Proteomic Analysis of Responsive Proteins Induced in Japanese Birch Plantlet Treated with Salicylic Acid
Abstract
:1. Introduction
2. Experimental
2.1. Plant Material
2.2. Salicylic Acid (SA) Treatment
2.3. Preparation of Protein Samples
2.4. Two-Dimensional Electrophoresis (2-DE)
2.5. Staining 2-DE Gel
2.6. Image Analysis of 2-DE Gels
2.7. In-Gel Digestion
2.8. LC/MS/MS of the Peptide Sample
2.9. Database Search
3. Results and Discussion
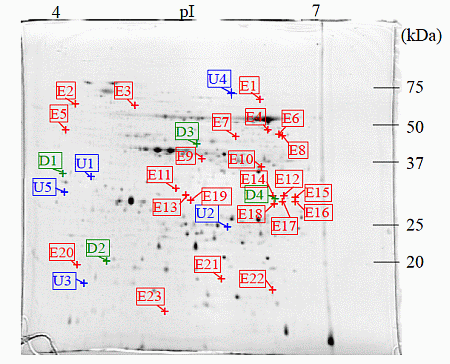
 : Specifically expressed protein spots in C1 gel.
: Specifically expressed protein spots in C1 gel.
 : Specifically expressed protein spots in C1 gel.
: Specifically expressed protein spots in C1 gel.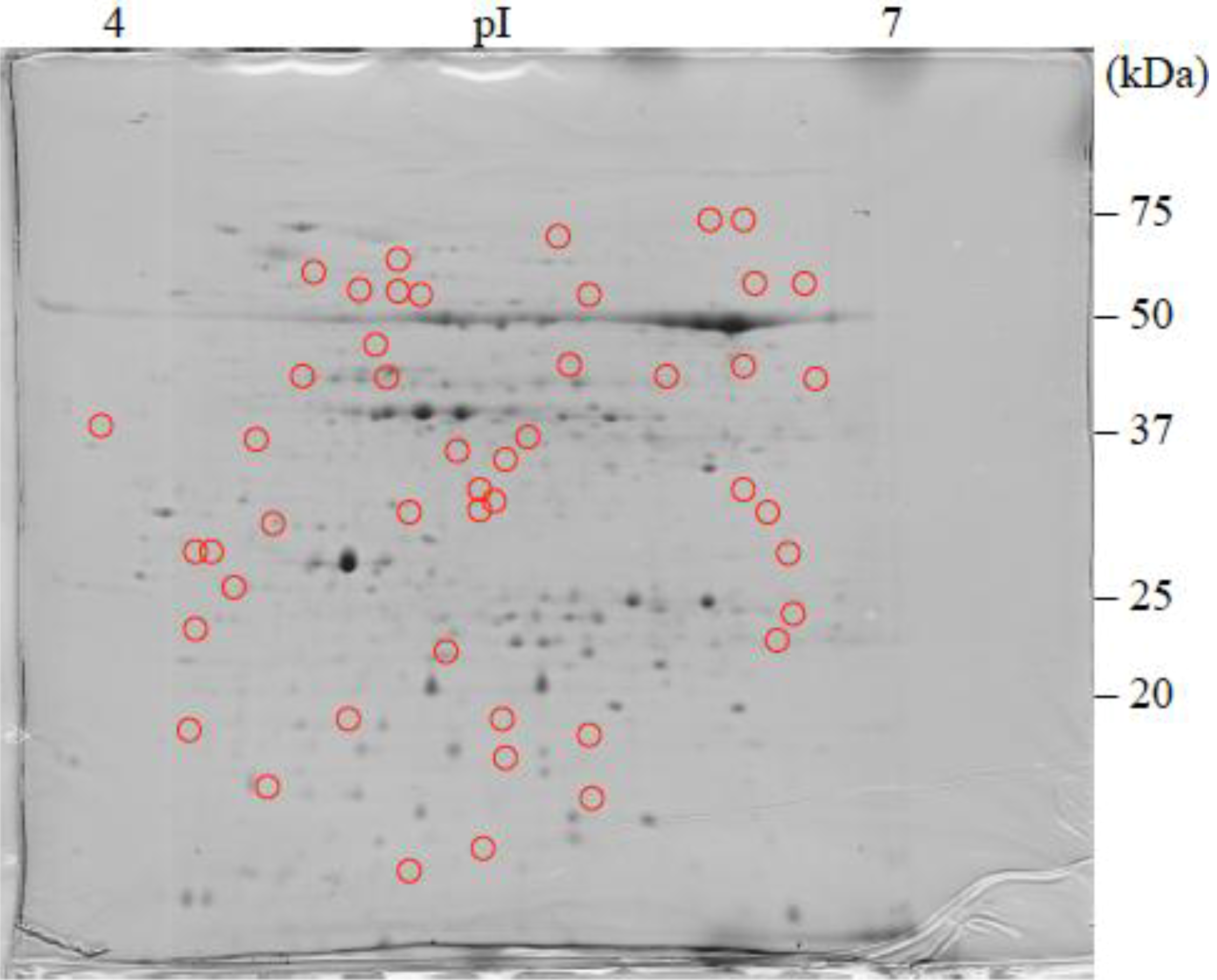
 : Specifically expressed protein spots in C2SA gel.
: Specifically expressed protein spots in C2SA gel.
 : Specifically expressed protein spots in C2SA gel.
: Specifically expressed protein spots in C2SA gel.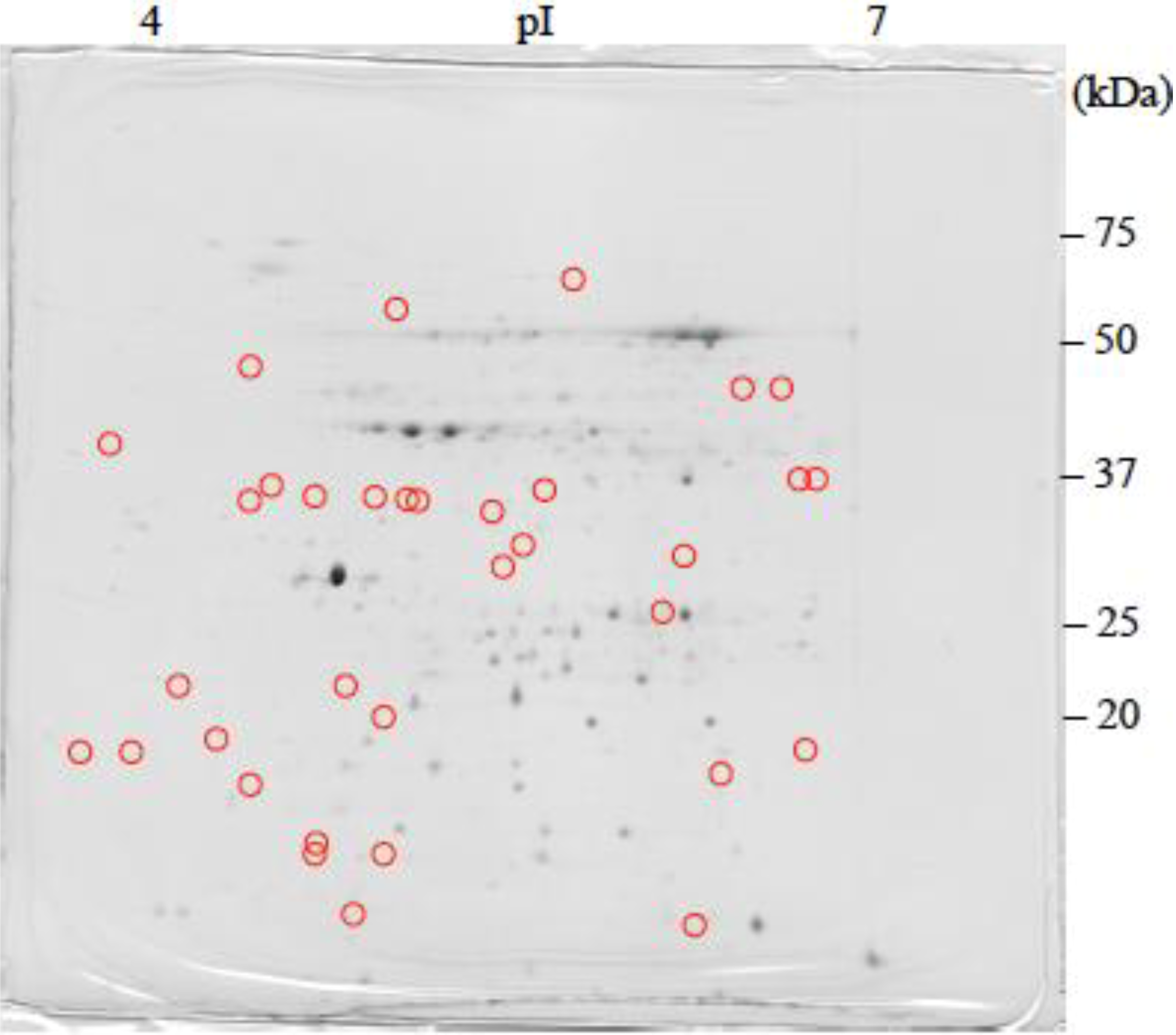
 : Specifically expressed protein spot in TSA gel.
: Specifically expressed protein spot in TSA gel.  : Significantly increased protein spots in TSA gel.
: Significantly increased protein spots in TSA gel.  : Significantly decreased protein spots in TSA gel.
: Significantly decreased protein spots in TSA gel.
 : Specifically expressed protein spot in TSA gel.
: Specifically expressed protein spot in TSA gel.  : Significantly increased protein spots in TSA gel.
: Significantly increased protein spots in TSA gel.  : Significantly decreased protein spots in TSA gel.
: Significantly decreased protein spots in TSA gel.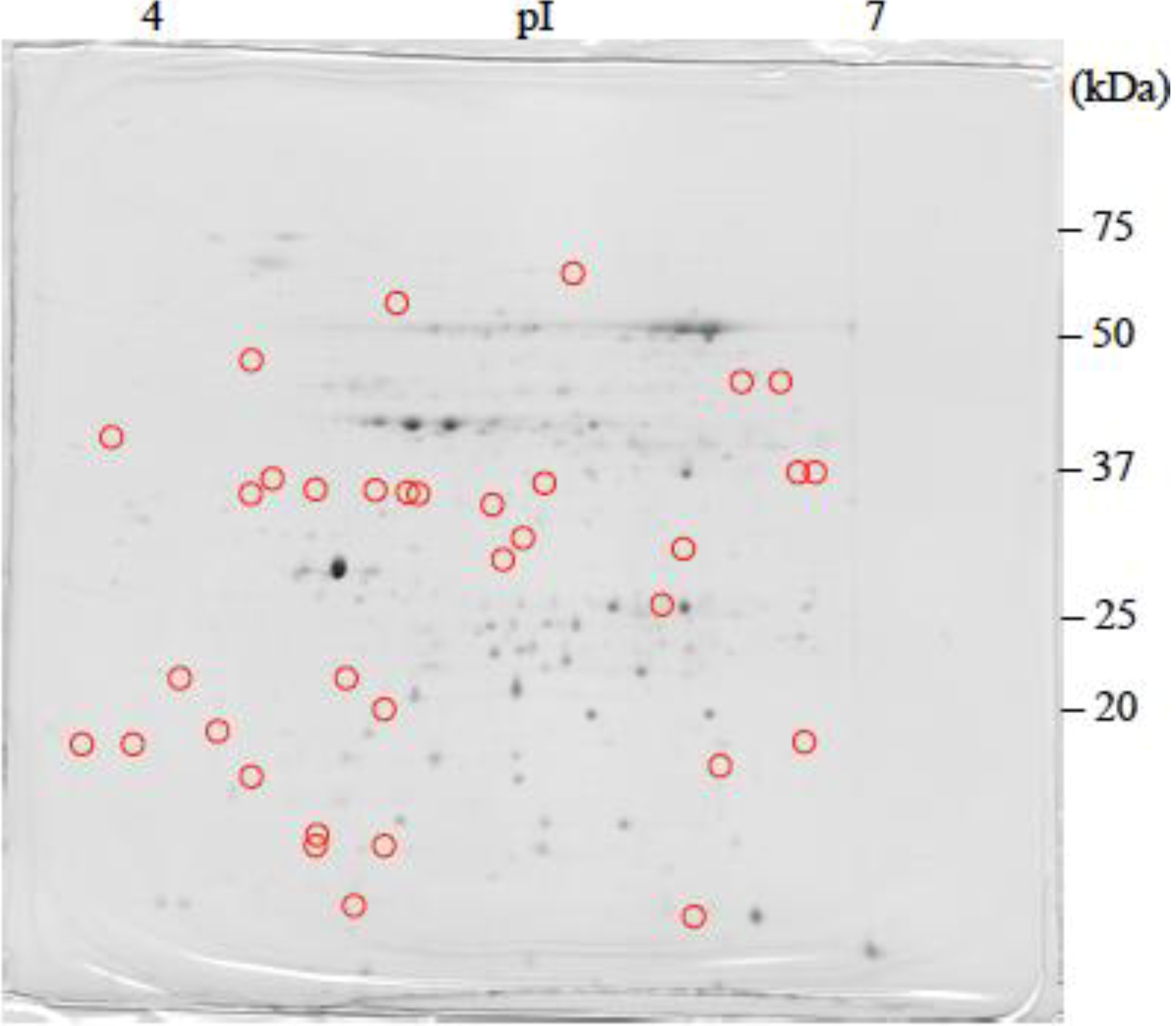
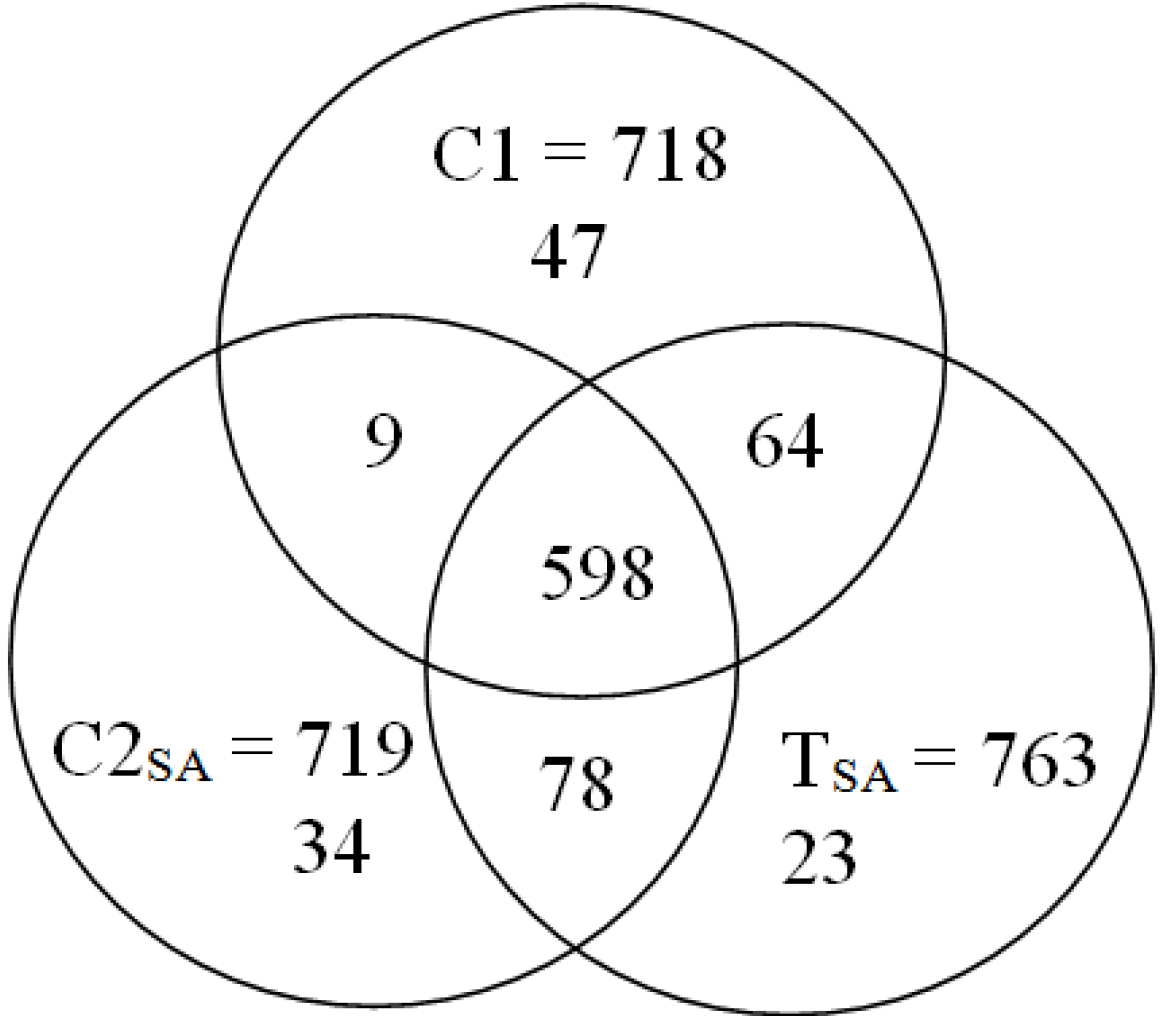
 : Specifically expressed protein spots in TSA gel;
: Specifically expressed protein spots in TSA gel;  : Significantly increased protein spots in TSA gel;
: Significantly increased protein spots in TSA gel;  : Significantly decreased protein spots in TSA gel.
: Significantly decreased protein spots in TSA gel.
 : Specifically expressed protein spots in TSA gel;
: Specifically expressed protein spots in TSA gel;  : Significantly increased protein spots in TSA gel;
: Significantly increased protein spots in TSA gel;  : Significantly decreased protein spots in TSA gel.
: Significantly decreased protein spots in TSA gel.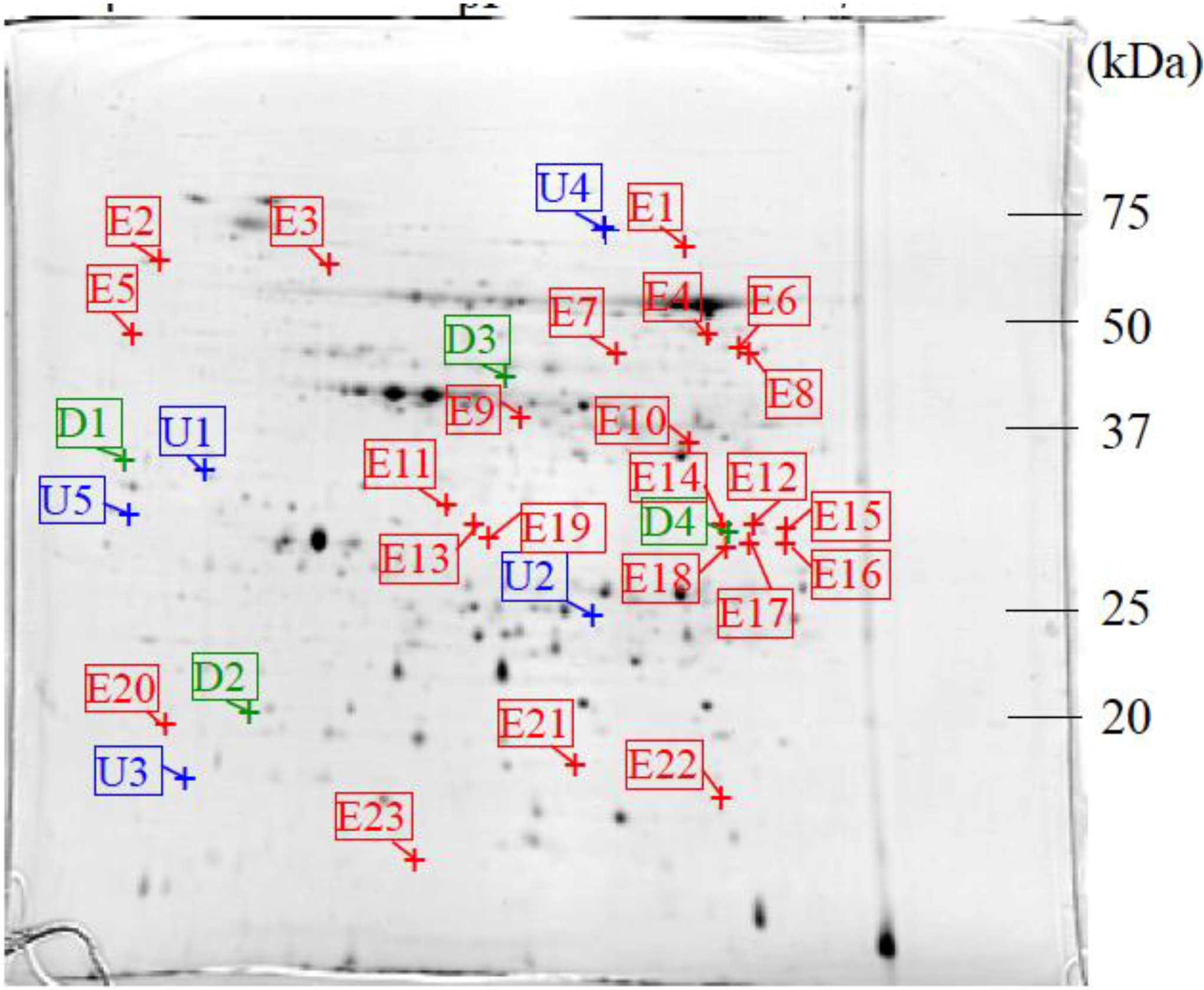
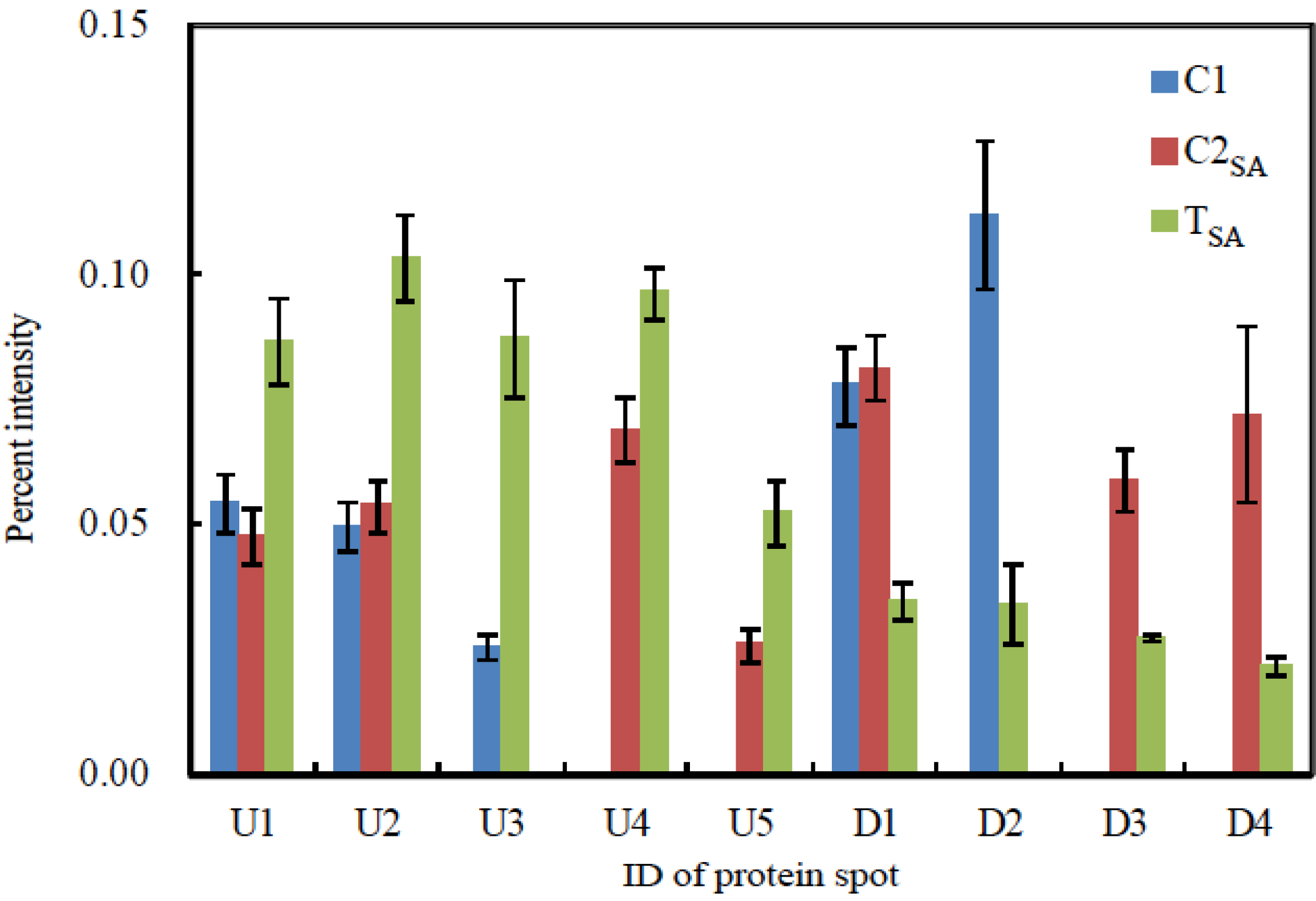
3.2. Protein Identification
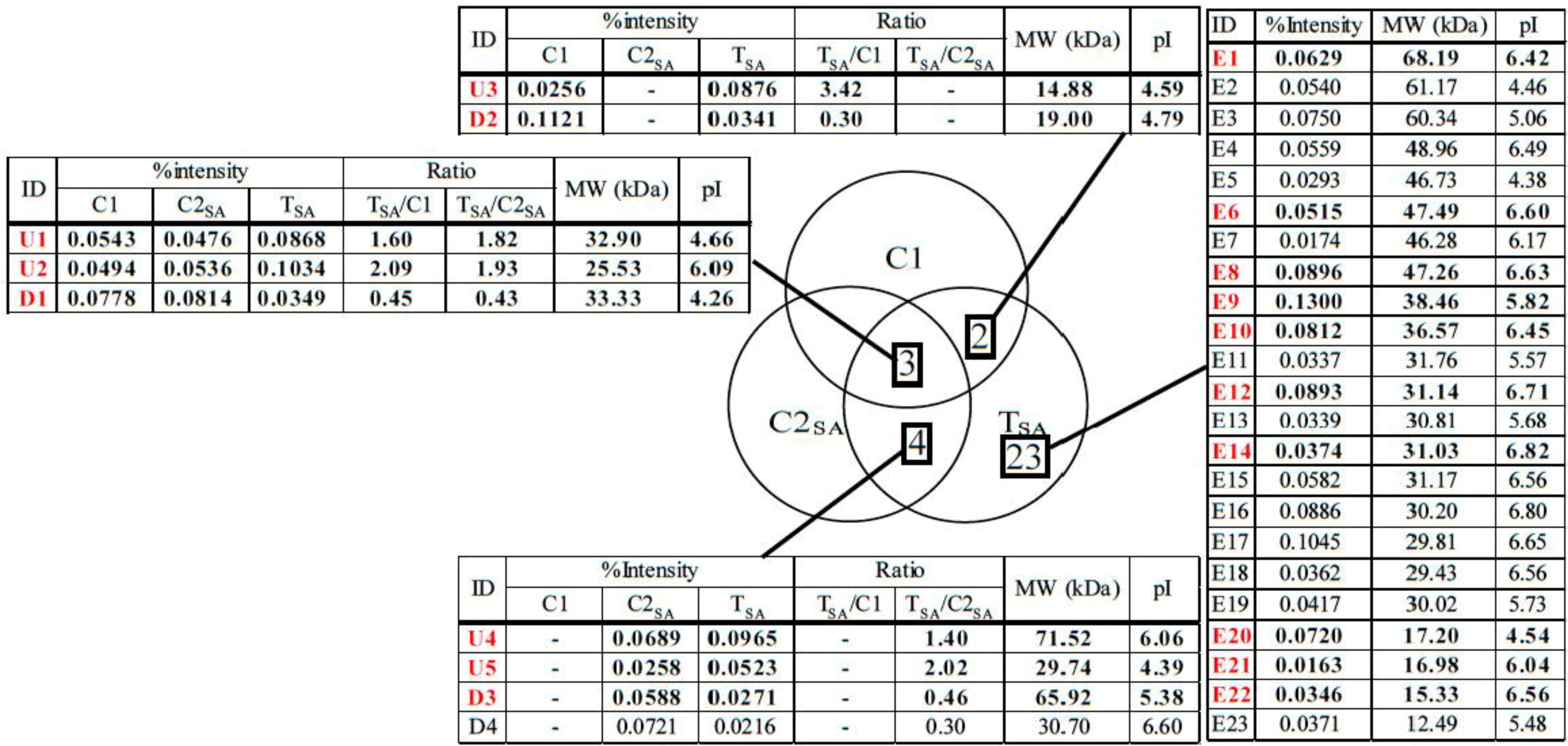
| ID | Protein | No. in NCBI | Score | % cover | Mw (kDa) Observed/Theoretical | pI Observed/Theoretical | Ratio | Plant | |
|---|---|---|---|---|---|---|---|---|---|
| TSA/C1 | TSA/C2SA | ||||||||
| Energy production | |||||||||
| E10 | Malate dehydrogenase | gi|255585544 | 139 | 14 | 36.57/28.84 | 6.45/5.44 | - | - | Ricinus communis |
| U4 | SDH1-1; ATP binding/succinate dehydrogenase | gi|15240075 | 43 | 2 | 71.52/70.24 | 6.06/5.86 | - | 1.40 | Arabidopsis thaliana |
| D3 | Phosphoglycerate kinase, chloroplastic; Flags; Precursor | gi|2499497 | 85 | 11 | 65.92/50.31 | 5.38/8.48 | - | 0.46 | Nicotiana tabacum |
| Metabolism | |||||||||
| E6 | Diaminopimelate decarboxylase putative | gi|255543757 | 79 | 3 | 47.34/54.55 | 6.62/6.58 | - | - | Ricinus communis |
| E9 | Arginase | gi|148828535 | 440 | 21 | 42.29/37.15 | 5.76/6.14 | - | - | Malus hupehensis |
| U1 | Chorismate mutase precursor | gi|429153 | 39 | 2 | 32.90/38.05 | 4.66/5.90 | 1.60 | 1.82 | Arabidopsisthaliana |
| Protein synthesis | |||||||||
| E22 | Peptidylprolyl isomerase (cyclophilin) | gi|21886603 | 44 | 8 | 15.33/18.51 | 6.56/8.68 | - | - | Betula pendula |
| U5 | Aminopeptidase | gi|255080640 | 35 | 1 | 29.74/63.62 | 4.39/7.97 | - | 2.02 | Micromonas sp. RCC299 |
| Unknown | |||||||||
| E12 | Predicted protein | gi|168008150 | 23 | 1 | 31.14/71.46 | 6.71/8.15 | - | - | Physcomitrella patens subsp. Patens |
| D1 | Hypothetical protein | gi|2342730 | 16 | 1 | 33.33/54.76 | 4.26/6.28 | 0.45 | 0.43 | Arabidopsis thaliana |
| D2 | Predicted: hypothetical protein | gi|225432620 | 101 | 14 | 19.00/22.52 | 4.79/9.73 | 0.30 | - | Vitis vinifera |
3.2.1. The Proteins Related to Energy Production
3.2.2. The Proteins Related to Metabolism
3.2.3. The Proteins Related to Protein Synthesis
3.2.4. Unknown Function Protein
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hosoi, Y. Betula platyphylla var. japonica. In Current Biotechnology Complete Book: Propagation and Breeding of Woody Plant; Ohyama, K., Saito, A., Eds.; Nougyo Tosyo: Tokyo, Japan, 1989; Volume 6; pp. 157–160. [Google Scholar]
- Ju, E.M.; Lee, S.E.; Hwang, H.J.; Kim, J.H. Antioxidant and anticancer activity of extract from Betula platyphylla var. japonica. Life Sci. 2004, 74, 1013–1026. [Google Scholar] [CrossRef]
- Sami, A.; Taru, M.; Salme, K.; Jari, Y.K. Pharmacological properties of the ubiquitous natural product betulin. Eur. J. Pharm. Sci. 2006, 29, 1–13. [Google Scholar] [CrossRef]
- Zabel, R.A. Basidiocarp development in Inonotus obliquus and its inhibition by stem treatment. Forest Sci. 1976, 22, 431–437. [Google Scholar]
- True, R.P.; King, J.F. Cankers and decays of birch associated with two Poria species. J. Forest 1995, 53, 412–415. [Google Scholar]
- Kim, Y.O.; Park, H.W.; Kim, J.H.; Lee, J.Y.; Moon, S.H.; Shin, C.S. Anti-cancer effect and structural characterization of endo-polysaccharide from cultivated mycelia of Inonotus obliquus. Life Sci. 2006, 79, 72–80. [Google Scholar] [CrossRef]
- Schumann, G.L.; D’Arcy, C.J. Induced (active) defenses. In Essential Plant Pathology; APS Press: St. Paul, MN, USA, 2006; pp. 203–206. [Google Scholar]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 23–61. [Google Scholar] [CrossRef]
- Lattanzio, V.; Lattanzio, V.M.T.; Cardinali, A. Role of phenolics in the resistance mechanisms of plants against fungal pathogens and insects. In Neural Proteoglycan; Maeda, N., Ed.; Research Signpost: Kerala, India, 2006; pp. 23–67. [Google Scholar]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef]
- Ferreira, R.B.; Monteiro, S.; Freitas, R.; Santos, C.N.; Chen, Z.; Batista, L.M.; Duarte, J.; Borges, A.; Teixeira, A.R. The role of plant defence proteins in fungal pathogenesis. Mol. Plant Pathol. 2007, 8, 677–700. [Google Scholar] [CrossRef]
- Mehdy, M.C. Active oxygen species in plant defense against pathogens. Plant Physiol. 1994, 105, 467–472. [Google Scholar]
- Siegrist, J.; Jeblick, W.; Kauss, H. Defense responses in infected and elicited cucumber (Cucumis sativus L.) hypocotyl segments exhibiting acquired resistance. Plant Physiol. 1994, 105, 1365–1374. [Google Scholar]
- Métraux, J.P.; Nawrath, C.; Genoud, T. Systemic acquired resistance. Euphytica 2002, 124, 237–243. [Google Scholar] [CrossRef]
- Gaffney, T.; Friedrich, L.; Vernooij, B.; Negrotto, D.; Nye, G.; Uknes, S.; Ward, E.; Kessmann, H.; Ryals, J. Requirement of salicylic acid for the induction of systemic acquired resistance. Science 1993, 261, 754–756. [Google Scholar]
- Rajjou, L.; Belghazi, M.; Huguet, R.; Robin, C.; Moreau, A.; Job, C.; Job, D. Proteomic investigation of the effect of salicylic acid on Arabidopsis seed germination and establishment of early defense mechanisms. Plant Physiol. 2006, 141, 910–923. [Google Scholar] [CrossRef]
- Kundu, S.; Chakraborty, D.; Pal, A. Proteomic analysis of salicylic acid induced resistance to mungbean yellow mosaic India virus in Vigna mungo. J. Proteomics 2011, 74, 337–349. [Google Scholar] [CrossRef]
- Wang, F.X.; Ma, Y.P.; Yang, C.L.; Zhao, P.M.; Yao, Y.; Jian, G.L.; Luo, Y.M.; Xia, G.X. Proteomic analysis of the sea-island cotton roots infected by wilt pathogen Verticillium dahliae. Proteomics 2011, 11, 4296–4309. [Google Scholar]
- Cao, H.; Bowling, S.A.; Gordon, A.S.; Dong, X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 1994, 6, 1583–1592. [Google Scholar] [CrossRef]
- Alverez, M.E.; Pennell, R.I.; Meijer, P.J.; Ishikawa, A.; Dixon, R.A.; Lamb, C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 1998, 92, 773–784. [Google Scholar]
- Martinez, C.; Baccou, J.C.; Bresson, E.; Baissac, Y.; Daniel, J.F.; Jalloul, A.; Montillet, J.L.; Geiger, J.P.; Assigbetsé, K.; Nicole, M. Salicylic acid mediated by the oxidative burst is a key molecule in local and systemic responses of cotton challenged by an avirulent race of Xanthomonas campestris pv malvacearum. Plant Physiol. 2000, 122, 757–766. [Google Scholar]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar]
- Blanco, F.; Salinas, P.; Cecchini, N.M.; Jordana, X.; van Hummelen, P.; Alvarez, M.E.; Holuigue, L. Early genomic responses to salicylic acid in Arabidopsis. Plant Mol. Biol. 2009, 70, 79–102. [Google Scholar]
- Park, S.W.; Kaimoyo, E.; Kumar, D.; Mosher, S.; Klessig, D.F. Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 2007, 318, 113–116. [Google Scholar] [CrossRef]
- Vernooij, B.; Friedrich, L.; Morse, A.; Reist, R.; Kolditz-Jawhar, R.; Ward, E.; Uknes, S.; Kessmann, H.; Ryals, J. Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell 1994, 6, 959–965. [Google Scholar] [CrossRef]
- Attaran, E.; Zeier, T.E.; Griebel, T.; Zeier, J. Methyl salicylate production and jasmonate signaling are not essential for systemic acquired resistance in Arabidopsis. Plant Cell 2009, 21, 954–971. [Google Scholar] [CrossRef]
- Maldonado, A.M.; Doerner, P.; Dixon, R.A.; Lamb, C.J.; Cameron, R.K. A putative lipid transfer protein involved in systemic resistance signaling in Arabidopsis. Nature 2002, 419, 399–403. [Google Scholar]
- Jung, H.W.; Tschaplinski, T.J.; Wang, L.; Glazebrook, J.; Greenberg, J.T. Priming in systemic plant immunity. Science 2009, 324, 89–91. [Google Scholar] [CrossRef]
- Takashima, Y.; Ishiguri, F.; Iizuka, K.; Yoshizawa, N.; Yokota, S. Proteome analysis of infection-specific proteins from Japanese birch (Betula platyphylla var. japonica) plantlet No.8 infected with Inonotus obliquus strain IO-U1. Plant Biotechnol. 2013, 30, 83–87. [Google Scholar] [CrossRef]
- Takashima, Y.; Suzuki, M.; Ishiguri, F.; Iizuka, K.; Yoshizawa, N.; Yokota, S. Cationic peroxidase related to basal resistance of Betula platyphylla var. japonica plantlet No.8 against canker-rot fungus Inonotus obliquus strain IO-U1. Plant Biotechnol. 2013, 30, 199–205. [Google Scholar] [CrossRef]
- Rahman, M.M.; Ishiguri, F.; Takashima, Y.; Azad, M.A.K.; Iizuka, K.; Yoshizawa, N.; Yokota, S. Anatomical and histochemical characteristics of Japanese birch (Tohoku) plantlets infected with the Inonotus obliquus IO-U1 strain. Plant Biotechnol. 2008, 25, 183–189. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- MS/MS Ions Search. Available online: http:www.matrixscience.com (accessed on 6 July 2011).
- Siedow, J.N.; Day, D.A. Respiration and photorespiration. In Biochemistry & Molecular Biology of Plants; Buchanan, B.B., Gruissem, W.G., Jones, R.L., Eds.; American Society of Plant Physiologists: Rockville, MD, USA, 2000; p. 685. [Google Scholar]
- Segarra, G.; Casanova, E.; Bellido, D.; Odena, M.A.; Oliveira, E.; Trillas, I. Proteome, salicylic acid, and jasmonic acid changes in cucumber plants inoculated with Trichoderma asperellum strain T34. Proteomics 2007, 7, 3943–3952. [Google Scholar] [CrossRef]
- Islam, M.A.; Sturrock, R.N.; Ekramoddoullah, A.K.M. A proteomics approach to identify proteins differentially expressed in Douglas-fir seedlings infected by Phellinus sulphurascens. J. Proteomics 2008, 71, 425–438. [Google Scholar] [CrossRef]
- Tomaz, T.; Bagard, M.; Pracharoenwattana, I.; Lindén, P.; Lee, C.P.; Carroll, A.J.; Ströher, E.; Smith, S.M.; Gardeström, P.; Millar, A.H. Mitochondrial malate dehydrogenase lowers leaf respiration and alters photorespiration and plant growth in Arabidopsis. Plant Physiol. 2010, 154, 1143–1157. [Google Scholar]
- Yao, Y.-X.; Dong, Q.-L.; Zhai, H.; You, C.-X.; Hao, Y.-J. The functions of an apple cytosolic malate dehydrogenase gene in growth and tolerance to cold and salt stresses. Plant Physiol. Biochem. 2011, 49, 257–264. [Google Scholar] [CrossRef]
- Siedow, J.N.; Day, D.A. Respiration and photorespiration. In Biochemistry & Molecular Biology of Plants; Buchanan, B.B., Gruissem, W.G., Jones, R.L., Eds.; American Society of Plant Physiologists: Rockville, MD, USA, 2000; p. 684. [Google Scholar]
- Sako, N.; Stahmann, M.A. Multiple molecular forms of enzymes in barley leaves infected with Erysiphe graminis f. sp. hordei. Physiol. Plant Pathol. 1972, 2, 217–226. [Google Scholar] [CrossRef]
- Dennis, D.T.; Blakeley, S.D. Carbohydrate metabolism. In Biochemistry & Molecular Biology of Plants; Buchanan, B.B., Gruissem, W.G., Jones, R.L., Eds.; American Society of Plant Physiologists: Rockville, MD, USA, 2000; p. 668. [Google Scholar]
- Azevedo, R.A.; Lea, P.J. Lysine metabolism in higher plants. Amino Acids 2001, 20, 261–279. [Google Scholar] [CrossRef]
- Jenkinson, C.P.; Grody, W.W.; Cederbaum, S.D. Comparative properties of arginases. Comp. Biochem. Physiol. 1996, 114B, 107–132. [Google Scholar] [CrossRef]
- Chen, H.; McCaig, B.C.; Melotto, M.; He, S.Y.; Howe, G.A. Regulation of plant arginase by wounding, jasmonate, and the phtotoxin coronatine. J. Biol. Chem. 2004, 279, 45998–46007. [Google Scholar]
- Jones, A.M.E.; Thomas, V.; Bennett, M.H.; Mansfield, J.; Grant, M. Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol. 2006, 142, 1603–1620. [Google Scholar] [CrossRef]
- Kusano, T.; Yamaguchi, K.; Berberich, T.; Takahashi, Y. Advances in polyamine research in 2007. J. Plant Res. 2007, 120, 345–350. [Google Scholar] [CrossRef]
- Walters, D.R. Polyamines and plant disease. Phytochemistry 2003, 64, 97–107. [Google Scholar] [CrossRef]
- Marina, M.; Maiale, S.J.; Rossi, F.R.; Romero, M.F.; Rivas, E.I.; Gárriz, A.; Ruiz, O.A.; Pieckenstain, F.L. Apoplastic polyamine oxidation plays different roles in local responses of tabacco to infection by necrotrophic fungus Sclerotinia sclerotiorum and the biotrophic bacterium Pseudomonas viridiflava. Plant Physiol. 2008, 147, 2164–2178. [Google Scholar] [CrossRef]
- Yoda, H.; Fujimura, K.; Takahashi, H.; Munemura, I.; Uchimiya, H.; Sano, H. Polyamines as a common source of hydrogen peroxide in host- and nonhost hypersensitive response during pathogen infection. Plant Mol. Biol. 2009, 70, 103–112. [Google Scholar] [CrossRef]
- Németh, M.; Janda, T.; Horváth, E.; Páldi, E.; Szalai, G. Exogenous salicylic acid increases polyamine content but may decrease drought tolerance in maize. Plant Sci. 2002, 162, 569–574. [Google Scholar]
- Tzin, V.; Galili, G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 2010, 3, 956–972. [Google Scholar] [CrossRef]
- Eberhard, J.; Ehrler, T.T.; Epple, P.; Felix, G.; Raesecke, H.R.; Amrhein, N.; Schmid, J. Cytosolic and plastidic chorismate mutase isozymes from Arabidopsis thaliana: Molecular characterization and enzymatic properties. Plant J. 1996, 10, 815–821. [Google Scholar]
- Dixon, R.A. Natural products and plant disease resistance. Nature 2001, 411, 843–847. [Google Scholar] [CrossRef]
- Pallas, J.A.; Paiva, N.L.; Lamb, C.; Dixon, R.A. Tobacco plants epigenetically suppressed in phenylalanine ammonia-lyase expression do not develop systemic acquired resistance in response to infection by tobacco mosaic virus. Plant J. 1996, 10, 281–293. [Google Scholar]
- Shirasu, K.; Nakajima, H.; Rajasekhar, V.K.; Dixon, R.A.; Lamb, C. Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 1997, 9, 261–270. [Google Scholar] [CrossRef]
- Smith-Becker, J.; Marois, E.; Huguet, E.J.; Midland, S.L.; Sims, J.J.; Keen, N.T. Accumulation of salicylic acid and 4-hydroxybenzoic acid in phloem fluids of cucumber during systemic acquired resistance is preceded by a transient increase in phenylalanine ammonia-lyase activity in petioles and stems. Plant Physiol. 1998, 116, 231–238. [Google Scholar] [CrossRef]
- Bächinger, H.P. The influence of peptidyl-prolyl cis-trans isomerase on the in vitro folding of type III collagen. J. Biol. Chem. 1987, 262, 17144–17148. [Google Scholar]
- Miernyk, J.A. Protein folding in the plant cell. Plant Physiol. 1999, 121, 695–703. [Google Scholar] [CrossRef]
- Marvet, J.; Margis-Pinheiro, M.; Frendo, P.; Burkard, G. Bean cyclophilin gene expression during plant development and stress conditions. Plant Mol. Biol. 1994, 26, 1181–1189. [Google Scholar] [CrossRef]
- Walling, L.L. Recycling or regulation? The role of amino-terminal modifying enzymes. Curr. Opin. Plant Biol. 2006, 9, 227–233. [Google Scholar] [CrossRef]
- Cheng, F.Y.; Blackburn, K.; Lin, Y.M.; Goshe, M.B.; Williamson, J.D. Absolute protein quantification by LC/MSn for global analysis of salicylic acid-induced plant protein secretion responses. J. Proteome Res. 2009, 8, 82–93. [Google Scholar] [CrossRef]
- Delaney, T.P.; Uknes, S.; Vernooij, B.; Friendrich, L.; Weymann, K.; Negrotto, D.; Gaffney, T.; Gut-Rella, M.; Kessmann, H.; Ward, E.; et al. A central role of salicylic acid in plant disease resistance. Science 1994, 266, 1247–1250. [Google Scholar]
- Solomon, M.; Belenghi, B.; Delledonne, M.; Menachem, E.; Levine, A. The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 1999, 11, 431–443. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Suzuki, H.; Takashima, Y.; Ishiguri, F.; Yoshizawa, N.; Yokota, S. Proteomic Analysis of Responsive Proteins Induced in Japanese Birch Plantlet Treated with Salicylic Acid. Proteomes 2014, 2, 323-340. https://doi.org/10.3390/proteomes2030323
Suzuki H, Takashima Y, Ishiguri F, Yoshizawa N, Yokota S. Proteomic Analysis of Responsive Proteins Induced in Japanese Birch Plantlet Treated with Salicylic Acid. Proteomes. 2014; 2(3):323-340. https://doi.org/10.3390/proteomes2030323
Chicago/Turabian StyleSuzuki, Hiromu, Yuya Takashima, Futoshi Ishiguri, Nobuo Yoshizawa, and Shinso Yokota. 2014. "Proteomic Analysis of Responsive Proteins Induced in Japanese Birch Plantlet Treated with Salicylic Acid" Proteomes 2, no. 3: 323-340. https://doi.org/10.3390/proteomes2030323