S.; Hochholdinger, F. Isolation; characterization; and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiol. 2005, 139, 1255–1267. [Google Scholar] [CrossRef] [PubMed]
- Dembinsky, D.; Woll, K.; Saleem, M.; Liu, Y.; Fu, Y.; Borsuk, L.A.; Lamkemeyer, T.; Fladerer, C.; Madlung, J.; Barbazuk, B.; et al. Transcriptomic and proteomic analyses of pericycle cells of the maize primary root. Plant Physiol. 2007, 145, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; von Behrens, I.; Muthreich, N.; Schütz, W.; Nordheim, A.; Hochholdinger, F. Regulation of the pericycle proteome in maize (Zea mays L.) primary roots by RUM1 which is required for lateral root initiation. Eur. J. Cell Biol. 2010, 89, 236–241. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fukao, Y.; Kobayashi, M.; Zargar, S.M.; Kurata, R.; Fukui, R.; Mori, I.C.; Ogata, Y. Quantitative Proteomic Analysis of the Response to Zinc, Magnesium, and Calcium Deficiency in Specific Cell Types of Arabidopsis Roots. Proteomes 2016, 4, 1. https://doi.org/10.3390/proteomes4010001
Fukao Y, Kobayashi M, Zargar SM, Kurata R, Fukui R, Mori IC, Ogata Y. Quantitative Proteomic Analysis of the Response to Zinc, Magnesium, and Calcium Deficiency in Specific Cell Types of Arabidopsis Roots. Proteomes. 2016; 4(1):1. https://doi.org/10.3390/proteomes4010001
Chicago/Turabian StyleFukao, Yoichiro, Mami Kobayashi, Sajad Majeed Zargar, Rie Kurata, Risa Fukui, Izumi C. Mori, and Yoshiyuki Ogata. 2016. "Quantitative Proteomic Analysis of the Response to Zinc, Magnesium, and Calcium Deficiency in Specific Cell Types of Arabidopsis Roots" Proteomes 4, no. 1: 1. https://doi.org/10.3390/proteomes4010001





