IPA Analysis of Cervicovaginal Fluid from Precancerous Women Points to the Presence of Biomarkers for the Precancerous State of Cervical Carcinoma
Abstract
:1. Introduction
1.1. Cervicovaginal Fluid (CVF): An Underestimated Source of Biomarkers for Pathologies of the Female Genital Tract
1.2. Cancer and the Necessity for Detecting Several Biomarkers Simultaneously
1.3. How Cervical Carcinoma Cells may Deliver Their Cancer Biomarkers into the CVF
2. Experimental
2.1. Study Design and Sample Collection
| Sample | Age | Genotype(s) | Viral load | Colposcopy | Cytology |
|---|---|---|---|---|---|
| A.1 | 57 | no HPV | Normal | Normal | |
| A.2 | 69 | no HPV | Normal | Normal | |
| A.3 | 52 | no HPV | Normal | Normal | |
| A.4 | 46 | no HPV | Normal | Normal | |
| A.5 | 66 | no HPV | Normal | Normal | |
| A.6 | 62 | no HPV | Normal | Normal | |
| B.1 | 61 | 31/52 | 335/2 | LSIL | CIN1 |
| B.2 | 53 | 16 | 417 | HSIL | CIN3 |
| B.3 | 47 | 39 | 2011 | LSIL | CIN2 |
| B.4 | 72 | 18/56 | 0.1322/1119 | LSIL | CIN1 |
| B.5 | 57 | 31 | 1148 | HSIL | CIN3 |
| B.6 | 60 | 31/52 | 258/25 | LSIL | CIN2 |
2.2. Proteomic Analysis
2.3. IPA Analysis
3. Results
3.1. IPA Analysis of CVF from Healthy Individuals versus CVF from Individuals in the Precancerous State: Rationale
3.2. Predicted Pathways Formed by CVF Proteins from Healthy or Precancerous Women Have a Similar Functional Distribution but Differ in Confidence
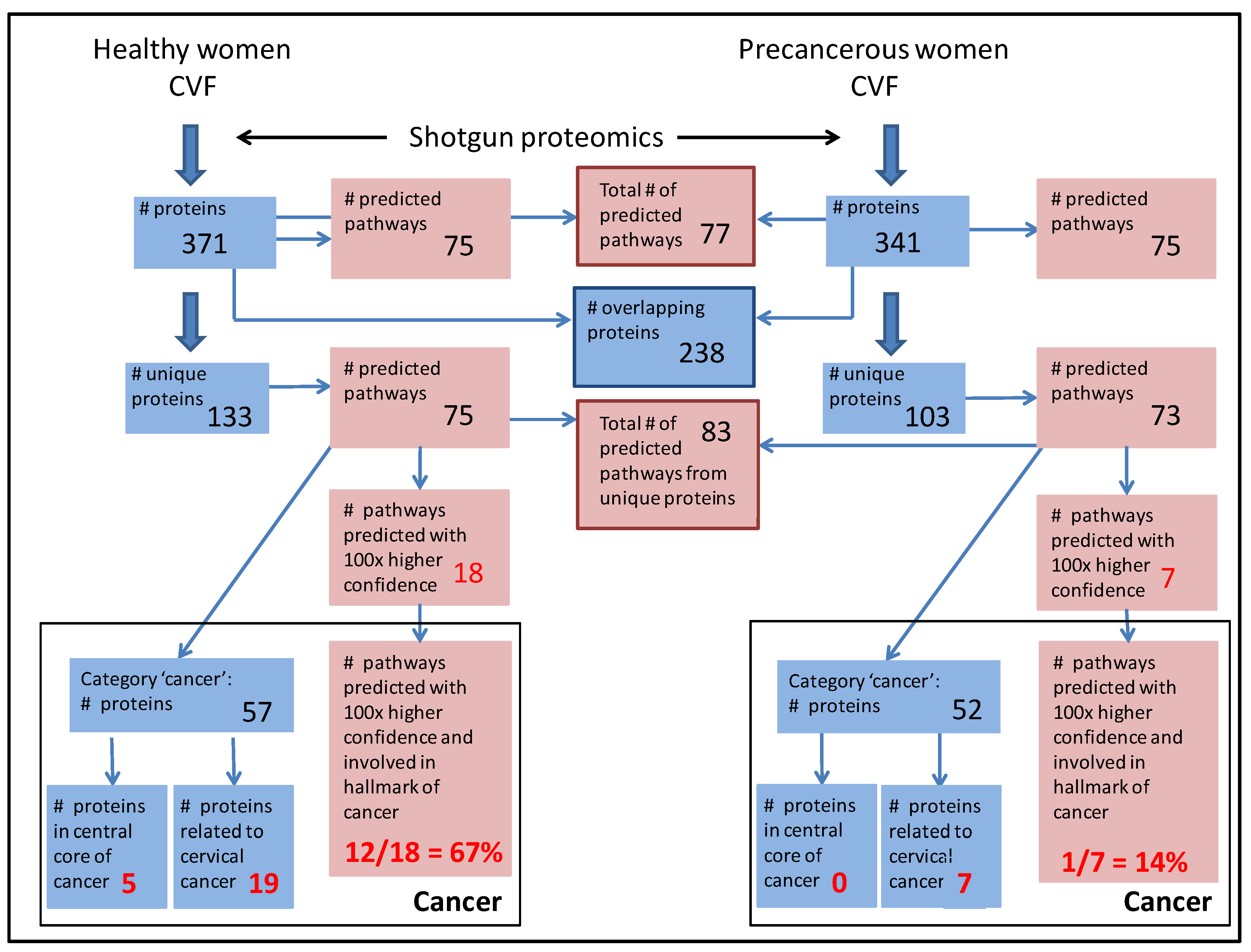
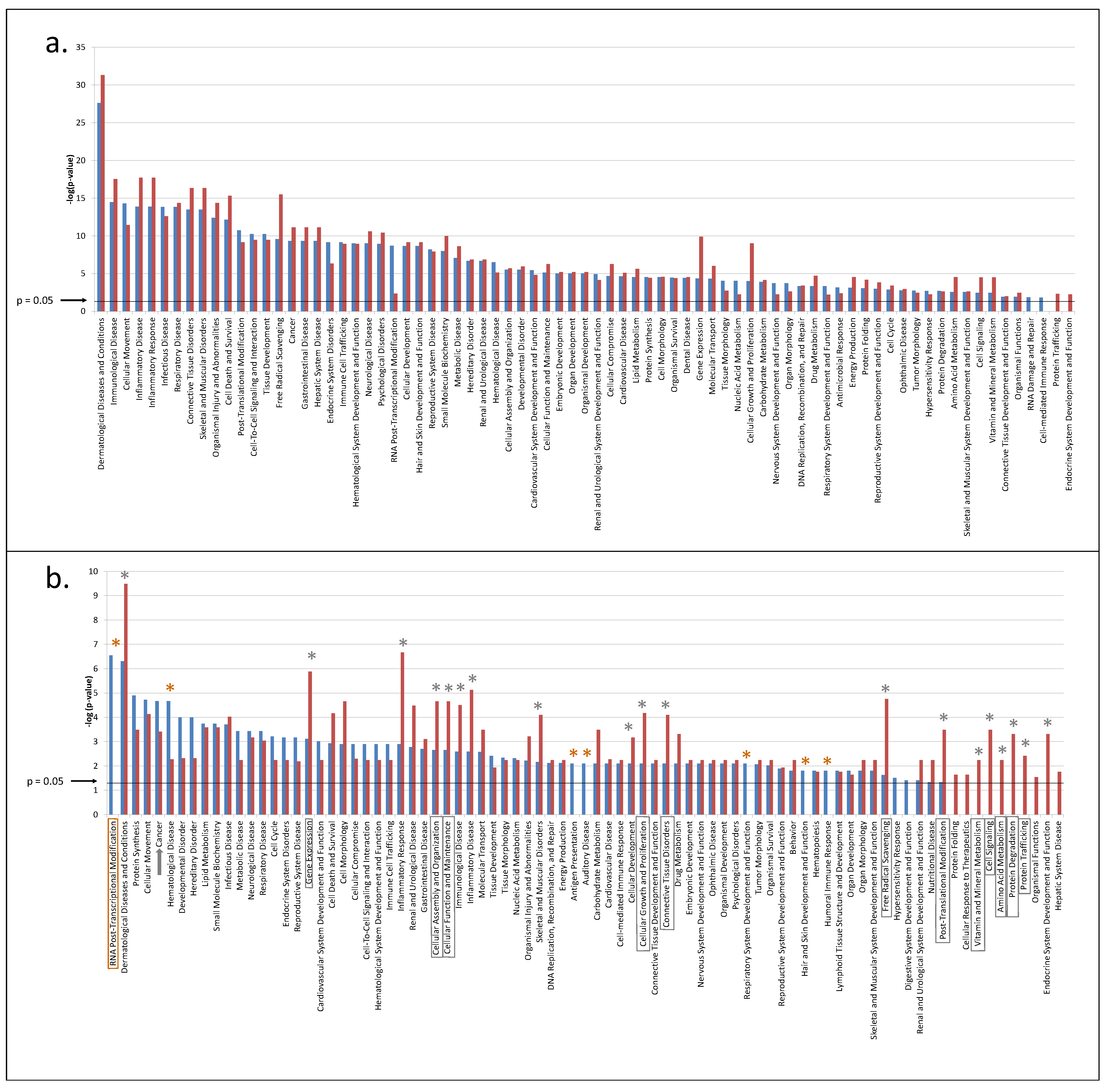
3.3. CVF from Precancerous Patients Contains Substantially More Unique Proteins That Are with Higher Confidence Involved in Pathways, Related to the Hallmarks of Cancer
3.3.1. CVF from Precancerous Patients Contains Substantially More ‘Cancer Proteins’ That Are Involved in Cervical Cancer and That Are Interconnected
| Precancerous-Category Cancer | Healthy-Category Cancer | |||
|---|---|---|---|---|
| Protein Name | Protein ID | Protein Name | Protein ID | |
| 14-3-3 protein epsilon | 1433E_HUMAN | 40S ribosomal protein S11 | RS11_HUMAN | |
| 14-3-3 protein theta | 1433T_HUMAN | 40S ribosomal protein S15 | RS15_HUMAN | |
| 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | PGDH_HUMAN | 40S ribosomal protein S16 | Q6IPX4_HUMAN | |
| 60S ribosomal protein L15 | RL15_HUMAN | 40S ribosomal protein S20 | RS20_HUMAN | |
| Actin-related protein 3 | ARP3_HUMAN | 40S ribosomal protein S6 | RS6_HUMAN | |
| Angiotensinogen | ANGT_HUMAN | 60S ribosomal protein L19 | RL19_HUMAN | |
| Annexin A11 | ANX11_HUMAN | 60S ribosomal protein L28 | RL28_HUMAN | |
| Annexin A4 | ANXA4_HUMAN | 60S ribosomal protein L34 | RL34_HUMAN | |
| Antithrombin-III | ANT3_HUMAN | 60S ribosomal protein L35 | RL35_HUMAN | |
| Argininosuccinate synthase | ASSY_HUMAN | 60S ribosomal protein L7 | RL7_HUMAN | |
| Calreticulin | CALR_HUMAN | Apolipoprotein C-1 | APOC1_HUMAN | |
| Carcinoembryonic antigen-related cell adhesion molecule 6 | CEAM6_HUMAN | Ataxin-1-like | ATX1_HUMAN | |
| Cathepsin B | CATB_HUMAN | ATP synthase-coupling factor 6, mitochondrial | ATP5J_HUMAN | |
| CD59 glycoprotein | CD59_HUMAN | Carcinoembryonic antigen-related cell adhesion molecule 7 | CEAM7_HUMAN | |
| Cellular retinoic acid-binding protein 2 | RABP2_HUMAN | Caspase recruitment domain-containing protein 11 | CAR11_HUMAN | |
| Centriolin | CNTRL_HUMAN | Collagen alpha-1 (I) chain | CO1A1_HUMAN | |
| Ceruloplasmin | CERU_HUMAN | Complement C4A/C4B | CO4A_HUMAN, CO4B_HUMAN | |
| Copine-7 | CPNE7_HUMAN | Complement factor B | CFAB_HUMAN | |
| Cytosolic non-specific dipeptidase | CNDP2_HUMAN | Complement factor H | CFAH_HUMAN | |
| F-actin-capping protein subunit alpha-2 | CAZA2_HUMAN | Cytochrome c oxidase subunit 6B1 | CX6B1_HUMAN | |
| Ferritin light chain | FRIL_HUMAN | Dystonin | DYST_HUMAN | |
| Filaggrin | FILA_HUMAN | Elongation factor 1-alpha 2 | EF1A2_HUMAN | |
| Fructose-biphosphate aldolase C | ALDOC_HUMAN | Far upstream element binding protein 1 | FUBP1_HUMAN | |
| Gelsolin | GELS_HUMAN | Heat shoch protein beta-8 | HSPB8_HUMAN | |
| Guanylate-binding protein 6 | GBP6_HUMAN | High mobility group protein B1 | HMGB1_HUMAN | |
| Heat shock 70 Da protein 1-like | HS71L_HUMAN | Histone H2B type 2-E | H2B2E_HUMAN | |
| Hemoglobin subunit epsilon | HBE_HUMAN | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial | IDH3A_HUMAN | |
| High mobility group protein B2 | HMGB2_HUMAN | Kalikrein-11 | KLK11_HUMAN | |
| Inter-alpha-trypsin inhibitor heavy chain H2 | ITIH2_HUMAN | Kallikrein-6 | KLK6_HUMAN | |
| Interleukin-18 | IL18_HUMAN | Kallikrein-8 | KLK8_HUMAN | |
| Junction Plakoglobin | PLAK_HUMAN | LIM domain only protein 7 | LMO7_HUMAN | |
| Lyphocyte antigen 6D | LY6D_HUMAN | Lymphocyte-specific protein 1 | LSP1_HUMAN | |
| Macrophage migration inhibitory factor | MIF_HUMAN | Membrane-associated transporter protein | S45A2_HUMAN | |
| Macrophage-capping protein | CAPG_HUMAN | Metalloproteinase inhibitor 2 | TIMP2_HUMAN | |
| Mucin-5B | MUC5B_HUMAN | Mesencephalic astrocyte-derived neurotrophic factor | MANF_HUMAN | |
| Myosin light polypeptide 6 | MYL6_HUMAN | Myosin-9 | MYH9_HUMAN | |
| Myristoylated alanine-rich C-kinase substrate | MARCS_HUMAN | Non-secretory ribonuclease | RNAS2_HUMAN | |
| Nicotinamide phosphoribosyltransferase | NAMPT_HUMAN | Nuclease-sensitive element binding protein 1 | YBOX1_HUMAN | |
| Olfactomedin-4 | OLFM4_HUMAN | Plasminogen | PLMN_HUMAN | |
| PHD finger protein | PHF1_HUMAN | Protein disulfide isomerase A6 | PDIA6_HUMAN | |
| Phosphoglycerate mutase 1 | PGAM1_HUMAN | Protein S100-A11 | S10AB_HUMAN | |
| Protein disulfide isomerase A3 | PDIA3_HUMAN | Protein S100-A2 | S10A2_HUMAN | |
| Protein S100-A14 | S10AE_HUMAN | Protein S100-A7 | S10A7_HUMAN | |
| Protein S100-P | S100P_HUMAN | Ras-related C3 botulinum toxin substrate 1 | RAC1_HUMAN | |
| Purine nucleoside phosphorylase | PNPH_HUMAN | Serine/arginine-rich splicing factor 1 | SRSF1_HUMAN | |
| Puromycin-sensitive aminopeptidase | PSA_HUMAN | Small nuclear ribonucleoprotein Sm D3 | SMD3_HUMAN | |
| Putative hydroxypyruvate isomerase | HYI_HUMAN | Src substrate cortactin | SRC8_HUMAN | |
| Rho GDP-dissociation inhibitor 1 | GDIR1_HUMAN | Stathmin | STMN1_HUMAN | |
| Serpin B12 | SPB12_HUMAN | THO complex subunit 4 | THOC4_HUMAN | |
| Serpin B13 | SPB13_HUMAN | Thymidine phosphorylase | TYPH_HUMAN | |
| Sororin | CDCA5_HUMAN | Voltage-dependent P/Q-type calcium channel subunit-1alpha | CAC1A_HUMAN | |
| Superoxide dismutase [Cu-Zn] | SODC_HUMAN | Zink-finger and BTB domain-containing protein 4 | ZBTB4_HUMAN | |
| Superoxide dismutase[Mn] | SODM_HUMAN | |||
| Transgelin-2 | TAGL2_HUMAN | |||
| Transmembrane protease serine 11B | TM11B_HUMAN | |||
| Ubiquitin-conjugating enzyme E2 variant 1 | UB2V1_HUMAN | |||
| Ubiquitin-like modifier-activating enzyme 1 | UBA1_HUMAN | |||
| No. | Gene Name | Protein ID | Protein Name | Cell Line, Tissue or Patient | Effect | Ref. |
|---|---|---|---|---|---|---|
| 1. | AGT | ANGT_HUMAN | Angiotensinogen | IHC was used for investigation of angiotensin receptor levels in normal and neoplastic cervical tissues and Siha cells. Invasion assay was examined in Siha cells and vascular endothelial growth factor levels were assayed by ELISA. | Precursor of Angiotensin II. Ang II is involved in the progression of cervical carcinoma via induction of VEGF secretion through angiotensin II type I receptor. This results in increased invasiveness of carcinoma cells. Correlation of angiotensin II type I R expression with progression from precancerous to invasive cervical carcinoma. | [32] |
| 2. | ANXA4 | ANXA4_HUMAN | Annexin A4 | Differential proteomics of HeLa cells versus non-tumorigenic cell line HaCaT | Annexin A4 is overexpressed in HeLa cells as compared to HaCaT | [26] |
| 3. | CAPG | CAPG_HUMAN | Macrophage-capping protein | Differential proteomics of HeLa cells versus non-tumorigenic cell line HaCaT | CAPG is overexpressed in HeLa cells as compared to HaCaT | [26] |
| 4. | CD59 | CD59_HUMAN | CD59 glycoprotein | CD59 expression was examined in normal, precancerous and cervical squamous carcinomas samples. | CD 59 is abundantly present in cervical carcinomas although staining of tumor cells appears less intense than staining of adjacent stroma. | [33] |
| IHC of CD59 was performed on cervical carcinomas, normal cervical epithelial cells, and the surrounding stroma. | CD59 expression was shown on cervical carcinomas, normal cervical epithelial cells, and the surrounding stromal cells. CD59 was a potent inhibitor of classical pathway-mediated lysis on cervical cancer cell lines. | [34] | ||||
| 5. | CTSB | CATB_HUMAN | Cathepsin B | Investigation of mRNA expression profiles of 8 thermoradiosensitive and 11 thermoradioresistant cervical tumors obtained by punch biopsy before treatment using a cDNA microarray. | A gene-expression profile of 35 genes, including CTSB, can predict the outcome of thermoradiotherapy for SCC. | [35] |
| ELISA and qPCR was used to measure cathepsin B expression in HeLa cells and 169 tissue samples from invasive carcinomas, precancerous and normal tissues (p < 0.01). | Cathepsin B expression in invasive carcinomas was significantly higher as compared to precancerous tissue and normal tissue. Cathepsin B expression in the invasive carcinomas was positively correlated to tumor invasion depth and lymphatic metastasis. Significant regression of HeLa tumor growth in nude mice, which received HeLa cells treated with siRNA for CTSB. | [36] | ||||
| 6. | GSN | GELS_HUMAN | Gelsolin | Differential proteomics of HeLa cells versus non-tumorigenic cell line HaCaT | Gelsolin is overexpressed in HeLa cells as compared to HaCaT | [26] |
| Differential expression of genes in tumor tissues and adjacent noncancerous mucosacancerous tissues was studied by microarray. Confirmation by Western blot and IHC. Plasma gelsolin measured by ELISA. HeLa gelsolin knockdown. | Gelsolin levels were significantly upregulated in 78% of patients with cervical cancer. Levels were higher in cervical tumor tissues than in the surrounding noncancerous tissues. Gelsolin abundance in the plasma of cervical cancer patients was increased 2.2-fold compared to healthy controls and was significantly different in the early and late stages. Survival and recurrence-free survival rates were significantly higher for the low-expression group. Cancer cells with reduced gelsolin expression exhibited reduced migration and proliferation. | [37] | ||||
| Plasma of patients with different grades of SCC | Correlation of serum gelsolin expression with CIN3 and different stages of SCC | [38] | ||||
| 7. | HMGB2 | HMGB2_HUMAN | High mobility group protein B2 | Microarray for identification of differentially expressed genes between untreated and TNF-treated cells. Confirmation by RT-PCR. | TNF treatment upregulated HMGB2 in E7-expressing cells but not in E7 negative cells. | [39] |
| 8. | IL18 | IL18_HUMAN | Interleukin-18 | Secretome of cervical cancer cell lines SiHa and CaSki were investigated by ELISA. | E6 and E7 reside in the extracellular fluid of HPV-containing cervical cancer cell lines and inhibit IL-18-induced IFN-gamma production locally in HPV lesions through inhibition of IL-18 binding to its alpha-chain receptor. This effect does not occur with the IL-18 mutant E42A. | [40,41] |
| HaCaT and C-33A, (HPV-negative cervical cancer cell line) stabile expressing E6, E6 mutant, E6E7, or E7 were investigated by RT-PCR. | E6 downregulated IL-18 mRNA expression, independent of p53 degradation, in HaCaT cells expressing a mutated p53 form. | [42] | ||||
| TaqMan Allelic Discrimination Assay was used to genotype IL-18 polymorphisms for women with SCC and healthy control women. | IL-18 1297 T/C, 607 C/A, 380 C/G, 137 G/C, and +105 A/C polymorphisms are not associated with susceptibility to SCC in Taiwanese women. | [43] | ||||
| 9. | MIF | MIF_HUMAN | Macrophage migration inhibitory factor | MIF protein and mRNA expression was analyzed by Western blotting, ELISA and RT-PCR in uterine cervical cancer cell lines SiHa and CaSki and their supernatant, and on 80 biopsies (cervical dysplasias, in situ carcinomas and invasive carcinomas) of uterine cervical tissue. | MIF is overexpressed in invasive cervical cancer as compared to cervical dysplasias. MIF is overexpressed in SiHA and CaSki cells and these cells also secrete the protein. | [44] |
| 250 patients with cervical cancer (49 cases with and 201 cases without lymphatic metastasis) were analyzed by RFLP and their serum by ELISA | Patients with GC and CC genotypes and C allele exhibited a lower degree of differentiation and a higher degree of malignancy. Early cervical cancer, lymphatic metastasis and poorly differentiated carcinomas exhibited higher MIF levels in serum. Moreover, patients with the CC genotype exhibited higher MIF serum concentration, which could increase the risk of early stage cervical cancer and lymphatic metastasis. | [45] | ||||
| Differential proteomics of HeLa cells versus non-tumorigenic cell line HaCaT | MIF is overexpressed in HeLa cells as compared to HaCaT | [26] | ||||
| MIF knockdown in HeLa cells | MIF is crucial for proliferation and tumorigenesis of human HeLa cells | [46] | ||||
| IHC on 209 tissue samples (40 normal cervical epithelia, 43 CIN 1, 41 CIN 2 to 3, and 85 SCC) and on cervical cancer cell lines SiHa and C-33A. Semi-quantitative PCR and Western blot on SiHa and C-33A | Overexpression of MIF in SCC and its precancerous lesions (CIN1-3) and in SiHa and C-33A indicates that MIF may play an important role in the pathogenesis of cervical cancer. | [47] | ||||
| 10. | MUC5B | MUC5B_HUMAN | Mucin 5B | Slot blot on normal and malignant cervical tissues | Increased expression of MUC5B in endometrial tumors but not in cervical tumors. | [48] |
| 11. | MYL6 | MYL6_HUMAN | myosin light chain 6 | Subtractive hybridization and semi-quantitative PCR on 48A9 cells (subclone from Caski cells that were cultivated during space flight and had lower tumorigenic potential) and Caski cells. | MYL6 expression was >3-fold increased in cells exposed to spaceflight and with lower tumorigenic potential. | [49] |
| 12. | OLFM4 | OLFM4_HUMAN | OLFM4 expression and distribution was tested by IHC and RT-PCR on cervical intraepithelial neoplasia (CIN) and invasive SCC. | OLFM4 expression correlated with progression of CIN and differentiation of cervical cancer | [50] | |
| 13. | PDIA3 | PDIA3_HUMAN | Protein disulfide-isomerase A3 | Differential 2D-PAGE proteomics on paired adjacent normal and tumor tissues from 4 patients, comprising 2 SCC in stage IB1 and IIA, one adenocarcinoma (AD) in stage IB2, and one adenosquamous cell carcinoma in stage IB1. Western blot, IHC and shRNA konckdown were performed to confirm the results and to estimate clinical significance. | PDIA3 was overexpressed in 73% of cancers. PDIA3 expression was significantly higher in patients with AD compared with SCC. Staining was intense in AD with a penetration depth greater than half of the cervical stroma. High expression was associated with low overall survival and recurrence-free survival (RFS) rates. Patients exhibiting both high PDIA3 expression and lymph node metastasis displayed poorer outcomes than other patient groups. Knockdown of PDIA3 in HeLa cells decreased cell invasiveness and inhibited lung metastasis in a xenograft mouse model. | [51] |
| Differential proteomics of HeLa cells versus non-tumorigenic cell line HaCaT | Protein Disulfide isomerase A3 is overexpressed in HeLa cells as compared to HaCaT | [26] | ||||
| 14. | PGAM1 | PGAM1_HUMAN | Phosphoglycerate mutase 1 | Differential 2D-PAGE proteomics on HeLa cells treated or not with suberonylanilide hydroxamic acid (SAHA). Confirmation on HeLa and CaSki cells by Western blot. | PGAM1 was significantly downregulated in HeLa and CaSki cells after treatment with HDAC1 inhibitor and antitumor agent SAHA. | [52] |
| Differential proteomics of HeLa cells versus non-tumorigenic cell line HaCaT | Differential proteomics study shows that Phosphoglycerate mutase 1 is overexpressed in HeLa cells as compared to HaCaT | [26] | ||||
| 15. | S100P | S100P_HUMAN | Protein S100-P | Differential-display PCR for identification of differentially expressed mRNAs in cells containing inducible E7. Verification of S100P mRNA and protein expression by Northern and Western blot, respectively. | S100P mRNA and protein expression was down-regulated in the E7-expressing cells. | [53] |
| HeLa, CGL3 and SiHa carcinoma cells as well as HCE16/3 immortalized cells were investigated for S100P expression. DNA methylation was inhibited by 5-aza-dC in S100P-negative cell lines CGL1 and Caski and the SP100 expressing SiHa cells. Suppression subtractive PCR between two HeLa cell lines with different tumerigenic capacity. | Expression of S100P gene in cervical carcinoma cells is not regulated by E7. S100P correlates with and contributes to the tumorigenic capacity of HeLa cells. Inhibition of DNA methylation resulted in induced S100P transcription in CGL1 and Caski but did not change S100P expression in SiHa cells. Suppression subtractive PCR identified differentially expressed genes, including S100P, with possible relevance for control of tumorigenic potential using two cervical carcinoma cell lines of the common HeLa origin, but of different capacity to generate tumors in nude mice. | [54,55] | ||||
| 16. | SERPINB13 | SPB13_HUMAN | SERPINB13 protein | Microarray analysis on primary cervical cancer samples with or without lymph node metastasis and confirmation by semi-quantitative PCR. | SERPINB13 was involved in a set of proteins that could be used for prediction of lymph node metastasis. | [56] |
| 17. | SOD1 | SODC_HUMAN | Superoxide dismutase [Cu-Zn] | siRNA knowkdown and RT-PCR of Cervical cancer cell line SiHa. | Knockdown of SOD1 expression in SiHa cells significantly enhanced lipid peroxidation and cytotoxicity on exposure to docosahexaenoic acid | [57] |
| 18. | SOD2 | SODM_HUMAN | Superoxide dismutase 2 | qPCR on SCC tissue from patients with or without lymph node metastasis | Correlation of SOD2 with lymph node metastasis in patients with early stage cervical carcinoma | [58] |
| Microarray on HPV16/18 immortalized keratinocytes and confirmation by Northern blot. Immunohistochemistry on 331 cervical histological samples. | Correlation of SOD2 expression with differential NF-kB activation by TNF in HPV16/18 immortalized keratinocytes. Abnormal expression of SOD 2 correlates with different stages of cervical neoplasia. | [59,60] | ||||
| 19. | YWHAQ | 1433T_HUMAN | 14-3-3 protein theta | Differential proteomics on normal versus cervical SCC tissues. | Decreased abundance of 14-3-3 epsilon in SCC tissues | [28] |
| 20. | YWHAE | 1433E_HUMAN | 14-3-3 protein epsilon | Immunostaining on 297 SCC tissues | Reduced immunostaining of 14-3-3 sigma protein in the cytoplasm and shuttle of 14-3-3 sigma protein into the nucleus may be two different mechanisms that determine the carcinogenesis of SCC | [61] |
| qPCR on SCC tissue from patients with or without lymph node metastasis | Correlation of 14-3-3 zeta protein expression with lymph node metastasis in patients with early stage cervical carcinoma | [58] | ||||
| Proteomics of HeLa cells versus non-tumorigenic cell line HaCaT, verification by Western Blot and Cytoscape pathway analysis. | Differential proteomics study and subsequent pathway analysis reveals that 14-3-3 zeta protein is key in the determination towards proliferation or cell death. The 14-3-3 family members emerge as important regulators in carcinogenesis and as possible clinical targets. | [26,27] | ||||
| Pull-down in HeLa cells | E6 oncoproteins from HR-HPVs interact with 14-3-3 zeta protein via PDZ domain | [62] |
3.3.2. Only CVF from Precancerous Patients Contains Proteins That Make up the Central Core of Cervical Cancer
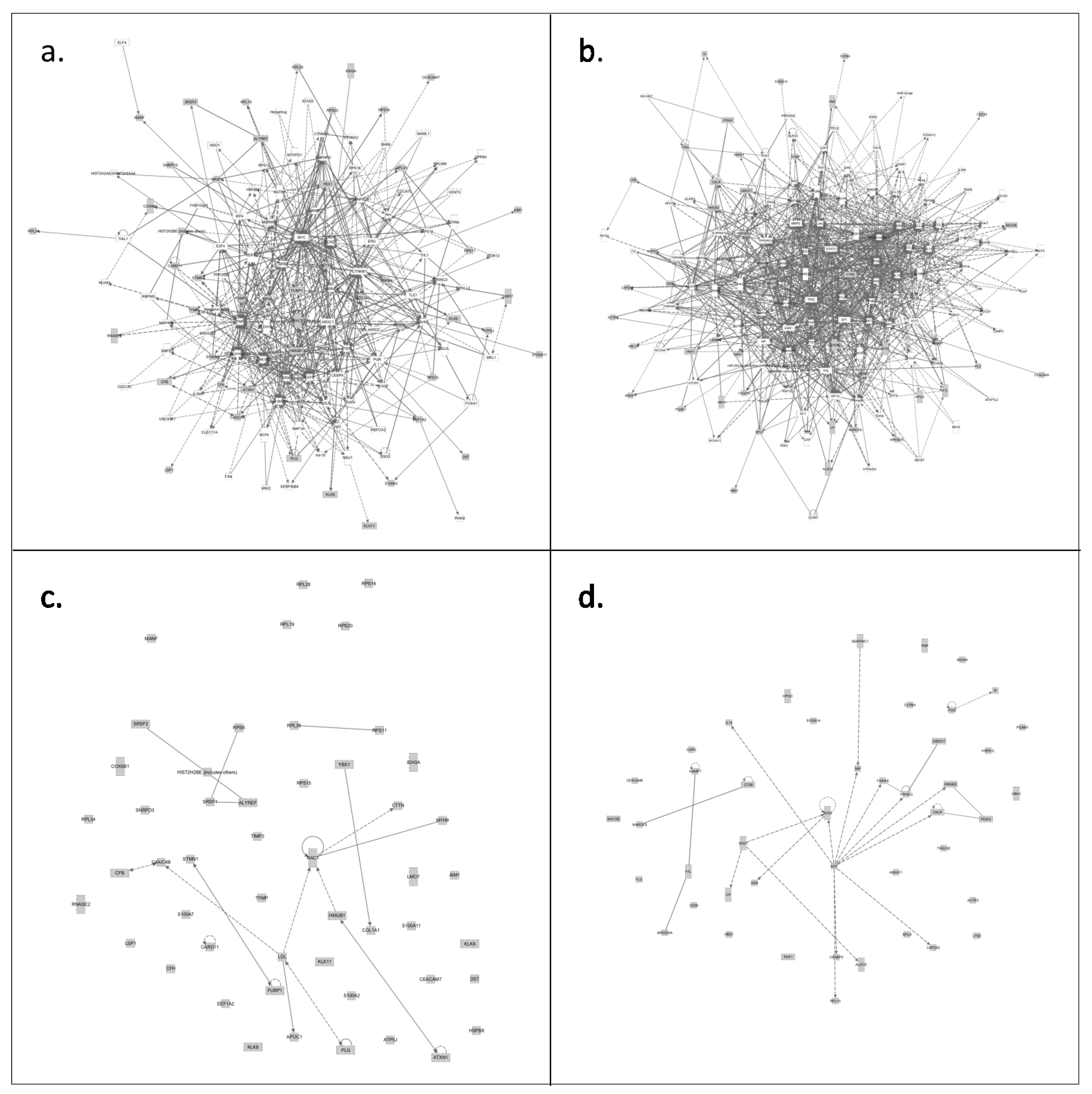
4. Conclusions
Supplementary Materials
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cole, A.M. Innate host defense of human vaginal and cervical mucosae. Curr. Top. Microbiol. Immunol. 2006, 306, 199–230. [Google Scholar]
- Zegels, G.; van Raemdonck, G.A.; Tjalma, W.A.; van Ostade, X.W. Use of cervicovaginal fluid for the identification of biomarkers for pathologies of the female genital tract. Proteome Sci. 2010, 8, e63. [Google Scholar] [CrossRef]
- Farage, M.A.; Miller, K.W.; Sobel, J.D. Dynamics of the vaginal ecosystem-hormonal influences. Infect. Dis. Res. Treat. 2010, 3, 1–15. [Google Scholar]
- Hickey, R.J.; Zhou, X.; Pierson, J.D.; Ravel, J.; Forney, L.J. Understanding vaginal microbiome complexity from an ecological perspective. Transl. Res. J. Lab. Clin. Med. 2012, 160, 267–282. [Google Scholar]
- Burgener, A.; Tjernlund, A.; Kaldensjo, T.; Abou, M.; McCorrister, S.; Westmacott, G.R.; Mogk, K.; Ambrose, E.; Broliden, K.; Ball, B. A systems biology examination of the human female genital tract shows compartmentalization of immune factor expression. J. Virol. 2013, 87, 5141–5150. [Google Scholar] [CrossRef]
- Birse, K.M.; Burgener, A.; Westmacott, G.R.; McCorrister, S.; Novak, R.M.; Ball, T.B. Unbiased proteomics analysis demonstrates significant variability in mucosal immune factor expression depending on the site and method of collection. PLoS One 2013, 8, e79505. [Google Scholar]
- Dezzutti, C.S.; Hendrix, C.W.; Marrazzo, J.M.; Pan, Z.; Wang, L.; Louissaint, N.; Kalyoussef, S.; Torres, N.M.; Hladik, F.; Parikh, U.; et al. Performance of swabs, lavage, and diluents to quantify biomarkers of female genital tract soluble mucosal mediators. PLoS One 2011, 6, e23136. [Google Scholar]
- Gok, M.; Heideman, D.A.; van Kemenade, F.J.; Berkhof, J.; Rozendaal, L.; Spruyt, J.W.; Voorhorst, F.; Belien, J.A.; Babovic, M.; Snijders, P.J.; et al. Hpv testing on self collected cervicovaginal lavage specimens as screening method for women who do not attend cervical screening: Cohort study. Br. Med. J. 2010, 340, c1040. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Shahneh, F.Z. Sensitive antibody-based CTCS detection from peripheral blood. Hum. Antibodies 2013, 22, 51–54. [Google Scholar]
- Alix-Panabieres, C.; Pantel, K. Circulating tumor cells: Liquid biopsy of cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef]
- Alix-Panabieres, C.; Pantel, K. Technologies for detection of circulating tumor cells: Facts and vision. Lab Chip 2014, 14, 57–62. [Google Scholar] [CrossRef]
- Pinto, A.P.; Degen, M.; Villa, L.L.; Cibas, E.S. Immunomarkers in gynecologic cytology: The search for the ideal ‘biomolecular papanicolaou test’. Acta Cytol. 2012, 56, 109–121. [Google Scholar]
- Stanley, M. Immunobiology of HPV and HPV vaccines. Gynecol. Oncol. 2008, 109, S15–S21. [Google Scholar] [CrossRef]
- Maglennon, G.A.; Doorbar, J. The biology of papillomavirus latency. Open Virol. J. 2012, 6, 190–197. [Google Scholar]
- Stanley, M. Potential mechanisms for HPV vaccine-induced long-term protection. Gynecol. Oncol. 2010, 118, S2–S7. [Google Scholar] [CrossRef]
- Kanodia, S.; Fahey, L.M.; Kast, W.M. Mechanisms used by human papillomaviruses to escape the host immune response. Curr. Cancer Drug Targets 2007, 7, 79–89. [Google Scholar] [CrossRef]
- Chang, Y.E.; Laimins, L.A. Microarray analysis identifies interferon-inducible genes and STAT-1 as major transcriptional targets of human papillomavirus type 31. J. Virol. 2000, 74, 4174–4182. [Google Scholar] [CrossRef]
- Nees, M.; Geoghegan, J.M.; Hyman, T.; Frank, S.; Miller, L.; Woodworth, C.D. Papillomavirus type 16 oncogenes downregulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-kappab-responsive genes in cervical keratinocytes. J. Virol. 2001, 75, 4283–4296. [Google Scholar] [CrossRef]
- Rosa, M.I.; Fachel, J.M.; Rosa, D.D.; Medeiros, L.R.; Igansi, C.N.; Bozzetti, M.C. Persistence and clearance of human papillomavirus infection: A prospective cohort study. Am. J. Obstetr. Gynecol. 2008, 199, e617. [Google Scholar]
- Van Raemdonck, G.A.A.; Tjalma, W.A.A.; Coen, E.P.; Depuydt, C.E.; van Ostade, X.W.M. Identification of protein biomarkers for cervical cancer using human cervicovaginal fluid. PLoS One 2014, in press. [Google Scholar]
- O’Meara, A.T. Present standards for cervical cancer screening. Curr. Opin. Oncol. 2002, 14, 505–511. [Google Scholar] [CrossRef]
- Zegels, G.; van Raemdonck, G.A.; Coen, E.P.; Tjalma, W.A.; van Ostade, X.W. Comprehensive proteomic analysis of human cervicovaginal fluid using colposcopy samples. Proteome Sci. 2009, 7, e17. [Google Scholar] [CrossRef]
- Ingenuity Pathway Analysis. Available online: http://www.ingenuity.com (accessed on 9 December 2013).
- Higareda-Almaraz, J.C.; Enriquez-Gasca Mdel, R.; Hernandez-Ortiz, M.; Resendis-Antonio, O.; Encarnacion-Guevara, S. Proteomic patterns of cervical cancer cell lines, a network perspective. BMC Syst. Biol. 2011, 5, e96. [Google Scholar]
- Higareda-Almaraz, J.C.; Valtierra-Gutierrez, I.A.; Hernandez-Ortiz, M.; Contreras, S.; Hernandez, E.; Encarnacion, S. Analysis and prediction of pathways in hela cells by integrating biological levels of organization with systems-biology approaches. PLoS One 2013, 8, e65433. [Google Scholar]
- Bae, S.M.; Lee, C.H.; Cho, Y.L.; Nam, K.H.; Kim, Y.W.; Kim, C.K.; Han, B.D.; Lee, Y.J.; Chun, H.J.; Ahn, W.S. Two-dimensional gel analysis of protein expression profile in squamous cervical cancer patients. Gynecol. Oncol. 2005, 99, 26–35. [Google Scholar] [CrossRef]
- Zhu, X.; Lv, J.; Yu, L.; Zhu, X.; Wu, J.; Zou, S.; Jiang, S. Proteomic identification of differentially-expressed proteins in squamous cervical cancer. Gynecol. Oncol. 2009, 112, 248–256. [Google Scholar]
- Song, J.Y.; Bae, H.S.; Koo, D.H.; Lee, J.K.; Jung, H.H.; Lee, K.W.; Lee, N.W. Candidates for tumor markers of cervical cancer discovered by proteomic analysis. J. Korean Med. Sci. 2012, 27, 1479–1485. [Google Scholar]
- De Marco, F.; Bucaj, E.; Foppoli, C.; Fiorini, A.; Blarzino, C.; Filipi, K.; Giorgi, A.; Schinina, M.E.; di Domenico, F.; Coccia, R.; et al. Oxidative stress in HPV-driven viral carcinogenesis: Redox proteomics analysis of HPV-16 dysplastic and neoplastic tissues. PLoS One 2012, 7, e34366. [Google Scholar]
- Kikkawa, F.; Mizuno, M.; Shibata, K.; Kajiyama, H.; Morita, T.; Ino, K.; Nomura, S.; Mizutani, S. Activation of invasiveness of cervical carcinoma cells by angiotensin II. Am. J. Obstetr. Gynecol. 2004, 190, 1258–1263. [Google Scholar]
- Simpson, K.L.; Jones, A.; Norman, S.; Holmes, C.H. Expression of the complement regulatory proteins decay accelerating factor (DAF, CD55), membrane cofactor protein (MCP, CD46) and CD59 in the normal human uterine cervix and in premalignant and malignant cervical disease. Am. J. Pathol. 1997, 151, 1455–1467. [Google Scholar]
- Gelderman, K.A.; Blok, V.T.; Fleuren, G.J.; Gorter, A. The inhibitory effect of CD46, CD55, and CD59 on complement activation after immunotherapeutic treatment of cervical carcinoma cells with monoclonal antibodies or bispecific monoclonal antibodies. Lab. Invest. 2002, 82, 483–493. [Google Scholar] [CrossRef]
- Harima, Y.; Togashi, A.; Horikoshi, K.; Imamura, M.; Sougawa, M.; Sawada, S.; Tsunoda, T.; Nakamura, Y.; Katagiri, T. Prediction of outcome of advanced cervical cancer to thermoradiotherapy according to expression profiles of 35 genes selected by cdna microarray analysis. Int. J. Radiat. Oncol. Biol. Phys. 2004, 60, 237–248. [Google Scholar] [CrossRef]
- Wu, D.; Wang, H.; Li, Z.; Wang, L.; Zheng, F.; Jiang, J.; Gao, Y.; Zhong, H.; Huang, Y.; Suo, Z. Cathepsin B may be a potential biomarker in cervical cancer. Histol. Histopathol. 2012, 27, 79–87. [Google Scholar]
- Liao, C.J.; Wu, T.I.; Huang, Y.H.; Chang, T.C.; Wang, C.S.; Tsai, M.M.; Hsu, C.Y.; Tsai, M.H.; Lai, C.H.; Lin, K.H. Overexpression of gelsolin in human cervical carcinoma and its clinicopathological significance. Gynecol. Oncol. 2011, 120, 135–144. [Google Scholar] [CrossRef]
- Lokamani, I.; Looi, M.L.; Ali, S.A.M.; Dali, A.Z.H.M.; Annuar, M.A.A.; Jamal, R. Gelsolin and ceruloplasmin as potential predictive biomarkers for cervical cancer by 2D-DIGE proteomics analysis. Pathol. Oncol. Res. 2014, 20, 119–129. [Google Scholar]
- Boccardo, E. HPV-mediated genome instability: At the roots of cervical carcinogenesis. Cytogenet. Genome Res. 2010, 128, 57–65. [Google Scholar] [CrossRef]
- Lee, K.A.; Cho, K.J.; Kim, S.H.; Shim, J.H.; Lim, J.S.; Cho, D.H.; Song, M.S.; Dinarello, C.A.; Yoon, D.Y. IL-18 E42A mutant is resistant to the inhibitory effects of HPV-16 E6 and E7 oncogenes on the IL-18-mediated immune response. Cancer Lett. 2005, 229, 261–270. [Google Scholar]
- Lee, S.J.; Cho, Y.S.; Cho, M.C.; Shim, J.H.; Lee, K.A.; Ko, K.K.; Choe, Y.K.; Park, S.N.; Hoshino, T.; Kim, S.; et al. Both E6 and E7 oncoproteins of human papillomavirus 16 inhibit IL-18-induced IFN-gamma production in human peripheral blood mononuclear and NK cells. J. Immunol. 2001, 167, 497–504. [Google Scholar] [CrossRef]
- Cho, Y.S.; Kang, J.W.; Cho, M.; Cho, C.W.; Lee, S.; Choe, Y.K.; Kim, Y.; Choi, I.; Park, S.N.; Kim, S.; et al. Down modulation of IL-18 expression by human papillomavirus type 16 E6 oncogene via binding to IL-18. FEBS Lett. 2001, 501, 139–145. [Google Scholar] [CrossRef]
- Yang, Y.C.; Chang, T.Y.; Chen, T.C.; Chang, S.C.; Lin, W.S.; Lee, Y.J. Genetic variants in interleukin-18 gene and risk for cervical squamous cell carcinoma. Hum. Immunol. 2013, 74, 882–887. [Google Scholar] [CrossRef]
- Krockenberger, M.; Engel, J.B.; Kolb, J.; Dombrowsky, Y.; Hausler, S.F.; Kohrenhagen, N.; Dietl, J.; Wischhusen, J.; Honig, A. Macrophage migration inhibitory factor expression in cervical cancer. J. Cancer Res. Clin. Oncol. 2010, 136, 651–657. [Google Scholar]
- Wu, S.; Lian, J.; Tao, H.; Shang, H.; Zhang, L. Correlation of macrophage migration inhibitory factor gene polymorphism with the risk of early-stage cervical cancer and lymphatic metastasis. Oncol. Lett. 2011, 2, 1261–1267. [Google Scholar]
- Xiao, D.Z.; Dai, B.; Chen, J.; Luo, Q.; Liu, X.Y.; Lin, Q.X.; Li, X.H.; Huang, W.; Yu, X.Y. Loss of macrophage migration inhibitory factor impairs the growth properties of human hela cervical cancer cells. Cell Prolif. 2011, 44, 582–590. [Google Scholar] [CrossRef]
- Cheng, R.J.; Deng, W.G.; Niu, C.B.; Li, Y.Y.; Fu, Y. Expression of macrophage migration inhibitory factor and cd74 in cervical squamous cell carcinoma. Int. J. Gynecol. Cancer 2011, 21, 1004–1012. [Google Scholar] [CrossRef]
- Hebbar, V.; Damera, G.; Sachdev, G.P. Differential expression of muc genes in endometrial and cervical tissues and tumors. BMC Cancer 2005, 5, e124. [Google Scholar]
- Zhang, Z.J.; Tong, Y.Q.; Wang, J.J.; Yang, C.; Zhou, G.H.; Li, Y.H.; Xie, P.L.; Hu, J.Y.; Li, G.C. Spaceflight alters the gene expression profile of cervical cancer cells. Chin. J. Cancer 2011, 30, 842–852. [Google Scholar]
- Yu, L.; He, M.; Yang, Z.; Chen, G.; Li, M.; Wang, L.; Chen, S. Olfactomedin 4 is a marker for progression of cervical neoplasia. Int. J. Gynecol. Cancer 2011, 21, 367–372. [Google Scholar] [CrossRef]
- Liao, C.J.; Wu, T.I.; Huang, Y.H.; Chang, T.C.; Wang, C.S.; Tsai, M.M.; Lai, C.H.; Liang, Y.; Jung, S.M.; Lin, K.H. Glucose-regulated protein 58 modulates cell invasiveness and serves as a prognostic marker for cervical cancer. Cancer Sci. 2011, 102, 2255–2263. [Google Scholar] [CrossRef]
- He, J.; Huang, C.; Tong, A.; Chen, B.; Zeng, Z.; Zhang, P.; Wang, C.; Wei, Y. Proteomic analysis of cervical cancer cells treated with suberonylanilide hydroxamic acid. J. Biosci. 2008, 33, 715–721. [Google Scholar] [CrossRef]
- Schonning, B.H.; Bevort, M.; Mikkelsen, S.; Andresen, M.; Thomsen, P.; Leffers, H.; Norrild, B. Human papillomavirus type 16 E7-regulated genes: Regulation of S100P and ADP/ATP carrier protein genes identified by differential-display technology. J. Gener. Virol. 2000, 81, 1009–1015. [Google Scholar]
- Jakubickova, L.; Barathova, M.; Pastorekova, S.; Pastorek, J.; Gibadulinova, A. Expression of s100p gene in cervical carcinoma cells is independent of E7 human papillomavirus oncogene. Acta Virol. 2005, 49, 133–137. [Google Scholar]
- Jakubickova, L.; Biesova, Z.; Pastorekova, S.; Kettmann, R.; Pastorek, J. Methylation of the CA9 promoter can modulate expression of the tumor-associated carbonic anhydrase IX in dense carcinoma cell lines. Int. J.Oncol. 2005, 26, 1121–1127. [Google Scholar]
- Kim, T.J.; Choi, J.J.; Kim, W.Y.; Choi, C.H.; Lee, J.W.; Bae, D.S.; Son, D.S.; Kim, J.; Park, B.K.; Ahn, G.; et al. Gene expression profiling for the prediction of lymph node metastasis in patients with cervical cancer. Cancer Sci. 2008, 99, 31–38. [Google Scholar]
- Ding, W.Q.; Vaught, J.L.; Yamauchi, H.; Lind, S.E. Differential sensitivity of cancer cells to docosahexaenoic acid-induced cytotoxicity: The potential importance of down-regulation of superoxide dismutase 1 expression. Mol. Cancer Ther. 2004, 3, 1109–1117. [Google Scholar]
- Huang, L.; Zheng, M.; Zhou, Q.M.; Zhang, M.Y.; Jia, W.H.; Yun, J.P.; Wang, H.Y. Identification of a gene-expression signature for predicting lymph node metastasis in patients with early stage cervical carcinoma. Cancer 2011, 117, 3363–3373. [Google Scholar] [CrossRef]
- Termini, L.; Boccardo, E.; Esteves, G.H.; Hirata, R., Jr.; Martins, W.K.; Colo, A.E.; Neves, E.J.; Villa, L.L.; Reis, L.F. Characterization of global transcription profile of normal and HPV-immortalized keratinocytes and their response to tnf treatment. BMC Med. Genomics 2008, 1, e29. [Google Scholar]
- Termini, L.; Filho, A.L.; Maciag, P.C.; Etlinger, D.; Alves, V.A.; Nonogaki, S.; Soares, F.A.; Villa, L.L. Deregulated expression of superoxide dismutase-2 correlates with different stages of cervical neoplasia. Dis. Mark. 2011, 30, 275–281. [Google Scholar]
- Holm, R.; Ali, T.; Svendsrud, D.H.; Nesland, J.M.; Kristensen, G.B.; Lyng, H. Expression of 14-3-3sigma in cervical squamous cell carcinomas: Relationship with clinical outcome. Oncol. Rep. 2009, 22, 11–15. [Google Scholar]
- Boon, S.S.; Banks, L. High-risk human papillomavirus E6 oncoproteins interact with 14-3-3zeta in a PDZ binding motif-dependent manner. J. Virol. 2013, 87, 1586–1595. [Google Scholar] [CrossRef]
- Gardino, A.K.; Yaffe, M.B. 14-3-3 proteins as signaling integration points for cell cycle control and apoptosis. Semin. Cell Dev. Biol. 2011, 22, 688–695. [Google Scholar] [CrossRef]
- Freeman, A.K.; Morrison, D.K. 14-3-3 proteins: Diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 2011, 22, 681–687. [Google Scholar] [CrossRef]
- Lemoine, J.; Fortin, T.; Salvador, A.; Jaffuel, A.; Charrier, J.P.; Choquet-Kastylevsky, G. The current status of clinical proteomics and the use of MRM and MRM(3) for biomarker validation. Expert Rev. Mol. Diagn. 2012, 12, 333–342. [Google Scholar] [CrossRef]
- Picotti, P.; Aebersold, R. Selected reaction monitoring-based proteomics: Workflows, potential, pitfalls and future directions. Nat. Methods 2012, 9, 555–566. [Google Scholar] [CrossRef]
- Yassine, H.; Borges, C.R.; Schaab, M.R.; Billheimer, D.; Stump, C.; Reaven, P.; Lau, S.S.; Nelson, R. Mass spectrometric immunoassay and MRM as targeted MS-based quantitative approachesin biomarker development: Potential applications to cardiovascular disease and diabetes. Proteomics Clin. Appl. 2013, 7, 528–540. [Google Scholar] [CrossRef]
- Ndayisaba, G.; Verwijs, M.C.; van Eeckhoudt, S.; Gasarabwe, A.; Hardy, L.; Borgdorff, H.; Kestelyn, E.; Jespers, V.A.; van de Wijgert, J. Feasibility and acceptability of a novel cervicovaginal lavage self-sampling device among women in Kigali, Rwanda. Sex. Transm. Dis. 2013, 40, 552–555. [Google Scholar]
- Zhao, F.H.; Jeronimo, J.; Qiao, Y.L.; Schweizer, J.; Chen, W.; Valdez, M.; Lu, P.; Zhang, X.; Kang, L.N.; Bansil, P.; et al. An evaluation of novel, lower-cost molecular screening tests for human papillomavirus in rural China. Cancer Prev. Res. 2013, 6, 938–948. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Van Ostade, X.; Dom, M.; Van Raemdonck, G. IPA Analysis of Cervicovaginal Fluid from Precancerous Women Points to the Presence of Biomarkers for the Precancerous State of Cervical Carcinoma. Proteomes 2014, 2, 426-450. https://doi.org/10.3390/proteomes2030426
Van Ostade X, Dom M, Van Raemdonck G. IPA Analysis of Cervicovaginal Fluid from Precancerous Women Points to the Presence of Biomarkers for the Precancerous State of Cervical Carcinoma. Proteomes. 2014; 2(3):426-450. https://doi.org/10.3390/proteomes2030426
Chicago/Turabian StyleVan Ostade, Xaveer, Martin Dom, and Geert Van Raemdonck. 2014. "IPA Analysis of Cervicovaginal Fluid from Precancerous Women Points to the Presence of Biomarkers for the Precancerous State of Cervical Carcinoma" Proteomes 2, no. 3: 426-450. https://doi.org/10.3390/proteomes2030426




