3.1. Effects of 1 GeV/n 48Ti Ions on HSPC-Derived Myeloid Colonies Collected at One Week Post-irradiation
For all exposed groups, a total of 1988 unique and non-redundant proteins were identified with ≥90% confidence (
Supplementary data A). Among the 1988 identified, a total of 1107 proteins were differentially expressed based on the one-way analysis of variance (ANOVA,
p < 0.01). For the multiple comparisons among groups (the Holm-Sidak method, with significant differences at
p < 0.05), we found that: (a) HSPC-derived myeloid progenitors obtained from mice exposed to 0.1 Gy, a total of 976 proteins were down-regulated, while only one protein up-regulated; (b) HSPC-derived myeloid progenitors obtained from mice exposed to 0.25 Gy, a total of 33 proteins were down-regulated while 14 proteins were upregulated; and (c) HSPC-derived myeloid progenitors obtained from mice exposed to 0.5 Gy, a total of 58 proteins were down-regulated while 54 proteins were up-regulated. Details of these proteins can be found in the
Supplementary Data A.
Table 1 summarizes the Ingenuity Pathway Analyses (IPA) analyses of identified proteins in each treatment group, as compared to the non-irradiated sham control group. These are top diseases and disorders, top molecular and cellular functions, top physiological system development and function, and top network. The results indicated that proteins involved in different diseases and disorders were found in each treatment group. The IPA analyses also presented various proteins with different molecular and cellular functions. Moreover, the IPA analyses provided information on the top networks involved in responses to 1 GeV/n
48Ti ions. Although more than one network was found for each dose of 1 GeV/n at one week post-exposure, we are presenting only the network with the highest score for each dose of 1 GeV/n
48Ti ions (as shown in
Table 1).
Figure 2,
Figure 3 and
Figure 4 show the top molecular networks detected by the IPA analyses in HSPC-derived myeloid cells after exposure of mice to 0.1, or 0.25, or 0.5Gy of 1 GeV/n
48Ti ions, respectively. The description of each IPA symbol has been previously described [
35].
In each figure, only proteins that are considered to be focus proteins (those shown in filled symbols) will be discussed. The focus proteins are from the uploaded list and are available for generating networks. Other proteins appearing in the network are added by the IPA analysis software because the IPA database indicates that they interact with the focus proteins.
Table 1.
The summary obtained from IPA analyses of identified proteins in HSPC-derived myeloid cells collected at one week after exposure of mice to various doses of 1 GeV/n 48Ti ions.
Table 1.
The summary obtained from IPA analyses of identified proteins in HSPC-derived myeloid cells collected at one week after exposure of mice to various doses of 1 GeV/n 48Ti ions.
| Radiation Dose (Gy) | Top Diseases and Disorders | Top Molecular and Cellular Functions | Top Physiological System Development and Function | Top Network |
|---|
| 0.1 | Infectious disease | Cell death and survival | Organismal survival | Hereditary Disorder, Metabolic Disease, Neurological Disease (score = 53) |
| Neurological disease | RNA post-transcriptional modification | Immune cell trafficking |
| Psychological disorders | Cellular growth and proliferation | Hematological system development and function |
| Skeletal and muscular disorders | Protein Synthesis | Connective tissue development and function |
| Cancer | Nucleic acid metabolism | Tissue development |
| 0.25 | Connective tissue disorders | Lipid metabolism | Hematological system development and function | Lipid metabolism, small molecule biochemistry, connective tissue disorders (score = 36) |
| Dermatological diseases and conditions | Small molecule biochemistry | Immune cell trafficking |
| Developmental disorders | Cell death and survival | Skeletal and muscular system |
| Hereditary disorder | Cellular movement | Tissue development |
| Metabolic diseases | Amino acid metabolism | Digestive system development and function |
| 0.5 | Cancer | Post-translational modification | Organismal development development and function | Dermatological diseases and conditions, connective tissue disorders, developmental disorder (score = 52) |
| Hematological disease | Protein folding | Cardiovascular system |
| Immunological disease | Cell morphology | Connective tissue development and function |
| Dermatological diseases and conditions | Cellular assembly and organization | Embryonic development |
| Connective tissue disorders | Cell death and survival | Cell death and survival |
Figure 2 shows the IPA-analysis top network in HSPC-derived myeloid colonies collected at 1 week post-irradiation in response to 0.1 Gy of
48Ti ions. This network is related to heredity disorder, metabolic diseases, and neurological disease (score = 53). The solid lines represent the direct molecular interaction among proteins in the network. There are 35 focus proteins (those proteins that are from uploaded list and pass filters are applied). All of these proteins are shown in the filled IPA symbols. These proteins are associated with cell death and survival, RNA post-transcriptional modification, cellular growth and proliferation, protein synthesis, and nucleic acid metabolism). All of these 35 focus proteins shown in the network were down regulated and connected to ubiquitin C (UBC). Hence, the decreased levels of these proteins are likely to be due to ubiquination, the process known to be associated with protein degradation [
36]. Of note, it is known that ubiquitination is important for several biological processes. These include: DNA repair, cell cycle, cell survival, differentiation, kinase modification, endocytosis, and regulation of other cell signaling pathways [
37].
Figure 2.
Ingenuity Pathway Analysis showing the top molecular Network in HSPC-derived myeloid cells collected at 1 week after exposure of mice to 0.1 Gy of 48Ti ions. This network is involved hereditary disorder, metabolic diseases, and neurological disease. It is constructed by 35 focus proteins: AABRACL (ABRA C-Terminal Like), ADPGK (ADP-Dependent Glucokinase), CNDP2 (CNDP Dipeptidase 2 (Metallopeptidase M20 Family), COLGALT1 (Collagen Beta1-O-Galactosyltransferase 1), DMXL1 (Dmx-Like 1), DOPEY2 (Dopey Family Member 2), EMC4 (ER Membrane Protein Complex Subunit 4), FAM136A (Family With Sequence Similarity 136, Member A), FAM3C (Family With Sequence Similarity 3, Member C), FAM49A (Family With Sequence Similarity 49, Member A), GLOD4 (Glyoxalase Domain Containing 4), GORASP2 (Golgi Reassembly Stacking Protein 2, 55kDa), GSDMD (Gasdermin D), HEBP1 (Heme Binding Protein 1), HINT2 (Histidine Triad Nucleotide Binding Protein 2), HMHA1 (Histocompatibility- Minor-HA-1), MPST (Mercaptopyruvate Sulfurtransferase), NAA38 (N-Alpha-Acetyltransferase 38, NatC Auxiliary Subunit), NANS (N-Acetylneuraminic Acid Synthase), NCLN (Nicalin), NDUFC2 (NADH Dehydrogenase (Ubiquinone) 1, Subcomplex Unknown 2, 14.5kDa), NOMO1 (NODAL Modulator 1), PITRM1 (Pitrilysin Metallopeptidase 1), PROSC (Proline Synthetase Co-Transcribed Homolog Bacterial), PRR12 (Proline Rich 12), F3B5 (Splicing Factor 3b, Subunit 5, 10kDa), TM9SF3 (Transmembrane 9 Superfamily Member 3), TMED7 (Transmembrane Emp24 Protein Transport Domain Containing 7), TMX1 (Thioredoxin-related transmembrane protein 1), TMX4 (Thioredoxin-related transmembrane protein 4), TSFM (Ts Translation Elongation Factor, Mitochondrial), UBC (Ubiquitin C), VPS13D (Vacuolar Protein Sorting 13 Homolog D), VWA5A (Von Willebrand Factor A Domain Containing 5A), and ZNF407 (Zinc Finger Protein 407). All of the focus proteins are down-regulated (blue IPA symbols).
Figure 2.
Ingenuity Pathway Analysis showing the top molecular Network in HSPC-derived myeloid cells collected at 1 week after exposure of mice to 0.1 Gy of 48Ti ions. This network is involved hereditary disorder, metabolic diseases, and neurological disease. It is constructed by 35 focus proteins: AABRACL (ABRA C-Terminal Like), ADPGK (ADP-Dependent Glucokinase), CNDP2 (CNDP Dipeptidase 2 (Metallopeptidase M20 Family), COLGALT1 (Collagen Beta1-O-Galactosyltransferase 1), DMXL1 (Dmx-Like 1), DOPEY2 (Dopey Family Member 2), EMC4 (ER Membrane Protein Complex Subunit 4), FAM136A (Family With Sequence Similarity 136, Member A), FAM3C (Family With Sequence Similarity 3, Member C), FAM49A (Family With Sequence Similarity 49, Member A), GLOD4 (Glyoxalase Domain Containing 4), GORASP2 (Golgi Reassembly Stacking Protein 2, 55kDa), GSDMD (Gasdermin D), HEBP1 (Heme Binding Protein 1), HINT2 (Histidine Triad Nucleotide Binding Protein 2), HMHA1 (Histocompatibility- Minor-HA-1), MPST (Mercaptopyruvate Sulfurtransferase), NAA38 (N-Alpha-Acetyltransferase 38, NatC Auxiliary Subunit), NANS (N-Acetylneuraminic Acid Synthase), NCLN (Nicalin), NDUFC2 (NADH Dehydrogenase (Ubiquinone) 1, Subcomplex Unknown 2, 14.5kDa), NOMO1 (NODAL Modulator 1), PITRM1 (Pitrilysin Metallopeptidase 1), PROSC (Proline Synthetase Co-Transcribed Homolog Bacterial), PRR12 (Proline Rich 12), F3B5 (Splicing Factor 3b, Subunit 5, 10kDa), TM9SF3 (Transmembrane 9 Superfamily Member 3), TMED7 (Transmembrane Emp24 Protein Transport Domain Containing 7), TMX1 (Thioredoxin-related transmembrane protein 1), TMX4 (Thioredoxin-related transmembrane protein 4), TSFM (Ts Translation Elongation Factor, Mitochondrial), UBC (Ubiquitin C), VPS13D (Vacuolar Protein Sorting 13 Homolog D), VWA5A (Von Willebrand Factor A Domain Containing 5A), and ZNF407 (Zinc Finger Protein 407). All of the focus proteins are down-regulated (blue IPA symbols).
![Proteomes 03 00132 g002]()
Figure 3.
Ingenuity Pathway Analysis showing the top molecular network in HSPC-derived myeloid cells collected at 1 week after exposure of mice to 0.25 Gy of 48Ti ions. This network is involved lipid metabolism, small molecule biochemistry, and connective tissue disorders and developmental disorder. This network is constructed with 35 proteins: ALB (Albumin), ALOX5 (Arachidonate 5-Lipoxygenase), ANXA4 (Annexin A4), APOH (Apolipoprotein H-Beta-2-Glycoprotein I), ARHGAP1 (Rho GTPase Activating Protein 1), ARHGAP8 (Rho GTPase Activating Protein 8), PRR5-ARHGAP8 (PRR5-ARHGAP8 Readthrough), ATPIF1 (ATPase Inhibitory Factor 1), CA1 (Carbonic Anhydrase I), CEACAM8 (Carcinoembryonic Antigen-Related Cell Adhesion Molecule 8), CLIP2 (CAP-GLY Domain Containing Linker Protein 2), COL1A2 (Collagen, Type I, Alpha 2), CSE1L (CSE1 Chromosome Segregation 1-Like), CSTA (Cystatin A or known as Stefin A), DUSP8 (Dual Specificity Phosphatase 8), DYNLL1 (Dynein, Light Chain, LC8-Type 1), EHD4 (EH-Domain Containing 4), FLII (Flightless I Homolog), Hsp90 (Heat Shock Protein 90), IgG (Immunoglobulin G), IGHG2 (Immunoglobulin Heavy Constant Gamma 2-G2m Marker), IL19 (Interleukin 19), Il8r (Interleukin 8 Receptor beta), Jnk (c-Jun N-terminal protein kinases), MPO (Myeloperoxidase),MYLK3 (Myosin Light Chain Kinase 3), NF-κB (Nuclear Factor kappa B), MAPK (Mitogen-activated protein kinases), PF4 (Platelet Factor 4), PI3K (Phosphoinositide-3-kinase), SLC2A3 (Solute Carrier Family 2 (Facilitated Glucose Transporter), Member 3), SPTAN1 (Spectrin, Alpha, Non-Erythrocytic 1), VEGF (Vascular endothelial growth factor), WDR34 (WD Repeat Domain 34). Among these, there are only 15 focus proteins (those in bold and shown in filled IPA symbols in the figure). Seven of the 15 focus proteins are down-regulated (those shown in blue IPA symbols), while eight of the 15 focus proteins are up-regulated (those shown in red IPA symbols).
Figure 3.
Ingenuity Pathway Analysis showing the top molecular network in HSPC-derived myeloid cells collected at 1 week after exposure of mice to 0.25 Gy of 48Ti ions. This network is involved lipid metabolism, small molecule biochemistry, and connective tissue disorders and developmental disorder. This network is constructed with 35 proteins: ALB (Albumin), ALOX5 (Arachidonate 5-Lipoxygenase), ANXA4 (Annexin A4), APOH (Apolipoprotein H-Beta-2-Glycoprotein I), ARHGAP1 (Rho GTPase Activating Protein 1), ARHGAP8 (Rho GTPase Activating Protein 8), PRR5-ARHGAP8 (PRR5-ARHGAP8 Readthrough), ATPIF1 (ATPase Inhibitory Factor 1), CA1 (Carbonic Anhydrase I), CEACAM8 (Carcinoembryonic Antigen-Related Cell Adhesion Molecule 8), CLIP2 (CAP-GLY Domain Containing Linker Protein 2), COL1A2 (Collagen, Type I, Alpha 2), CSE1L (CSE1 Chromosome Segregation 1-Like), CSTA (Cystatin A or known as Stefin A), DUSP8 (Dual Specificity Phosphatase 8), DYNLL1 (Dynein, Light Chain, LC8-Type 1), EHD4 (EH-Domain Containing 4), FLII (Flightless I Homolog), Hsp90 (Heat Shock Protein 90), IgG (Immunoglobulin G), IGHG2 (Immunoglobulin Heavy Constant Gamma 2-G2m Marker), IL19 (Interleukin 19), Il8r (Interleukin 8 Receptor beta), Jnk (c-Jun N-terminal protein kinases), MPO (Myeloperoxidase),MYLK3 (Myosin Light Chain Kinase 3), NF-κB (Nuclear Factor kappa B), MAPK (Mitogen-activated protein kinases), PF4 (Platelet Factor 4), PI3K (Phosphoinositide-3-kinase), SLC2A3 (Solute Carrier Family 2 (Facilitated Glucose Transporter), Member 3), SPTAN1 (Spectrin, Alpha, Non-Erythrocytic 1), VEGF (Vascular endothelial growth factor), WDR34 (WD Repeat Domain 34). Among these, there are only 15 focus proteins (those in bold and shown in filled IPA symbols in the figure). Seven of the 15 focus proteins are down-regulated (those shown in blue IPA symbols), while eight of the 15 focus proteins are up-regulated (those shown in red IPA symbols).
![Proteomes 03 00132 g003]()
Figure 3 shows the IPA-analysis top network (score = 36) in HSPC-derived myeloid colonies collected at 1 week post-irradiation in response to 0.25 Gy of 1 GeV/n
48Ti ions. This network is involved lipid metabolism, small molecule biochemistry, and connective tissue disorders. The solid and dotted lines represent the direct and indirect molecular interaction among proteins in the network, respectively. Although there are 35 proteins in the network (
Figure 3), only 15 proteins (those in filled IPA symbols) are uploaded from the list (focus proteins). Eight of these 15 proteins (shown in red color symbols) showed higher levels of expression than the control level. These proteins are albumim (ALB), arachidonate 5-lipoxygenase (ALOX5), carbonic anhydrase 1 (CA1), cellular apoptosis susceptible 1 (CSE1), EH domain-containing protein 4 (EHD4), flightless-II (FLII), platelet factor 4 (PF4), and solute carrier family 2 member 3 (facilitated glucose transporter 3) (SLC2A3).
Highly expression of ALB and CA were previously reported in BM cells of CBA/Ca or C57BL/6 mice exposed to γ-rays [
15]. Our study is the first to report a high level of expression of ALOX5 protein in HSPC-derived myeloid colonies obtained from mice exposed to space radiation, although expression of ALOX5 is known to be critical regulator of chronic ML stem cells, both in humans and mice [
38]. Taken together, these findings suggest that expression of ALOX5 after irradiation may play an important role in radiation-induced late health effects.
Figure 3 also shows that an increased expression level of ALOX5 has a positive impact on the expression of heat shock protein 90 (Hsp90). Highly expressed SLC2A3 has been detected in several types of solid tumors such as colon [
39] and brain [
40] cancers, although its role in hematopoietic neoplasms has not been reported. It should be noted that these up-regulated proteins are associated, either directly or indirectly, to the nuclear factor-kappa B (NF-κB) complex, an important transcription factor involved in cell survival [
41] and cell death [
42] after exposure to radiation, depending on certain circumstances and cell type. Kinase activities from several sources facilitate such interaction: P38-mitogen-activated protein kinases (p38-MAPK), or phosphoinositide-3 (PI-3) kinase, or c-Jun N-terminal kinase (Jnk), or extracellular signal-regulated protein kinase (ERK).
Figure 3 also shows seven focused proteins that their levels of expression were down-regulated, in relation with the control level 9Those in blue color). These proteins are: Annexin A4 (ANXA4), RHO GTPase-activator protein 1 (ARHGP1 or RHO 1), ATPase inhibitory factor 1 (ATPIF1), collagen, Type I, Alpha 2 (COL1A2), cystatin A (CSTA), dynein light chain 1 (DYNLL1), and spectrin (SPTAN1). The majority of down-regulated proteins are involved in cell death, cell survival, and cell proliferation (e.g., ANXA4, COL1A2, and CSTA). The results from IPA analysis also show that NF-κB, Jnk, ERK, and Hsp90 are involved in the down-regulation of these proteins.
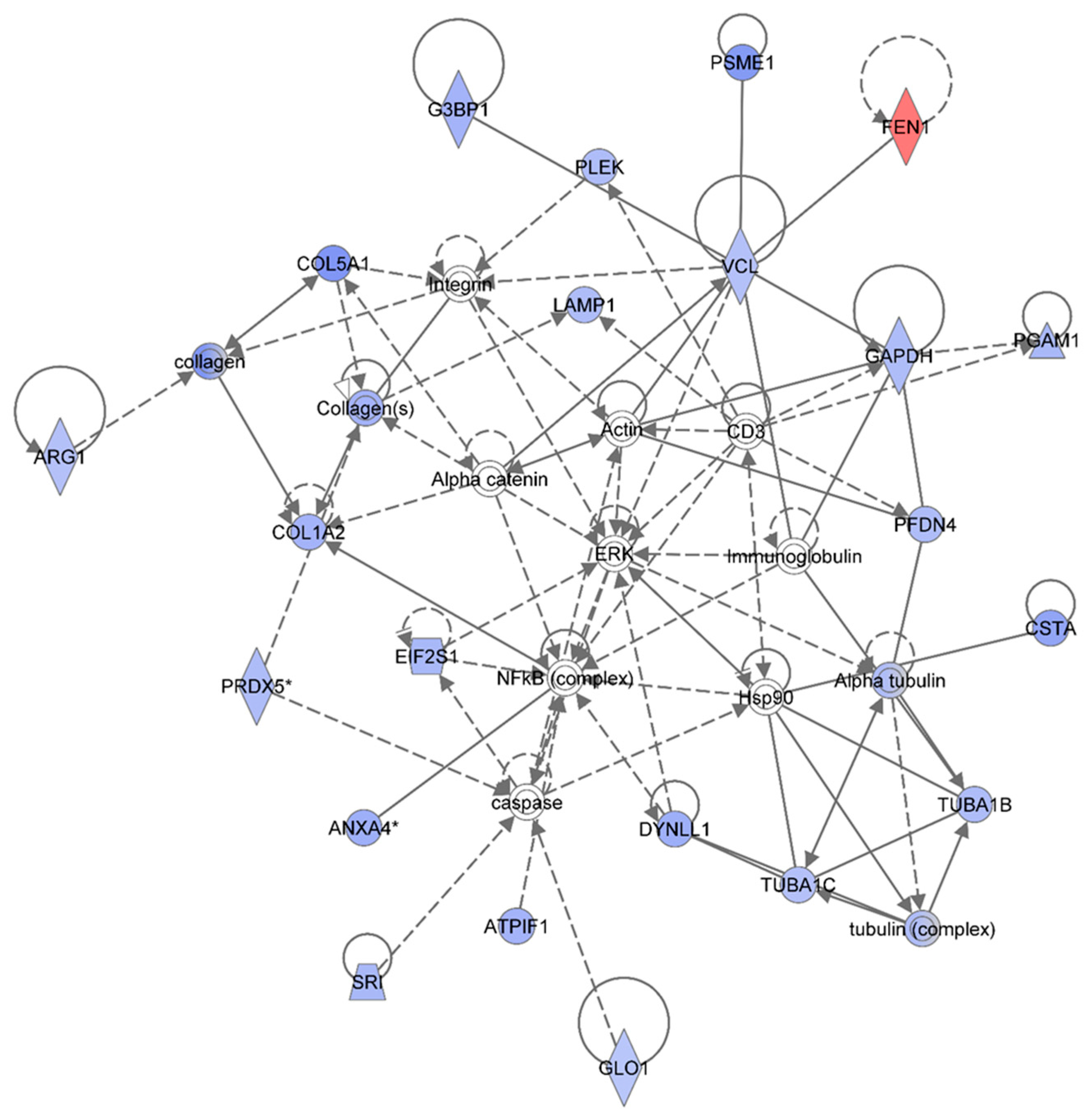
Figure 4 shows the IPA-analysis top network (score = 52) in HSPC-derived myeloid colonies collected at 1 wk post-exposure in response to 0.5 Gy of 1 GeV/n
48Ti ions. The IPA analysis indicated that proteins in this network are involved dermatological diseases and conditions, connective tissue disorders, developmental disorder. Similar to
Figure 3, the solid and dotted lines represent the direct and indirect molecular interaction among proteins in the network, respectively. Although there are 35 proteins shown in the network, only 21 proteins were the focus of this network (those in filled IPA symbols). Twenty of the focused proteins were down-regulated (those in blue symbols). Among these, five proteins associated with cell growth, cell survival and cell proliferation (
i.e., ANXA4, ATPIF1, COL1A2, CSTA, and DYNLL1) were also down-regulated in the 0.25 Gy-exposed mice previously shown in
Figure 3. Several of the other down-regulated proteins were linked to actin, connective tissue (collagens) and cytoskeleton (tubulin) complexes.
Figure 4.
Ingenuity Pathway Analysis showing the top molecular network in HSPC-derived myeloid cells collected at 1 week after exposure of mice to 0.25 Gy of 48Ti ions. This network is involved in dermatological diseases and conditions, connective tissue disorders, and development disorder. It consists of 35 proteins: Actin, Alpha catenin, Alpha tubulin, ANXA4 (Annexin A4), ARG1 (Arginase 1), ATPIF1 (ATPase Inhibitory Factor 1), COL1A2 (Collagen, Type I, Alpha 2), COL5A1 (Collagen, Type V, Alpha 1), CSTA (Cystatin A , also known as Stefin A), DYNLL1 (Dynein, Light Chain, LC8-Type 1, EIF2S1 (Eukaryotic Translation Initiation Factor 2, Subunit 1 Alpha, 35kDa), FEN1 (Flap Structure-Specific Endonuclease 1), G3BP1 (GTPase Activating Protein (SH3 Domain) Binding Protein 1), GAPDH (Glyceraldehyde-3-Phosphate Dehydrogenase), GLO1 (Glyoxalase I), LAMP1 (Lysosomal-Associated Membrane Protein 1), PFDN4 (Prefoldin Subunit 4), PGAM1 (Phosphoglycerate Mutase 1), PLEK (Pleckstrin), PRDX5 (Peroxiredoxin 5), PSME1 (Proteasome (Prosome, Macropain) Activator Subunit 1-PA28 Alpha), SRI (Sorcin), TUBA1B (Tubulin, Alpha 1b), TUBA1C (Tubulin, Alpha 1c), tubulin complex, and VCL (Vinculin). Among these, there are only 21 focus proteins (those in bold and shown in filled IPA symbols shown in the figure). Twenty out of 21 focus proteins are down-regulated (those in blue IPA symbols). Only one focus protein is up-regulated (shown in red IPA symbol).
Figure 4.
Ingenuity Pathway Analysis showing the top molecular network in HSPC-derived myeloid cells collected at 1 week after exposure of mice to 0.25 Gy of 48Ti ions. This network is involved in dermatological diseases and conditions, connective tissue disorders, and development disorder. It consists of 35 proteins: Actin, Alpha catenin, Alpha tubulin, ANXA4 (Annexin A4), ARG1 (Arginase 1), ATPIF1 (ATPase Inhibitory Factor 1), COL1A2 (Collagen, Type I, Alpha 2), COL5A1 (Collagen, Type V, Alpha 1), CSTA (Cystatin A , also known as Stefin A), DYNLL1 (Dynein, Light Chain, LC8-Type 1, EIF2S1 (Eukaryotic Translation Initiation Factor 2, Subunit 1 Alpha, 35kDa), FEN1 (Flap Structure-Specific Endonuclease 1), G3BP1 (GTPase Activating Protein (SH3 Domain) Binding Protein 1), GAPDH (Glyceraldehyde-3-Phosphate Dehydrogenase), GLO1 (Glyoxalase I), LAMP1 (Lysosomal-Associated Membrane Protein 1), PFDN4 (Prefoldin Subunit 4), PGAM1 (Phosphoglycerate Mutase 1), PLEK (Pleckstrin), PRDX5 (Peroxiredoxin 5), PSME1 (Proteasome (Prosome, Macropain) Activator Subunit 1-PA28 Alpha), SRI (Sorcin), TUBA1B (Tubulin, Alpha 1b), TUBA1C (Tubulin, Alpha 1c), tubulin complex, and VCL (Vinculin). Among these, there are only 21 focus proteins (those in bold and shown in filled IPA symbols shown in the figure). Twenty out of 21 focus proteins are down-regulated (those in blue IPA symbols). Only one focus protein is up-regulated (shown in red IPA symbol).
![Proteomes 03 00132 g004]()
Proteins that are linked to actin are: Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), GTPase activating protein SH3 domain (G3BP1), phosphoglycerate mutase 1 (PGAM1), prefoldin (PFDN4), pleckstrin (PLEK), proteasome activator complex subunit 1 (PMSE1), and vinculin (VCL). These actin-associated proteins are also associated with embryonic development. The IPA results display a direct link among VCL, alpha-catenin, and actin that regulates cell-cell adhesion to ensure the stabilization of cell structure for synchronization of normal development. Hence, a reduction in these proteins after irradiation would disturb the homeostasis of the function of cells. The IPA results also show an association of VCLwith alpha-catenin, actin, and the collagen compartment (connective tissue) of the network in which several proteins are involved. Proteins involved in the collagen compartment are: Arginase (ARG1), COL1A2, collagen Type V Alpha 1 (COL5A1), lysosomal-associated membrane protein 1 (LAMP1, a protein regulates translation process), and peroxiredoxin (PRDX5). Normally, PRDX5 protein (an antioxidant enzyme) protects cells from radiation-induced death by suppression of redox-sensitive caspase activation [
43]. Hence, a reduction in the level of PRDX5 in HSPC-derived myeloid colonies after exposure of mice to
48Ti ions is indicative of a higher level of cell death (via the caspase pathway). It is known that caspases are important for maintaining homeostasis through regulating cell death and inflammation [
44]. Hence, dysregulation of caspases would trigger the induction of diseases such as cancer and inflammatory disorders.
The IPA analysis also reveals the down-regulation of sorcin (SRI, a calcium-binding protein) and glyoxalase 1-defense-enzyme (GLO1). These two proteins are also involved in defense mechanisms or detoxification through suppression of caspase activity. Overall, the results indicate that exposure to 48Ti ions diminishes the expression of several proteins involved in cell death and survival, mediated through suppression of caspase activity. It should be noted that caspase is linked to Hsp90, which interacts with the tubulin family (the cytoskeleton protein) that are involved in maintaining intracellular organization and normal cell movement. The IPA analysis shows that expression levels of four types of tubulins in HSPC-derived myeloid cells were reduced after exposure of mice to 48Ti ions. These proteins are: Alpha tubulin which directly binds to cysteine protease inhibitor (CSTA), tubulin alpha 1 C (TUBA1C), tubulin alpha 1B (TUBA1B), and tubulin complex (a heterodimer of tubulins alpha and beta that constitutes the promoter for microtubule assembly). Importantly, it should be noted that these down-regulated proteins are associated (either directly or indirectly) with the NF-κB complex and ERK, indicating the importance of these two signaling pathways after exposure to radiation.
One striking finding is that only one protein is up-regulated in this network,
i.e., flap structure specific endonuclease 1 (FEN1), shown in the red symbol. This protein is an enzyme involved in the base excision repair system, and its abnormality has been shown to be associated with tumor progression of mouse models with microsatellite instability [
39]. Of note, FEN1 directly binds to VCL, an actin-binding, for its endonuclease activity. As mentioned previously, the level of VCL is decreased in HSPC-derived myeloid cells collected at 1 week after exposure of mice to
48Ti ions. Hence, it is possible that although the level of expression of FEN1 is high, its activity is defective since its protein partner,
i.e., VCL, is unavailable. This would result in an insufficient DNA repair process that, in turn, induces higher levels of damage.
3.2. Effects of 1 GeV/n 48Ti Ions on HSPC-Derived Myeloid Colonies Collected at 6 Months Post-irradiation
For all exposed groups, a total of 1988 unique and non-redundant proteins were identified with ≥90% confidence (
Supplemental data A). Among the 1988 identified, a total of 578 proteins were differentially expressed based on the one-way analysis of variance (ANOVA,
p < 0.01). For the multiple comparisons among groups (the Holm-Sidak method, with significant differences at
p < 0.05), we found that: (a) HSPC-derived myeloid progenitors obtained from mice exposed to 0.1 Gy, a total of 12 proteins are down-regulated, while no up-regulated protein; (b) HSPC-derived myeloid progenitors obtained from mice exposed to 0.25 Gy, three proteins are down-regulated while 10 proteins are up-regulated; and (c) HSPC-derived myeloid progenitors obtained from mice exposed to 0.5 Gy, a total of two proteins were down-regulated while 80 proteins are up-regulated. Details of up- and down-regulated proteins found in each treatment group can be found in the
Supplemental Data B.
Table 2 summarizes the Ingenuity Pathway Analyses (IPA) of identified proteins in each treatment group, as compared to the non-irradiated sham control group. These are top diseases and disorders, top molecular and cellular functions, top physiological system development and function, and top network. Similar to the data from the 1-week harvest time-point, the results indicate different proteins were found in each treatment group. The IPA analyses also presented proteins with different molecular and cellular functions. Moreover, the IPA analyses provided information on the top molecular networks involved in responses to different doses of 1 GeV/n
48Ti ions. Similar to the data for the 1-week harvest time-point, we are presenting only the molecular network with the highest score for each dose of 1 GeV/n
48Ti ions.
Figure 5,
Figure 6 and
Figure 7 show the top molecular networks detected by the IPA analyses in HSPC-derived myeloid cells collected at 6 mos after exposure of mice to 0.1, or 0.25, or 0.5Gy of 1 GeV/n
48Ti ions, respectively. As previously mentioned, the description of each IPA symbol has been described [
35].
Table 2.
The summary obtained from IPA analyses of identified proteins in HSPC-derived myeloid cells collected at six months after exposure of mice to various doses of 1GeV/n 48Ti ions.
Table 2.
The summary obtained from IPA analyses of identified proteins in HSPC-derived myeloid cells collected at six months after exposure of mice to various doses of 1GeV/n 48Ti ions.
| Radiation Dose (Gy) | Top Diseases and Disorders | Top Molecular and Cellular Functions | Top Physiological System Development and Function | Top Networks |
|---|
| 0.1 | Connective tissue disorders | Cellular movement | Connective tissue development and function | Cancer, tumor morphology, cardiac necrosis/cell death (score = 29) |
| Dermatological diseases and conditions | Cellular growth and proliferation | Embryonic development |
| Hereditary disorder | Lipid metabolism | Hematological system development and function |
| Immunological disease | Small molecule biochemistry | Organismal development |
| Inflammatory disease | Cell to cell signaling and interaction | Tissue development |
| 0.25 | Cancer | Carbohydrate metabolism | Cardiovascular system development and function | Cell death and survival, cell cycle, connective tissue development and function (score = 25) |
| Connective tissue disorders | Cell cycle | Hair and skin development and function |
| Dermatological diseases and conditions | Cell death and survival | Hepatic system development and function |
| Developmental disorders | Cell morphology | Tissue development |
| Hereditary disorder | Cell-to-cell signaling and interaction | Tissue morphology |
| 0.5 | Cancer | Ribonucleic acid (RNA) post-transcriptional modification | Digestive system development and function | Cancer, hematological disease, immunological disease (score = 48) |
| Hematological disease | Cellular development | Embryonic development |
| Immunological disease | Cellular growth and proliferation | Hair and skin development and function |
| Organismal injury and abnormalities | Cell death and survival | Hematological system development and function |
| Respiratory disease | Gene expression | Hematopoiesis |
The description of the focus proteins has previously been presented in samples collected at 1 week post-irradiation. Similarly, only proteins that are considered to be the focus protein by the IPA analysis (those in filled IPA symbols) will be discussed.
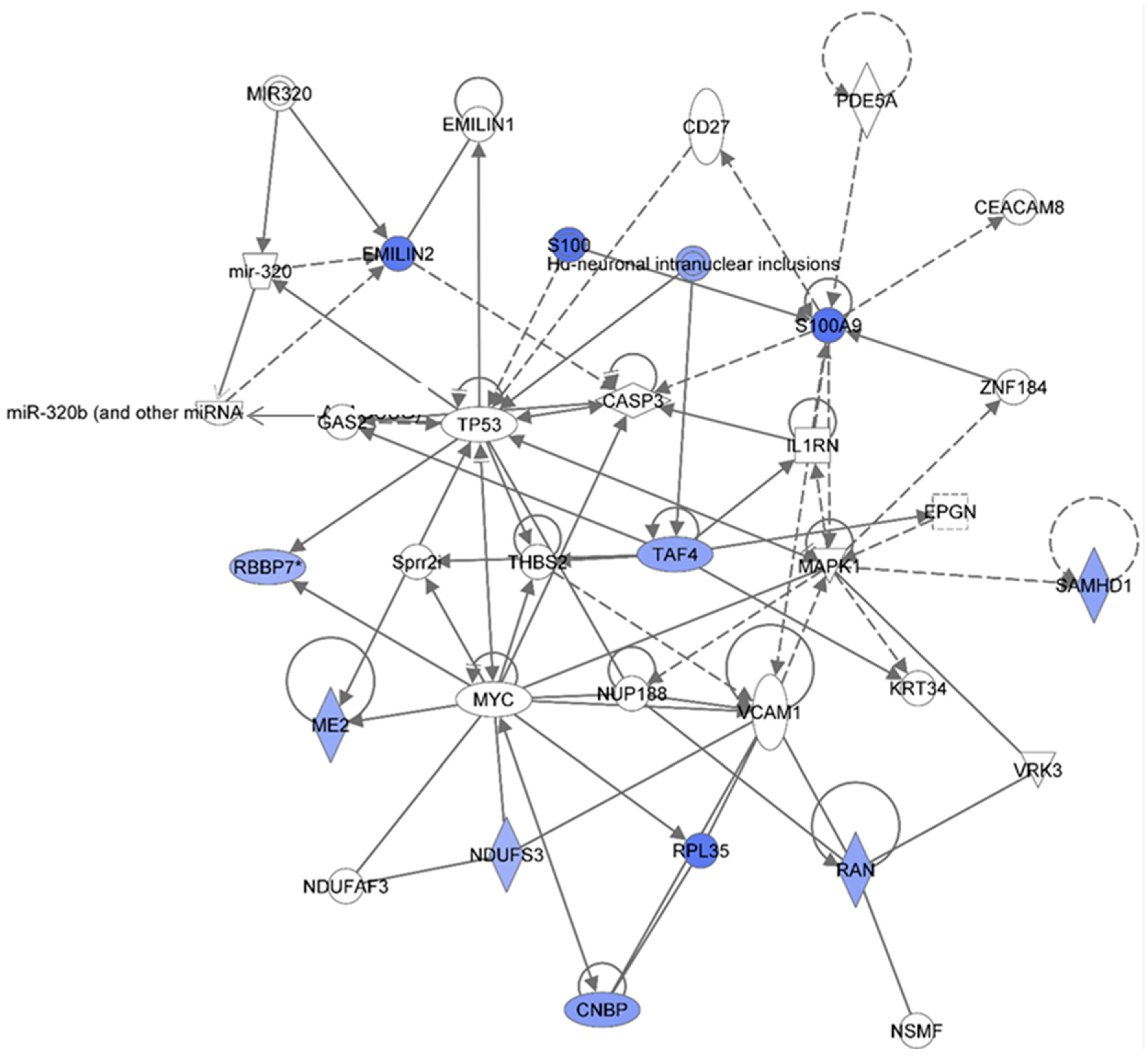
Figure 5 shows the IPA-analysis of identified proteins HSPC-derived myeloid colonies collected at 6 months post-irradiation in response to 0.1 Gy of
48Ti ions. The results show that this network is involved in cancer, tumor morphology, and cardiac necrosis/cell death (score = 29). The solid and dotted lines represent the direct and indirect molecular interaction among proteins in the network, respectively. Although this network is constructed with 34 proteins, there are only 12 focus proteins (those in filled IPA symbols). All of the focus proteins are down-regulated (blue symbols). The majority of these proteins are enzymes and transcription regulators.
Down-regulation of elastin microfibril inter-located protein 2 (EMILIN2) was detected in HSPC-derived myeloid cells obtained at 6 months after exposure of CBA/CaJ mice to 1GeV/n
48Ti ions. EMILIN2 is an extracellular matrix (ECM) glycoprotein in the EMILIN family. Of note, tumor suppressor activity of EMILIN has been suggested [
45]. Expression of EMILIN2 has been detected in several tissues of mice (
i.e., lung, heart, aorta and bone marrow), with the highest expression in bone marrow. It is also known that EMILIN2 is a binding partner of EMILIN1 [
46]. Recently, it has been found that EMILIN2 regulates platelet activation, thrombus formation and clot retraction [
47]. Hence, deficiency or reduction in EMILIN2 would impair platelet aggregation response. Of note, the IPA analyses indicate that TP53 and microRNA320 and microRNA320b directly regulate the expression of proteins in the EMILIN family.
The IPA analysis also shows that the expression of S100 and S100A9 are down-regulated. The S100 proteins (a multigenic-family of Ca
2+ binding proteins, e.g., S100A1, S100A2, S100A8, and S100A9) are involved in several intracellular and extracellular regulatory activities, e.g., enzyme activities, dynamics of cytoskeleton constituents, cell growth and differentiation, cell cycle, and Ca
2+ homeostasis [
48]. It has been reported that some proteins in the S100 family (
i.e., S100A1, and S100A2) bind to TP53 proteins for proper transcriptional activity of tumor suppressor protein 53 (TP53) [
49]. Hence, down-regulation of the S100 proteins would affect the normal activity of TP53. The S00A9 (also known as MRP14, migration inhibitory factor-related protein 14) is tightly regulated during differentiation of myeloid cells and is essential for myeloid cell function [
50]. Hence, a reduction in S100A9 after exposure to 1 GeV/n
48Ti ions suggests dysregulation of the function of myeloid cells such as cytokines production in response inflammatory stimulation. It is shown in the network that the activity of S100A9 is dependent to MAPK and zinc finger proteins 184 (ZNF184).
Low levels of cellular nucleic acid protein (CNBP) may reduce rate of global protein synthesis, thereby reducing proliferation and increasing apoptosis. Further, the expression level of Ras-related nuclear protein (RAN), a nuclear transport protein, is also down-regulated, that may reflect a reduced cell proliferation. Down-regulation of a mitochondrial NAD-dependent maleic enzyme 2 (ME2) is detected. An increased in the level of reactive oxygen species (ROS) has been found to be linked to a reduction in the level of ME2 [
51]. Hence, down-regulated ME2 may reflects the persistent increases in the level of ROS in HSPC-derived myeloid cells collected at 6 mos after exposure of mice to 0.1 Gy of 1 GeV/n
48Ti ions that, in turn, lead to late-occurring damage such as chromosome aberrations or aberrant pattern of DNA methylation [
52]. Further, similar to the HSPC-derived myeloid cells collected at 1 wk post-irradiation), down-regulation of proteins in the family of NADH dehydrogenase (ubiquinone) Fe-S protein (NDUFS), a protein involved in ROS regulation [
53] is found. The IPA analyses reveal that expression of ME2, NDUFS3, RAN, retinoblastoma binding protein 7 (RBBP7, involved in cell proliferation and differentiation), and ribosomal protein L35 (RPL35) are modulated by the MYC protein. Of note, expression of the RBBP7 protein is directly regulated by TP53 protein.
Figure 5.
Ingenuity Pathway Analysis showing the top molecular Network in HSPC-derived myeloid cells collected at 6 mos after exposure of mice to 0.1 Gy of 48Ti ions. This network is involved cancer, tumor morphology, cardiac necrosis/cell death (score = 29). It is constructed by 35 proteins: CASP3 (Caspase 3, Apoptosis-Related Cysteine Peptidase), CD27 (CD27 Molecule), CEACAM8 (Carcinoembryonic Antigen-Related Cell Adhesion Molecule 8), CNBP (CCHC-Type Zinc Finger, Nucleic Acid Binding Protein), EMILIN1 (Elastin Microfibril Interfacer 1), EMILIN2 (Elastin Microfibril Interfacer 2), EPGN (Epithelial Mitogen), GAS2 (Growth Arrest-Specific 2), Hu (histone-like protein protein)-neuronal intranuclear inclusions, IL1RN (Interleukin 1 Receptor Antagonist), KRT34 (Keratin 34, Type I), MAPK1 (Mitogen-Activated Protein Kinase 1), ME2 (Malic Enzyme 2, NAD(+)-Dependent, Mitochondrial), miR320 (microRNA 320) and others, MYC (V-Myc Avian Myelocytomatosis Viral Oncogene Homolog), NDUFAF3 (NADH Dehydrogenase Ubiquinone Complex I, Assembly Factor 3), NDUFS3 (NADH Dehydrogenase Ubiquinone Fe-S Protein 3, 30kDa NADH-Coenzyme Q Reductase), NSMF (NMDA Receptor Synaptonuclear Signaling And Neuronal Migration Factor), NUP188 (Nucleoporin 188 kDa), PDE5A (Phosphodiesterase 5A, CGMP-Specific), RAN (ras-related nuclear protein), RBBP7 (Retinoblastoma Binding Protein 7), RPL35 (Ribosomal Protein L35), S100 (S100 Calcium Binding Protein), S100A9 (S100 Calcium Binding Protein A9), SAMHD1 (SAM Domain And HD Domain 1), Sprr2i (Small proline-rich protein 2I), TAF4 (TAF4 RNA Polymerase II, TATA Box Binding Protein (TBP)-Associated Factor, 135kDa), THBS2 (Thrombospondin 2), TP53 (Tumor Suppression Protein P53), VCAM1 (Vascular Cell Adhesion Molecule 1), VRK3 (Vaccinia Related Kinase 3), and ZNF184 (Zinc Finger Protein 184). Among these proteins, there are only 12 focus proteins (those in bold and shown in the filled IPA symbols in the figure). All of the focus proteins are down-regulated (IPA symbols filled with blue color).
Figure 5.
Ingenuity Pathway Analysis showing the top molecular Network in HSPC-derived myeloid cells collected at 6 mos after exposure of mice to 0.1 Gy of 48Ti ions. This network is involved cancer, tumor morphology, cardiac necrosis/cell death (score = 29). It is constructed by 35 proteins: CASP3 (Caspase 3, Apoptosis-Related Cysteine Peptidase), CD27 (CD27 Molecule), CEACAM8 (Carcinoembryonic Antigen-Related Cell Adhesion Molecule 8), CNBP (CCHC-Type Zinc Finger, Nucleic Acid Binding Protein), EMILIN1 (Elastin Microfibril Interfacer 1), EMILIN2 (Elastin Microfibril Interfacer 2), EPGN (Epithelial Mitogen), GAS2 (Growth Arrest-Specific 2), Hu (histone-like protein protein)-neuronal intranuclear inclusions, IL1RN (Interleukin 1 Receptor Antagonist), KRT34 (Keratin 34, Type I), MAPK1 (Mitogen-Activated Protein Kinase 1), ME2 (Malic Enzyme 2, NAD(+)-Dependent, Mitochondrial), miR320 (microRNA 320) and others, MYC (V-Myc Avian Myelocytomatosis Viral Oncogene Homolog), NDUFAF3 (NADH Dehydrogenase Ubiquinone Complex I, Assembly Factor 3), NDUFS3 (NADH Dehydrogenase Ubiquinone Fe-S Protein 3, 30kDa NADH-Coenzyme Q Reductase), NSMF (NMDA Receptor Synaptonuclear Signaling And Neuronal Migration Factor), NUP188 (Nucleoporin 188 kDa), PDE5A (Phosphodiesterase 5A, CGMP-Specific), RAN (ras-related nuclear protein), RBBP7 (Retinoblastoma Binding Protein 7), RPL35 (Ribosomal Protein L35), S100 (S100 Calcium Binding Protein), S100A9 (S100 Calcium Binding Protein A9), SAMHD1 (SAM Domain And HD Domain 1), Sprr2i (Small proline-rich protein 2I), TAF4 (TAF4 RNA Polymerase II, TATA Box Binding Protein (TBP)-Associated Factor, 135kDa), THBS2 (Thrombospondin 2), TP53 (Tumor Suppression Protein P53), VCAM1 (Vascular Cell Adhesion Molecule 1), VRK3 (Vaccinia Related Kinase 3), and ZNF184 (Zinc Finger Protein 184). Among these proteins, there are only 12 focus proteins (those in bold and shown in the filled IPA symbols in the figure). All of the focus proteins are down-regulated (IPA symbols filled with blue color).
![Proteomes 03 00132 g005]()
Down-regulation of the SAM domain-and HD domain-containing protein 1 (SAMHD1) was detected. The SAMHD1 has deoxynucleoside triphosphate triphosphohydrolase and 3′→5′ exonuclease activity. It is known that SAMHD1 protects host cells from viral infection and DNA damage. It also has been reported that deficiency in SAMHD1 expression results in a complex inherited autoimmune inflammatory encephalopathy disease namely Aicardi-Goutières syndrome [
54]. Further, it is known that SAMHD1 a major regulator of DNA precursor pools in mammalian cells [
55]. Recently, it has been found that SAMHD1 plays a role in DNA repair [
56]. Taken together, the reduction in expression level of SAMHD1 would interfere with normal homeostasis of the cells. The IPA analysis indicates that MAPK affects the normal function of SAMHD1. Of note, the IPA analysis also indicates a reduction in the level of Hu-neuronal intranuclear inclusions in HSPC-derived myeloid cells after exposure of CBA/Ca mice to 0.1 Gy of 1GeV/n
48Ti ions. This protein directly binds to transcription initiation factor TFIID subunit 4 (TAF4), a protein involved in transcription initiation. The role of these two proteins in response to
48Ti-ion -exposure is unclear.
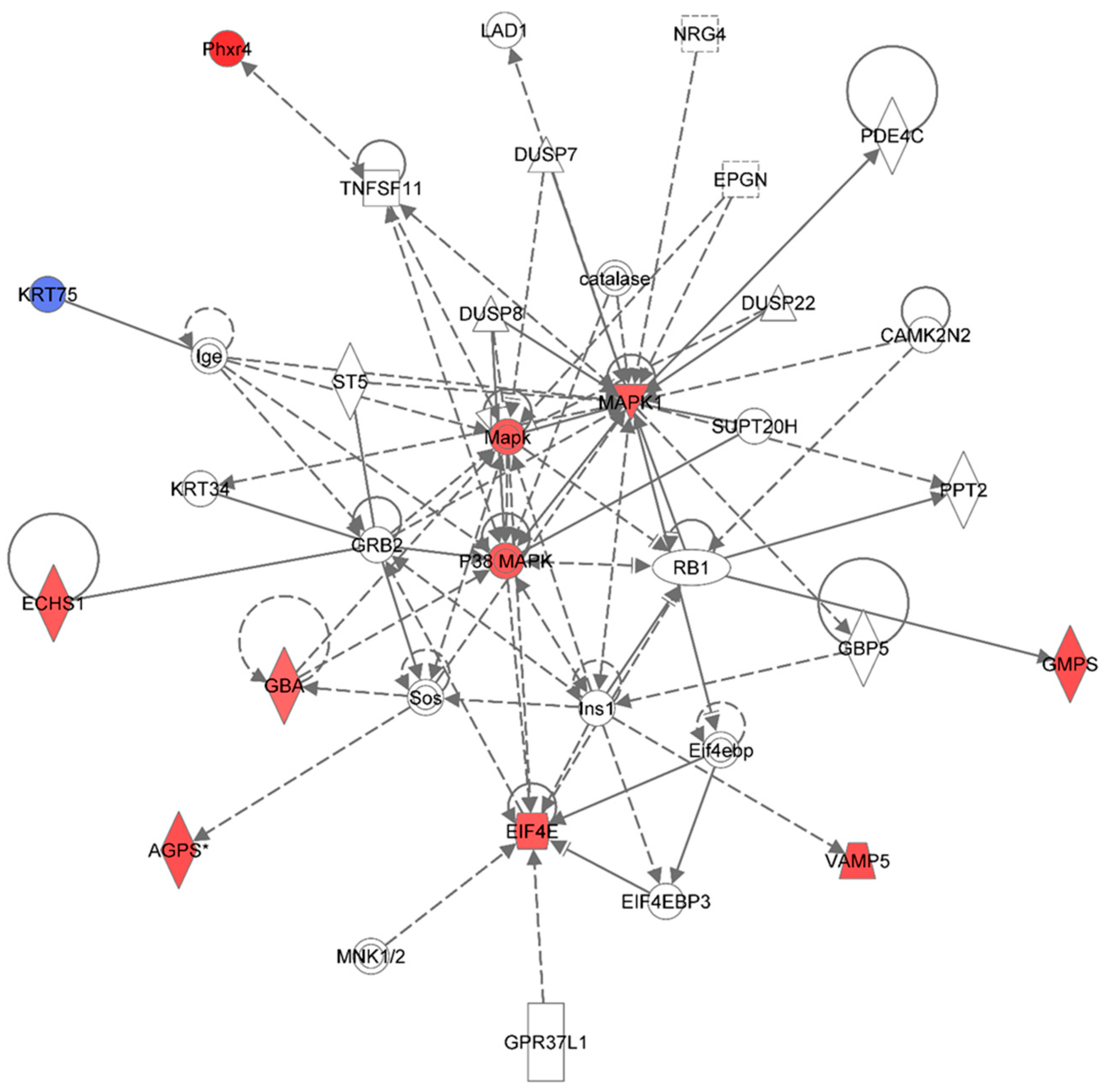
Figure 6 shows the top network resulting from the IPA-analysis of identified proteins in HSPC-derived myeloid cells collected at 6 months post-irradiation in response to 0.25 Gy of
48Ti ions. As shown in
Table 2, this network is involved cell death and survival, cell cycle, connective tissue development and function (score = 25). The solid and dotted lines represent the direct and indirect molecular interactions among proteins in the network, respectively. Although this network is constructed with 35 proteins, there are only 11 focus proteins (those in filled IPA symbols). Ten focus proteins are up-regulated (those shown in symbols filled with red color). Only one focus protein (keratin 75 or KRT75), which is shown in the symbol filled with blue color, is down-regulated. It is known that keratin is expressed in hair follicles and that forms major cytoskeleton in all embryonic and adult epithelia [
57]. However, increasing evidence suggests that keratins can act as scaffolds to regulate cell growth and survival in epithelial cells. It has been found that KRT75 is involved in the differentiation of hematopoietic-progenitor cells through a combination of mechanical and signaling mechanisms [
58]. Hence, it is possible that down-regulation of KT75 may be attributed to the abnormal differentiation process of hematopoietic-progenitor cells.
Figure 6.
Ingenuity Pathway Analysis showing the top molecular network in HSPC-derived myeloid cells collected at 6 months after exposure of mice to 0.25 Gy of 48Ti ions. This network is involved cell death and survival, cell cycle, and connective tissue development/function. It is constructed with 35 proteins: AGPS (Alkylglycerone phosphate synthase), CAMK2N2 (Calcium/Calmodulin-Dependent Protein Kinase II Inhibitor 2), catalase (Catalase), DUSP7 (dual specificity phosphatase 7), DUSP8 (dual specificity phosphatase 8), DUSP22 (dual specificity phosphatase 22), ECHS1 (Enoyl CoA hydratase, short chain, 1, mitochondrial), EIF4E (Eukaryotic translation initiation factor 4E), EIF4EBP3 (Eukaryotic Translation Initiation Factor 4E Binding Protein 3), EPGN (Epithelial Mitogen Homologue), GBA (Glucosidase, beta, acid), GBP5 (Guanine Nucleotide-Binding Protein 5), GMPS (Guanine monphosphate synthase), GPR37L1 (G Protein-Coupled Receptor 37 Like 1), GRB2 (Growth factor receptor-bound protein 2), Ige (Immunoglobulin E), Ins1 (Insulin 1), KRT34, KRT75 (keratin 75), MAPK or Mapk (Mitogen-activated protein kinase), p38 MAPK (p38 MAPK-activated protein kinases), MAPK1 (mitogen-activated protein kinase 1), PDE4C (Phosphodiesterase 4C, cAMP-specific), Phxr4 (Per-hexamer repeat gene 4), PPT2 (Palmitoyl-Protein Thioesterase 2), RB1 (retinoblastoma 1), Sos (Son of Sevenless-encoding guanine nucleotide exchange factors, ST5 (Suppression of tumorigenicity 5), SUPT20H (Transcription factor SPT20 homolog), TNFSF11 (Tumor Necrosis Factor (Ligand) Superfamily, Member 11), and VAMP5 (vesicle-associated membrane protein 5). Among these proteins, there are only 11 focus proteins (those in bold and shown in the filled IPA symbols in the Figure). Ten out of 11 focus proteins are down-regulated (shown blue IPA symbols). Only one focus protein is up-regulated (shown in red IPA symbols).
Figure 6.
Ingenuity Pathway Analysis showing the top molecular network in HSPC-derived myeloid cells collected at 6 months after exposure of mice to 0.25 Gy of 48Ti ions. This network is involved cell death and survival, cell cycle, and connective tissue development/function. It is constructed with 35 proteins: AGPS (Alkylglycerone phosphate synthase), CAMK2N2 (Calcium/Calmodulin-Dependent Protein Kinase II Inhibitor 2), catalase (Catalase), DUSP7 (dual specificity phosphatase 7), DUSP8 (dual specificity phosphatase 8), DUSP22 (dual specificity phosphatase 22), ECHS1 (Enoyl CoA hydratase, short chain, 1, mitochondrial), EIF4E (Eukaryotic translation initiation factor 4E), EIF4EBP3 (Eukaryotic Translation Initiation Factor 4E Binding Protein 3), EPGN (Epithelial Mitogen Homologue), GBA (Glucosidase, beta, acid), GBP5 (Guanine Nucleotide-Binding Protein 5), GMPS (Guanine monphosphate synthase), GPR37L1 (G Protein-Coupled Receptor 37 Like 1), GRB2 (Growth factor receptor-bound protein 2), Ige (Immunoglobulin E), Ins1 (Insulin 1), KRT34, KRT75 (keratin 75), MAPK or Mapk (Mitogen-activated protein kinase), p38 MAPK (p38 MAPK-activated protein kinases), MAPK1 (mitogen-activated protein kinase 1), PDE4C (Phosphodiesterase 4C, cAMP-specific), Phxr4 (Per-hexamer repeat gene 4), PPT2 (Palmitoyl-Protein Thioesterase 2), RB1 (retinoblastoma 1), Sos (Son of Sevenless-encoding guanine nucleotide exchange factors, ST5 (Suppression of tumorigenicity 5), SUPT20H (Transcription factor SPT20 homolog), TNFSF11 (Tumor Necrosis Factor (Ligand) Superfamily, Member 11), and VAMP5 (vesicle-associated membrane protein 5). Among these proteins, there are only 11 focus proteins (those in bold and shown in the filled IPA symbols in the Figure). Ten out of 11 focus proteins are down-regulated (shown blue IPA symbols). Only one focus protein is up-regulated (shown in red IPA symbols).
![Proteomes 03 00132 g006]()
In this network, the MAPK is the major kinase signaling pathway involved in up-regulation of several proteins with various functions such as enzymes and regulators of the translational process. The enzymes that are up-regulated are: Alkylglycerone phosphate synthase (AGPS), enoyl co-enzyme A hydratase (ECHST, a mitochondrial enzyme), glucosidase (GBA), and guanine monophosphate synthase (GMPS), and vesicle-associated membrane protein 5 (VAMP5) and putative per hexamer repeat protein 4 (Phxr4).
One of the important regulators of the translational process that is up-regulated in HSPC-myeloid cells by the MAPK signaling pathway after exposure of mice to 1 GeV/n 48Ti ions is eukaryotic translation initiation factor 4E (EIF4E, the rate-limiting component in protein synthesis). The EIF4E protein plays an important role in translational regulation in response to stress and apoptosis that may lead to cell/tissue damage [
59]. Over-expression of EIF4E is linked to cancer progression and drives cellular transformation, tumorigenesis, and metastatic progression [
60]. Hence, EIF4E is frequently overexpressed in several types of human cancers, both solid and hematopoietic neoplasms [
60,
61]. Such oncogenic potential of EIF4E arises from its ability to bind the 7-methyl guanosine (m7G) cap on mRNAs, thereby selectively enhancing EIF4E-dependent nuclear mRNA export and translation [
62]. Hence, it is plausible to speculate that over-expression of EIF4E is associated with the induction of genomic instability after exposure to
48Ti ions. Since targeting the expression of EIF4E for cancer therapy is being actively investigated, it is possible to search for EIF4E inhibitors to serve as radiation countermeasures to protect cells/tissues from radiation-induced damage.
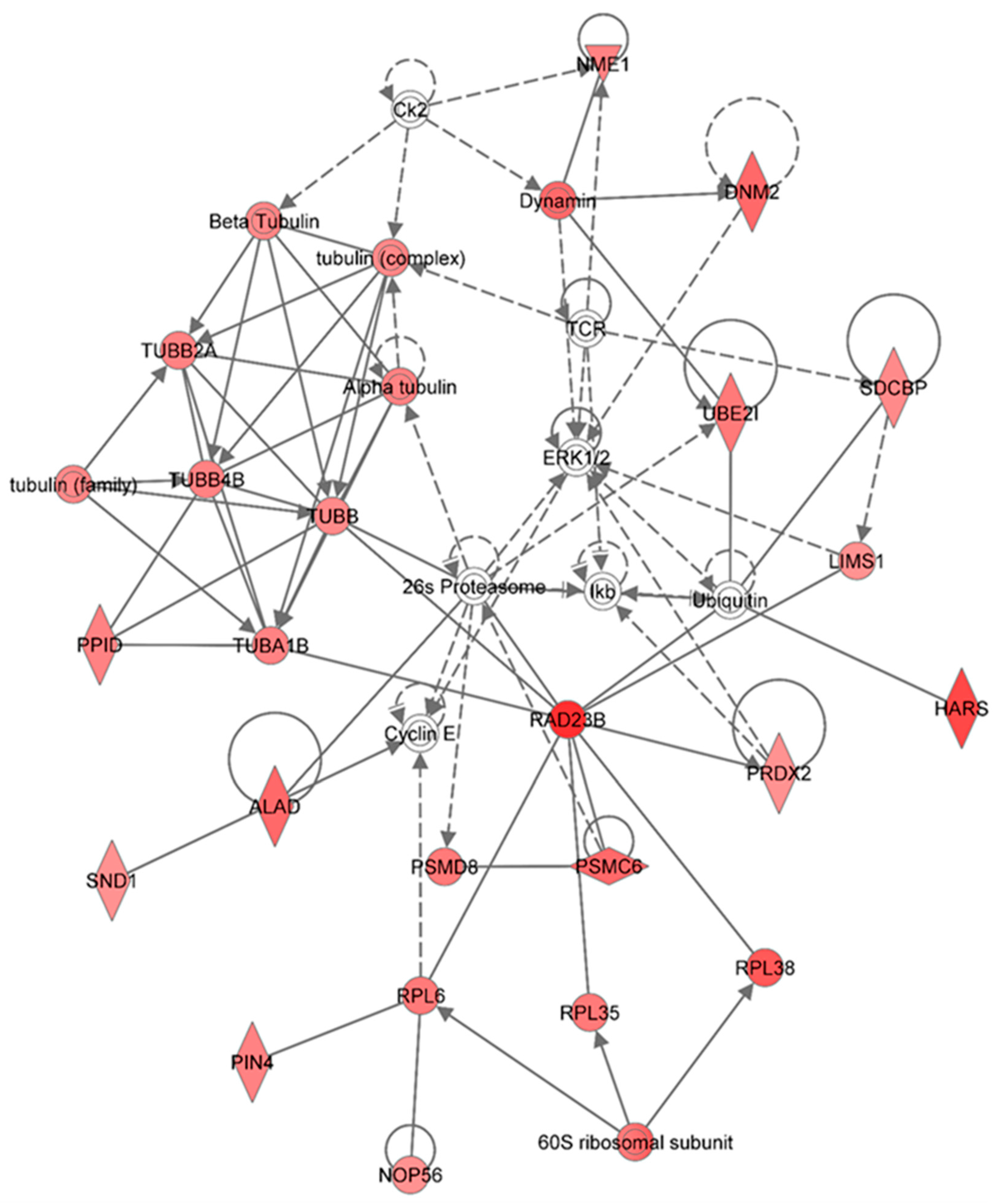
Figure 7 shows the top network resulting from the IPA-analysis of identified proteins in HSPC-derived myeloid cells collected at 6 months in response to 0.5 Gy of 1 GeV/n
48Ti ions. As shown in
Table 2, the results show that this network is involved in cancer, hematological disease, and immunological disease (score = 48). As with other networks, the solid and dotted lines represent the direct and indirect molecular interactions among proteins in the network, respectively. This network is constructed with 35 proteins, but only 28 of these are focus proteins (shown in filled IPA symbols with red color). All of the focus proteins are up-regulated. The majority of proteins involved in this network are the tubulin family, proteasome/ubiquination members, kinases (ERK or I kappa B, normally known as IκB which is a regulator of NF-κB activation). Intriguingly, proteins in the tubulin family are down-regulated in HSPC-derived myeloid cells collected at an early time-point (1 week post-exposure, as shown previously in
Figure 2) but they are up-regulated in these cells collected at 6 months post-irradiation, at which time genomic instability has been detected [
52]. Hence, it is possible that up-regulation of tubulin proteins (which are the component of microtubules) has a role in the induction of genomic instability after exposure to 1 GeV/n
48Ti ions. This statement is supported by previous reports showing a link between deregulation of microtubules/tubulins (such as an increased microtubule assembly) and genomic instability [
63] and cancer [
64]. It should be noted that the proteasome/ubiquination pathway is the major pathway to regulate the level of tubulin proteins, which is also directly linked to the inhibitor-kappa B (IκB), a regulator of NF-κB activation.
Figure 7.
Ingenuity Pathway Analysis showing the top molecular network in HSPC-derived myeloid cells collected at 6 months after exposure of mice to 0.5 Gy of 48Ti ions. This network is involved cancer, hematological disease, and immunological diseases. It is constructed with 35 proteins: 26s Proteasome, 60S ribosomal subunit, ALAD (Aminolevulinate dehydratase), Alpha tubulin, Beta Tubulin, Ck2 (Casein Kinase 2), Cyclin E, DNM2 (Dynamin 2), Dynamin, ERK1/2 (Extracellular-signal-regulated kinases 1/2), HARS (Histidyl-tRNA synthetase), Ikb (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor), LIMS1 ((LIN-11, ISL-1, MEC-3 and Senescent Cell Antigen-Like Domains 1), NME1 (Non-Metastatic Cells 1), NOP56 (Nucleolar protein 56), PIN4 (Peptidylprolyl Cis/Trans Isomerase, NIMA-Interacting 4), PPID (peptidylprolyl isomerase), PRDX2 (Peroxiredoxin 2), PSMC6 (Proprotein Convertase Subtilisin/Kexin Type 6), PSMD8 (Proteasome or Prosome or Macropain, Subunit, Beta Type 8), RAD23B (RAD23 Homolog B), RPL6 (Ribosomal Protein L 6), RPL35 (Ribosomal Protein L 35) , RPL38 (Ribosomal Protein L 38), SDCBP (Syndecan Binding Protein or Syntenin), SND1 (Staphylococcal Nuclease And Tudor Domain Containing 1), TCR (T cell receptor), TUBA1B (Tubulin, Alpha 1b), TUBB (Tubulin, Beta), TUBB2A (Tubulin, Beta 2A Class IIa), TUBB4B (Tubulin, Beta 4A Class IV b), tubulin (complex), tubulin (family), UBE2I (Ubiquitin-Conjugating Enzyme E2I), and Ubiquitin. Among these proteins, there are only 28 focus proteins (those in bold and shown in the filled IPA symbols in the Figure). All of the focus proteins are up-regulated (IPA symbols filled with red color).
Figure 7.
Ingenuity Pathway Analysis showing the top molecular network in HSPC-derived myeloid cells collected at 6 months after exposure of mice to 0.5 Gy of 48Ti ions. This network is involved cancer, hematological disease, and immunological diseases. It is constructed with 35 proteins: 26s Proteasome, 60S ribosomal subunit, ALAD (Aminolevulinate dehydratase), Alpha tubulin, Beta Tubulin, Ck2 (Casein Kinase 2), Cyclin E, DNM2 (Dynamin 2), Dynamin, ERK1/2 (Extracellular-signal-regulated kinases 1/2), HARS (Histidyl-tRNA synthetase), Ikb (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor), LIMS1 ((LIN-11, ISL-1, MEC-3 and Senescent Cell Antigen-Like Domains 1), NME1 (Non-Metastatic Cells 1), NOP56 (Nucleolar protein 56), PIN4 (Peptidylprolyl Cis/Trans Isomerase, NIMA-Interacting 4), PPID (peptidylprolyl isomerase), PRDX2 (Peroxiredoxin 2), PSMC6 (Proprotein Convertase Subtilisin/Kexin Type 6), PSMD8 (Proteasome or Prosome or Macropain, Subunit, Beta Type 8), RAD23B (RAD23 Homolog B), RPL6 (Ribosomal Protein L 6), RPL35 (Ribosomal Protein L 35) , RPL38 (Ribosomal Protein L 38), SDCBP (Syndecan Binding Protein or Syntenin), SND1 (Staphylococcal Nuclease And Tudor Domain Containing 1), TCR (T cell receptor), TUBA1B (Tubulin, Alpha 1b), TUBB (Tubulin, Beta), TUBB2A (Tubulin, Beta 2A Class IIa), TUBB4B (Tubulin, Beta 4A Class IV b), tubulin (complex), tubulin (family), UBE2I (Ubiquitin-Conjugating Enzyme E2I), and Ubiquitin. Among these proteins, there are only 28 focus proteins (those in bold and shown in the filled IPA symbols in the Figure). All of the focus proteins are up-regulated (IPA symbols filled with red color).
![Proteomes 03 00132 g007]()
Other proteins that directly interact with proteasome/ubiquinition pathways are RAD23B (a nucleotide excision repair enzyme) and several enzymes that are that are required for the maintenance of basic cellular function. These enzymes include: Peptidyl-prolyl
cis–
trans isomerase D (PPID, also known as Cyclophilin-type PPIase, known to accelerate protein folding), Peptidyl-prolyl
cis–
trans isomerase NIMA-interacting 4 (PIN4), and aminolevulinate dehydratase (ALAD, known to response to oxidative stress induced by toxix agents such as radiation or lead), staphylococcal nuclease domain-containing protein 1 (SND1, known to regulate gene expression and promote tumor angiogenesis. In addition to proteasome/ubiquitin pathway, the RAD23B protein also interacts directly with several molecules involved in protein synthesis,
i.e., 60S ribosomal subunit, ribosomal protein L (RPL) 6, 35, and 38, including nucleolar protein 56. As well as molecules involved in proteasome/ubiquitinin pathway and in protein synthesis, RAD23B interact with peroxiredoxin 2 (PRDX2, an antioxidant enzyme), which is also up-regulated. Similar to other anti-oxidant proteins (e.g., manganese superoxide dismutase, MnSOD [
65]), a high level of PRDX2 has been found in breast cancer cells [
66], conceivably due to the anti-apoptotic and enhance cell proliferation of these enzymes plausibly. Further, a high level of MnSOD also plays a role in radiation-induced genomic instability [
67]. Hence, it is plausibly to speculate that a high level of PRDX2 expression detected in HSPC-derived myeloid cells has a role in the induction of genomic instability after exposure of mice to 1GeV/n
48Ti ions. Recently, highly expressed PRDX2 has been observed in neutrophils of myelodysplastic syndrome (MDS) patients with refractory cytopenia and multilineage dysplasia, in relation to that of healthy control subjects [
68]. It is known that MDS patients are at a high risk for developing acute ML. Hence, it is reasonable to speculate that the expression level of PRDX2 protein would be high in acute ML patients. However, it has been reported that PRDX2 is suppressed in acute ML patients [
69]. Of note, it is unknown if radiation is the causal factor for the induction of acute ML in these patients. Hence, these findings warrant further investigation of the role of PRDX2 in radiation-induced acute ML.
Other proteins that are up-regulated and interact with proteasome/ubiquitin and kinase pathways include: Proteins in the dynamin family (a GTPase enzyme responsible for endocytosis of cells), histidyl-tRNA synthetase (HARS), LIM and sebescent cell and antigen-like domain 1 (LIMS1), nucleoside diphosphate kinase A (NME1), and syntenin or syndecan binding protein (SDCBP).