Surviving a Dry Future: Abscisic Acid (ABA)-Mediated Plant Mechanisms for Conserving Water under Low Humidity
Abstract
:1. Introduction
2. ABA-Mediated Humidity Responses in Angiosperms
3. Possible Candidates for the Angiosperm Pathway for Rapid VPD Responses
3.1. Candidates for the Plant Water Status Sensor that Triggers VPD Responses
3.2. Candidate Transcriptional Regulators for Key NCED Genes during VPD Responses
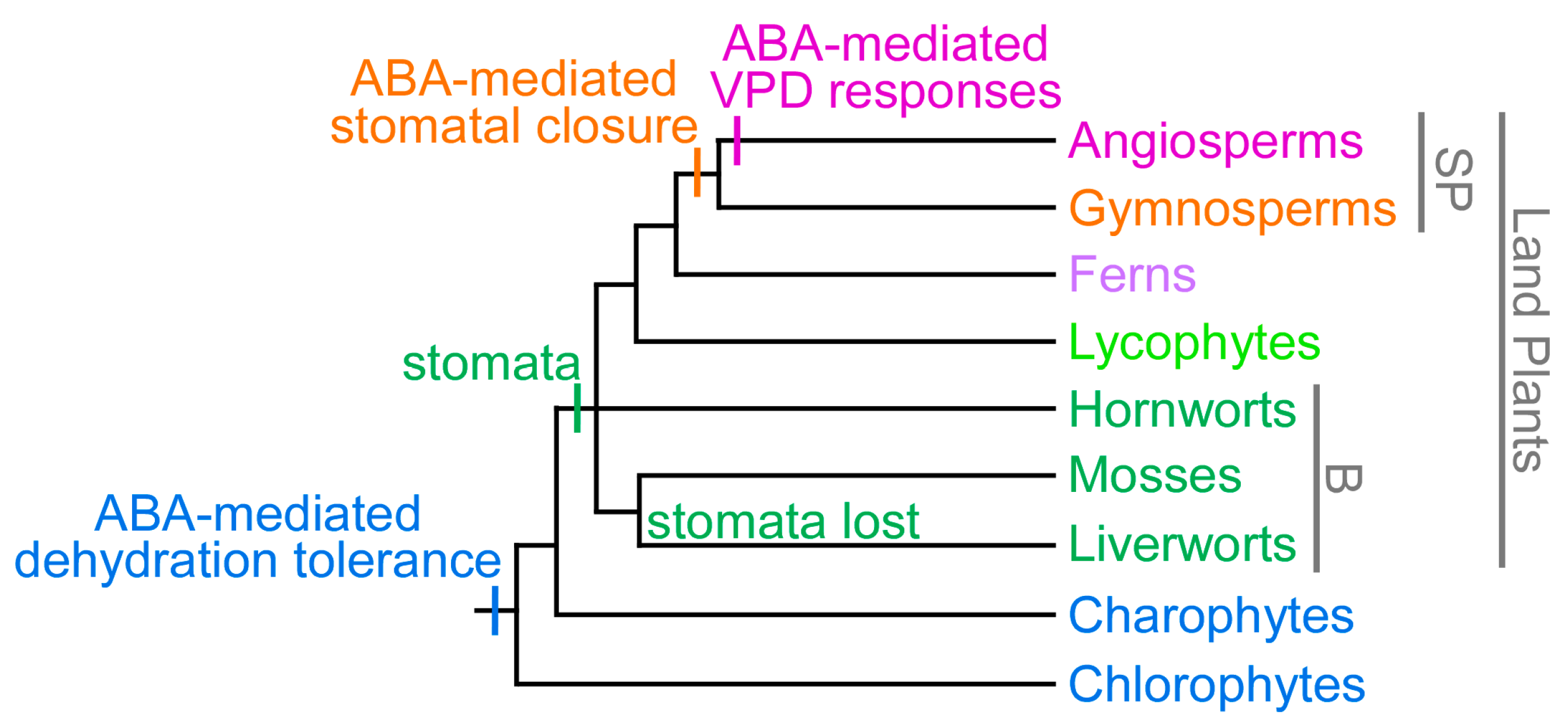
4. The Evolution of ABA-Mediated Plant Humidity Responses
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
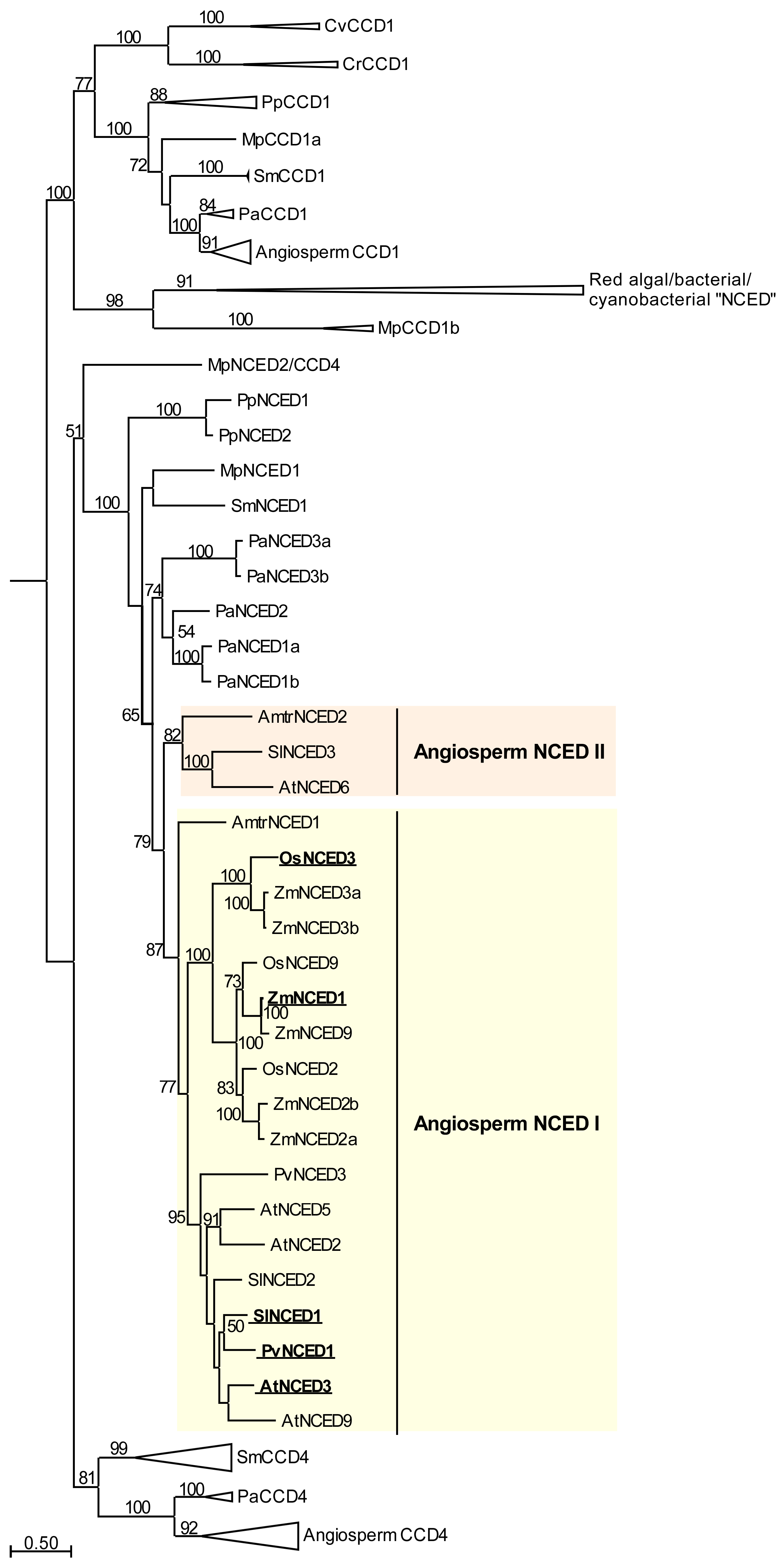
Appendix A
| Protein/Collapsed Subclade Name | Species | Accession | Source | Reference |
|---|---|---|---|---|
| Angiosperm CCD1 | Amborella trichopoda | evm_27.model.AmTr_v1.0_scaffold00022.400 evm_27.model.AmTr_v1.0_scaffold00022.401 | v1.0; Phytozome | This study |
| Arabidopsis thaliana | AT3G63520 (AtCCD1) | TAIR10; Phytozome | [230] | |
| Oryza sativa | LOC_Os12g44310 | v7; Phytozome | [231] | |
| Phaseolus vulgaris | Phvul.011G211200 | v2.1; Phytozome | This study | |
| Solanum lycopersicum | Solyc01g087250 Solyc01g087260 | iTAG2.4; Phytozome | [232] | |
| Zea mays | GRMZM2G057243 | v1.0; Phytozome | [231] | |
| Angiosperm CCD4 | Amborella trichopoda | evm_27.model.AmTr_v1.0_scaffold00011.172 | v1.0; Phytozome | This study |
| Arabidopsis thaliana | AT4G19170 (AtCCD4) | TAIR10; Phytozome | [230] | |
| Oryza sativa | LOC_Os02g47510 LOC_Os12g24800 | v7; Phytozome | [231] | |
| Phaseolus vulgaris | Phvul.002G120600 | v2.1; Phytozome | This study | |
| Solanum lycopersicum | Solyc08g075480 Solyc08g075490 | iTAG2.4; Phytozome | [232] | |
| Zea mays | GRMZM2G110192 GRMZM2G150363 | Ensembl-18; Phytozome | [231] | |
| AmtrNCED1 | Amborella trichopoda | evm_27.model.AmTr_v1.0_scaffold00092.158 | v1.0; Phytozome | [21] |
| AmtrNCED2 | evm_27.model.AmTr_v1.0_scaffold00039.158 | |||
| AtNCED2 | Arabidopsis thaliana | AT4G18350 | TAIR10; Phytozome | [45,230] |
| AtNCED3 | AT3G14440 | |||
| AtNCED5 | AT1G30100 | |||
| AtNCED6 | AT3G24220 | |||
| AtNCED9 | AT1G78390 | |||
| CrCCD1 | Chlamydomonas reinhardtii | Cre03.g149650 Cre08.g365825 | v5.5; Phytozome | This study: top BLASTp hits for AtNCED3 |
| CvCCD1 (prev. NCED) | Chlorella variabilis | EFN52762 | GenBank | [196] |
| MpCCD1a | Marchantia polymorpha | Mapoly0003s0307 | v3.1; Phytozome | This study |
| MpCCDb | Mapoly0012s0197 Mapoly0066s0017 | |||
| MpNCED1 | Mapoly0015s0066 | |||
| MpNCED2/CCD4 | Mapoly0149s0036 | |||
| OsNCED2 | Oryza sativa | LOC_Os12g42280 | v7; Phytozome | [231] |
| OsNCED3a | LOC_Os07g05940 | |||
| OsNCED9 | LOC_Os03g44380 | |||
| PaCCD1 | Picea abies | MA_10435932g0010 MA_210464g0010 MA_906445g0010 | v1.0; ConGenIE | This study |
| PaCCD4 | MA_10425950g0010 MA_90573g0010 MA_10428396g0010 | |||
| PaNCED1a | MA_10428505g0020 | |||
| PaNCED1b | MA_10428505g0010 comp88253_c0_seq1 | |||
| PaNCED2 | MA_10434448g0010 | |||
| PaNCED3a | MA_10174788g0010 | |||
| PaNCED3b | MA_198304g0010 | |||
| PpCCD1 | Physcomitrella patens | Pp3c12_22350V3.1 Pp3c18_17950V3.1 Pp3c21_12920V3.1 Pp3c22_6380V3.1 | v3.3; Phytozome | [182,231] |
| PpNCED1 | Pp3c16_17210V3.1 | |||
| PpNCED2 | Pp3c25_4816V3.1 | |||
| PvNCED1 | Phaseolus vulgaris | Phvul.005G051600 | v2.1; Phytozome | [41] |
| PvNCED3 | Phvul.007G198800 | [21] | ||
| Red algal/bacterial/cyanobacterial “NCED” | Cyanidioschyzon merolae | XP_005538977 (CMS362C) | GenBank | [194,195] |
| Phaeodactylum tricornutum | XP_002177588 | |||
| Trichodesmium erythraeum | WP_011612676 (Tery_3212) | |||
| SlNCED1 (NOT) | Solanum lycopersicum | Solyc07g056570 | iTAG2.4; Phytozome | [197,232] |
| SlNCED2 | Solyc08g016720 | |||
| SlNCED3 | Solyc05g053530 | |||
| SmCCD1 | Selaginella moellendorffii | 165469 272067 | v1.0; Phytozome | [155,231] |
| SmCCD4 (prev. SmNCEDa + c) | Selaginella moellendorffii | 11287 11289 11292 11304 75383 79628 80651 91815 94523 | v1.0; Phytozome | [155,231] |
| SmNCED1 (prev. SmNCEDb) | 233638 | |||
| ZmNCED1 (VVP1) | Zea mays | GRMZM2G014392 | Ensembl-18; Phytozome | This study; [199,231] |
| ZmNCED2a | GRMZM5G858784 | |||
| ZmNCED2b | GRMZM2G407181 | |||
| ZmNCED3a | GRMZM2G417954 | |||
| ZmNCED3b | GRMZM2G408158 | |||
| ZmNCED9 | GRMZM5G838285 |
References
- Mekonnen, M.M.; Hoekstra, A.Y. Four billion people facing severe water scarcity. Sci. Adv. 2016, 2, e1500323. [Google Scholar] [CrossRef] [PubMed]
- Schewe, J.; Heinke, J.; Gerten, D.; Haddeland, I.; Arnell, N.W.; Clark, D.B.; Dankers, R.; Eisner, S.; Fekete, B.M.; Colón-González, F.J.; et al. Multimodel assessment of water scarcity under climate change. Proc. Natl. Acad. Sci. USA 2014, 111, 3245–3250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trenberth, K.E.; Dai, A.; van der Schrier, G.; Jones, P.D.; Barichivich, J.; Briffa, K.R.; Sheffield, J. Global warming and changes in drought. Nat. Clim. Chang. 2014, 4, 17–22. [Google Scholar] [CrossRef]
- Elliott, J.; Deryng, D.; Müller, C.; Frieler, K.; Konzmann, M.; Gerten, D.; Glotter, M.; Flörke, M.; Wada, Y.; Best, N.; et al. Constraints and potentials of future irrigation water availability on agricultural production under climate change. Proc. Natl. Acad. Sci. USA 2014, 111, 3239–3244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerland, P.; Raftery, A.E.; Ševčíková, H.; Li, N.; Gu, D.; Spoorenberg, T.; Alkema, L.; Fosdick, B.K.; Chunn, J.; Lalic, N.; et al. World population stabilization unlikely this century. Science 2014, 346, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Blumwald, E. Genetic engineering for modern agriculture: Challenges and perspectives. Annu. Rev. Plant Biol. 2010, 61, 443–462. [Google Scholar] [CrossRef] [PubMed]
- Raven, J.A. Selection pressures on stomatal evolution. New Phytol. 2002, 153, 371–386. [Google Scholar] [CrossRef]
- Mittelheuser, C.J.; Van Steveninck, R.F.M. Stomatal closure and inhibition of transpiration induced by (RS)-abscisic acid. Nature 1969, 221, 281–282. [Google Scholar] [CrossRef]
- Kriedemann, P.E.; Loveys, B.R.; Fuller, G.L.; Leopold, A.C. Abscisic acid and stomatal regulation. Plant Physiol. 1972, 49, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Cowan, I.R.; Farquhar, G.D. Stomatal function in relation to leaf metabolism and environment. Symp. Soc. Exp. Biol. 1977, 31, 471–505. [Google Scholar] [PubMed]
- Brodribb, T.J.; McAdam, S.A.M.; Jordan, G.J.; Martins, S.C.V. Conifer species adapt to low-rainfall climates by following one of two divergent pathways. Proc. Natl. Acad. Sci. USA 2014, 111, 14489–14493. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, A.; Kusano, M.; Nakamichi, N.; Kobayashi, M.; Hayashi, N.; Sakakibara, H.; Mizuno, T.; Saito, K. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc. Natl. Acad. Sci. USA 2009, 106, 7251–7256. [Google Scholar] [CrossRef] [PubMed]
- Covington, M.F.; Maloof, J.N.; Straume, M.; Kay, S.A.; Harmer, S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008, 9, R130. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.-J.; Nakajima, M.; Suzuki, Y.; Yamaguchi, I. Cloning and characterization of the abscisic acid-specific glucosyltransferase gene from adzuki bean seedlings. Plant Physiol. 2002, 129, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.-Z.; Chen, X.; Xiang, C.-B.; Tang, N.; Zhang, Q.-F.; Xiong, L.-Z. Evaluation of seven function-known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions. Mol. Plant 2009, 2, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.-J.; Song, S.I.; Kim, Y.S.; Jang, H.-J.; Kim, S.Y.; Kim, M.; Kim, Y.-K.; Nahm, B.H.; Kim, J.-K. Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 2005, 138, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Li, J.; Liu, P.; Duan, J.; Zhao, Y.; Guo, X.; Li, Y.; Zhang, H.; Ali, J.; Li, Z. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE 2014, 9, e92913. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Zeevaart, J.A.D. Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 2002, 128, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Lou, D.; Wang, H.; Liang, G.; Yu, D. OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front. Plant Sci. 2017, 8, 993. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Running, S.W. Drought-induced reduction in global terrestrial net primary production from 2000 through 2009. Science 2010, 329, 940–943. [Google Scholar] [CrossRef] [PubMed]
- McAdam, S.A.M.; Sussmilch, F.C.; Brodribb, T.J. Stomatal responses to vapour pressure deficit are regulated by high speed gene expression in angiosperms. Plant Cell Environ. 2016, 39, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Wang, Y.; Williamson, L.; Holroyd, G.H.; Tagliavia, C.; Murchie, E.; Theobald, J.; Knight, M.R.; Davies, W.J.; Leyser, H.M.O.; et al. The identification of genes involved in the stomatal response to reduced atmospheric relative humidity. Curr. Biol. 2006, 16, 882–887. [Google Scholar] [CrossRef] [PubMed]
- McAdam, S.A.M.; Sussmilch, F.C.; Brodribb, T.J.; Ross, J.J. Molecular characterization of a mutation affecting abscisic acid biosynthesis and consequently stomatal responses to humidity in an agriculturally important species. AoB Plants 2015, 7, plv091. [Google Scholar] [CrossRef] [PubMed]
- McAdam, S.A.M.; Brodribb, T.J. The evolution of mechanisms driving the stomatal response to vapor pressure deficit. Plant Physiol. 2015, 167, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Bauerle, W.L.; Whitlow, T.H.; Setter, T.L.; Vermeylen, F.M. Abscisic acid synthesis in Acer rubrum L. leaves—A vapor-pressure-deficit-mediated response. J. Am. Soc. Hortic. Sci. 2004, 129, 182–187. [Google Scholar]
- McAdam, S.A.M.; Brodribb, T.J. Linking turgor with ABA biosynthesis: Implications for stomatal responses to vapour pressure deficit across land plants. Plant Physiol. 2016, 171, 2008–2016. [Google Scholar] [CrossRef] [PubMed]
- Sack, L.; John, G.P.; Buckley, T.N. ABA accumulation in dehydrating leaves is associated with decline in cell volume not turgor pressure. Plant Physiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Pierce, M.; Raschke, K. Correlation between loss of turgor and accumulation of abscisic acid in detached leaves. Planta 1980, 148, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Creelman, R.A.; Zeevaart, J.A. Abscisic acid accumulation in spinach leaf slices in the presence of penetrating and nonpenetrating solutes. Plant Physiol. 1985, 77, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Sussmilch, F.C.; Brodribb, T.J.; McAdam, S.A.M. Upregulation of NCED3 and ABA biosynthesis occurs within minutes of a decrease in leaf turgor but AHK1 is not required. J. Exp. Bot. 2017, 68, 2913–2918. [Google Scholar] [CrossRef] [PubMed]
- Merilo, E.; Laanemets, K.; Hu, H.; Xue, S.; Jakobson, L.; Tulva, I.; Gonzalez-Guzman, M.; Rodriguez, P.L.; Schroeder, J.I.; Broschè, M.; et al. PYR/RCAR receptors contribute to ozone-, reduced air humidity-, darkness-, and CO2-induced stomatal regulation. Plant Physiol. 2013, 162, 1652–1668. [Google Scholar] [CrossRef] [PubMed]
- Merilo, E.; Yarmolinsky, D.; Jalakas, P.; Parik, H.; Tulva, I.; Rasulov, B.; Kilk, K.; Kollist, H. Stomatal VPD response: There is more to the story than ABA. Plant Physiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Georgopoulou, Z.; Milborrow, B.V. Initiation of the synthesis of ‘stress’ ABA by (+)-[2H6]ABA infiltrated into leaves of Commelina communis. Physiol. Plant 2012, 146, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.-Y.; Lee, K.H.; Dong, T.; Jeong, J.C.; Jin, J.B.; Kanno, Y.; Kim, D.H.; Kim, S.Y.; Seo, M.; Bressan, R.A.; et al. A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 2012, 24, 2184–2199. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Piao, H.L.; Kim, H.-Y.; Choi, S.M.; Jiang, F.; Hartung, W.; Hwang, I.; Kwak, J.M.; Lee, I.-J.; Hwang, I. Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 2006, 126, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Ethier, G.; Pepin, S.; Dubé, P.; Desjardins, Y.; Gosselin, A. Persistent negative temperature response of mesophyll conductance in red raspberry (Rubus idaeus L.) leaves under both high and low vapour pressure deficits: A role for abscisic acid? Plant Cell Environ. 2017, 40, 1940–1959. [Google Scholar] [CrossRef] [PubMed]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Hirai, N.; Matsumoto, C.; Ohigashi, H.; Ohta, D.; Sakata, K.; Mizutani, M. Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 2004, 134, 1439–1449. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Okamoto, M.; Kushiro, T.; Nambara, E.; Oono, Y.; Seki, M.; Kobayashi, M.; Koshiba, T.; Kamiya, Y.; Shinozaki, K. CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J. 2006, 46, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Jackson, A.C.; Symonds, R.C.; Mulholland, B.J.; Dadswell, A.R.; Blake, P.S.; Burbidge, A.; Taylor, I.B. Ecotopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J. 2000, 23, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Zeevaart, J.A.D. The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. USA 1999, 96, 15354–15361. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.H.; Tan, B.C.; Gage, D.A.; Zeevaart, J.A.D.; McCarty, D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 1997, 276, 1872–1874. [Google Scholar] [CrossRef] [PubMed]
- Neuman, H.; Galpaz, N.; Cunningham, F.X.; Zamir, D.; Hirschberg, J. The tomato mutation nxd1 reveals a gene necessary for neoxanthin biosynthesis and demonstrates that violaxanthin is a sufficient precursor for abscisic acid biosynthesis. Plant J. 2014, 78, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Chernys, J.T.; Zeevaart, J.A.D. Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol. 2000, 124, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef] [PubMed]
- McAdam, S.A.M.; Manzi, M.; Ross, J.J.; Brodribb, T.J.; Gómez-Cadenas, A. Uprooting an abscisic acid paradigm: Shoots are the primary source. Plant Signal Behav. 2016, 11, e1169359. [Google Scholar] [CrossRef] [PubMed]
- McAdam, S.A.M.; Brodribb, T.J.; Ross, J.J. Shoot-derived abscisic acid promotes root growth. Plant Cell Environ. 2016, 39, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Manzi, M.; Lado, J.; Rodrigo, M.J.; Zacarías, L.; Arbona, V.; Gómez-Cadenas, A. Root ABA accumulation in long-term water-stressed plants is sustained by hormone transport from aerial organs. Plant Cell Physiol. 2015, 56, 2457–2466. [Google Scholar] [CrossRef] [PubMed]
- Bauer, H.; Ache, P.; Lautner, S.; Fromm, J.; Hartung, W.; Al-Rasheid, K.A.S.; Sonnewald, S.; Sonnewald, U.; Kneitz, S.; Lachmann, N.; et al. The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr. Biol. 2013, 23, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.-H.; Endo, A.; Zhou, L.; Penney, J.; Chen, H.-C.; Arroyo, A.; Leon, P.; Nambara, E.; Asami, T.; Seo, M.; et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 2002, 14, 2723–2743. [Google Scholar] [CrossRef] [PubMed]
- Koiwai, H.; Nakaminami, K.; Seo, M.; Mitsuhashi, W.; Toyomasu, T.; Koshiba, T. Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiol. 2004, 134, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Sawada, Y.; Takahashi, H.; Okamoto, M.; Ikegami, K.; Koiwai, H.; Seo, M.; Toyomasu, T.; Mitsuhashi, W.; Shinozaki, K.; et al. Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol. 2008, 147, 1984–1993. [Google Scholar] [CrossRef] [PubMed]
- Boursiac, Y.; Léran, S.; Corratgé-Faillie, C.; Gojon, A.; Krouk, G.; Lacombe, B. ABA transport and transporters. Trends Plant Sci. 2013, 18, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Hwang, J.-U.; Lee, M.; Kim, Y.-Y.; Assmann, S.M.; Martinoia, E.; Lee, Y. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. USA 2010, 107, 2355–2360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuromori, T.; Miyaji, T.; Yabuuchi, H.; Shimizu, H.; Sugimoto, E.; Kamiya, A.; Moriyama, Y.; Shinozaki, K. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. USA 2010, 107, 2361–2366. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.F.; et al. Abscisic acid inhibits PP2Cs via the PYR/PYL family of ABA-binding START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.; Dupeux, F.; Round, A.; Antoni, R.; Park, S.-Y.; Jamin, M.; Cutler, S.R.; Rodriguez, P.L.; Marquez, J.A. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 2009, 462, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef] [PubMed]
- Vlad, F.; Rubio, S.; Rodrigues, A.; Sirichandra, C.; Belin, C.; Robert, N.; Leung, J.; Rodriguez, P.L.; Laurière, C.; Merlot, S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 2009, 21, 3170–3184. [Google Scholar] [CrossRef] [PubMed]
- Soon, F.-F.; Ng, L.-M.; Zhou, X.E.; West, G.M.; Kovach, A.; Tan, M.H.E.; Suino-Powell, K.M.; He, Y.; Xu, Y.; Chalmers, M.J.; et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 2012, 335, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Acharya, B.R.; Jeon, B.W.; Zhang, W.; Assmann, S.M. Open Stomata 1 (OST1) is limiting in abscisic acid responses of Arabidopsis guard cells. New Phytol. 2013, 200, 1049–1063. [Google Scholar] [CrossRef] [PubMed]
- Melcher, K.; Ng, L.-M.; Zhou, X.E.; Soon, F.-F.; Xu, Y.; Suino-Powell, K.M.; Park, S.-Y.; Weiner, J.J.; Fujii, H.; Chinnusamy, V.; et al. A gate-latch-lock mechanism for hormone signaling by abscisic acid receptors. Nature 2009, 462, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Miyazono, K.-I.; Miyakawa, T.; Sawano, Y.; Kubota, K.; Kang, H.-J.; Asano, A.; Miyauchi, Y.; Takahashi, M.; Zhi, Y.; Fujita, Y.; et al. Structural basis of abscisic acid signalling. Nature 2009, 462, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, N.; Hitomi, K.; Arvai, A.S.; Rambo, R.P.; Hitomi, C.; Cutler, S.R.; Schroeder, J.I.; Getzoff, E.D. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 2009, 326, 1373–1379. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Fan, H.; Hao, Q.; Yuan, X.; Wu, D.; Pang, Y.; Yan, C.; Li, W.; Wang, J.; Yan, N. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat. Struct. Mol. Biol. 2009, 16, 1230–1236. [Google Scholar] [CrossRef] [PubMed]
- Tischer, S.V.; Wunschel, C.; Papacek, M.; Kleigrewe, K.; Hofmann, T.; Christmann, A.; Grill, E. Combinatorial interaction network of abscisic acid receptors and coreceptors from Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 144, 10280–10285. [Google Scholar] [CrossRef] [PubMed]
- Geiger, D.; Scherzer, S.; Mumm, P.; Stange, A.; Marten, I.; Bauer, H.; Ache, P.; Matschi, S.; Liese, A.; Al-Rasheid, K.A. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. USA 2009, 106, 21425–21430. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Lan, W.; Buchanan, B.B.; Luan, S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. USA 2009, 106, 21419–21424. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.-M.; Soon, F.-F.; Zhou, X.E.; West, G.M.; Kovach, A.; Suino-Powell, K.M.; Chalmers, M.J.; Li, J.; Yong, E.-L.; Zhu, J.-K.; et al. Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. Proc. Natl. Acad. Sci. USA 2011, 108, 21259–21264. [Google Scholar] [CrossRef] [PubMed]
- Imes, D.; Mumm, P.; Böhm, J.; Al-Rasheid, K.A.S.; Marten, I.; Geiger, D.; Hedrich, R. Open stomata 1 (OST1) kinase controls R–type anion channel QUAC1 in Arabidopsis guard cells. Plant J. 2013, 74, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Ache, P.; Becker, D.; Ivashikina, N.; Dietrich, P.; Roelfsema, M.R.G.; Hedrich, R. GORK, a delayed outward rectifier expressed in guard cells of Arabidopsis thaliana, is a K+-selective, K+-sensing ion channel. FEBS Lett. 2000, 486, 93–98. [Google Scholar] [CrossRef]
- Hosy, E.; Vavasseur, A.; Mouline, K.; Dreyer, I.; Gaymard, F.; Porée, F.; Boucherez, J.; Lebaudy, A.; Bouchez, D.; Véry, A.-A.; et al. The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc. Natl. Acad. Sci. USA 2003, 100, 5549–5554. [Google Scholar] [CrossRef] [PubMed]
- Geiger, D.; Scherzer, S.; Mumm, P.; Marten, I.; Ache, P.; Matschi, S.; Liese, A.; Wellmann, C.; Al-Rasheid, K.A.S.; Grill, E.; et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. USA 2010, 107, 8023–8028. [Google Scholar] [CrossRef] [PubMed]
- Demir, F.; Horntrich, C.; Blachutzik, J.O.; Scherzer, S.; Reinders, Y.; Kierszniowska, S.; Schulze, W.X.; Harms, G.S.; Hedrich, R.; Geiger, D.; et al. Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc. Natl. Acad. Sci. USA 2013, 110, 8296–8301. [Google Scholar] [CrossRef] [PubMed]
- Maierhofer, T.; Diekmann, M.; Offenborn, J.N.; Lind, C.; Bauer, H.; Hashimoto, K.; Al-Rasheid, K.A.S.; Luan, S.; Kudla, J.; Geiger, D.; et al. Site- and kinase-specific phosphorylation-mediated activation of SLAC1, a guard cell anion channel stimulated by abscisic acid. Sci. Signal 2014, 7, ra86. [Google Scholar] [CrossRef] [PubMed]
- Scherzer, S.; Maierhofer, T.; Al-Rasheid, K.A.S.; Geiger, D.; Hedrich, R. Multiple calcium-dependent kinases modulate ABA-activated guard cell anion channels. Mol. Plant 2012, 5, 1409–1412. [Google Scholar] [CrossRef] [PubMed]
- Mori, I.C.; Murata, Y.; Yang, Y.; Munemasa, S.; Wang, Y.-F.; Andreoli, S.; Tiriac, H.; Alonso, J.M.; Harper, J.F.; Ecker, J.R.; et al. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol. 2006, 4, e327. [Google Scholar] [CrossRef] [PubMed]
- Corratgé-Faillie, C.; Ronzier, E.; Sanchez, F.; Prado, K.; Kim, J.-H.; Lanciano, S.; Leonhardt, N.; Lacombe, B.; Xiong, T.C. The Arabidopsis guard cell outward potassium channel GORK is regulated by CPK33. FEBS Lett. 2017, 591, 1982–1992. [Google Scholar] [CrossRef] [PubMed]
- Brandt, B.; Brodsky, D.E.; Xue, S.; Negi, J.; Iba, K.; Kangasjärvi, J.; Ghassemian, M.; Stephan, A.B.; Hu, H.; Schroeder, J.I. Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. USA 2012, 109, 10593–10598. [Google Scholar] [CrossRef] [PubMed]
- Brandt, B.; Munemasa, S.; Wang, C.; Nguyen, D.; Yong, T.; Yang, P.G.; Poretsky, E.; Belknap, T.F.; Waadt, R.; Alemán, F.; et al. Calcium specificity signaling mechanisms in abscisic acid signal transduction in Arabidopsis guard cells. eLife 2015, 4, e03599. [Google Scholar] [CrossRef]
- Lefoulon, C.; Boeglin, M.; Moreau, B.; Véry, A.-A.; Szponarski, W.; Dauzat, M.; Michard, E.; Gaillard, I.; Chérel, I. The Arabidopsis AtPP2CA protein phosphatase inhibits the GORK K+ efflux channel and exerts a dominant suppressive effect on phosphomimetic-activating mutations. J. Biol. Chem. 2016, 291, 6521–6533. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Ren, H.-M.; Tan, Y.-Q.; Qi, G.-N.; Yao, F.-Y.; Wu, G.-L.; Yang, L.-W.; Hussain, J.; Sun, S.-J.; Wang, Y.-F. S-type anion channels SLAC1 and SLAH3 function as essential negative regulators of inward K+ channels and stomatal opening in Arabidopsis. Plant Cell 2016, 28, 949–965. [Google Scholar] [CrossRef]
- Sato, A.; Sato, Y.; Fukao, Y.; Fujiwara, M.; Umezawa, T.; Shinozaki, K.; Hibi, T.; Taniguchi, M.; Miyake, H.; Goto, D.B.; et al. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 2009, 424, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Barrero, J.M.; Rodriguez, P.L.; Quesada, V.; Piqueras, P.; Ponce, M.R.; Micol, J.L. Both abscisic acid (ABA)-dependent and ABA-independent pathways govern the induction of NCED3, AAO3 and ABA1 in response to salt stress. Plant Cell Environ. 2006, 29, 2000–2008. [Google Scholar] [CrossRef] [PubMed]
- Burbidge, A.; Grieve, T.; Jackson, A.; Thompson, A.; Taylor, I. Structure and expression of a cDNA encoding a putative neoxanthin cleavage enzyme (NCE), isolated from a wilt-related tomato (Lycopersicon esculentum Mill.) library. J. Exp. Bot. 1997, 48, 2111–2112. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 2000, 123, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Jackson, A.C.; Parker, R.A.; Morpeth, D.R.; Burbidge, A.; Taylor, I.B. Abscisic acid biosynthesis in tomato: Regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol. Biol. 2000, 42, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Urao, T.; Yakubov, B.; Satoh, R.; Yamaguchi-Shinozaki, K.; Seki, M.; Hirayama, T.; Shinozaki, K. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 1999, 11, 1743–1754. [Google Scholar] [CrossRef] [PubMed]
- Wohlbach, D.J.; Quirino, B.F.; Sussman, M.R. Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell 2008, 20, 1101–1117. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.-S.P.; Urao, T.; Qin, F.; Maruyama, K.; Kakimoto, T.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 20623–20628. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.N.; Jane, W.-N.; Verslues, P.E. Role of the putative osmosensor Arabidopsis Histidine Kinase1 in dehydration avoidance and low-water-potential response. Plant Physiol. 2013, 161, 942–953. [Google Scholar] [CrossRef] [PubMed]
- Lahr, W.; Raschke, K. Abscisic-acid contents and concentrations in protoplasts from guard cells and mesophyll cells of Vicia faba L. Planta 1988, 173, 528–531. [Google Scholar] [CrossRef] [PubMed]
- Loveys, B.R.; Robinson, S.P. Abscisic acid synthesis and metabolism in barley leaves and protoplasts. Plant Sci. 1987, 49, 23–30. [Google Scholar] [CrossRef]
- Christmann, A.; Grill, E.; Huang, J. Hydraulic signals in long-distance signaling. Curr. Opin. Plant Biol. 2013, 16, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.-S.P. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Haswell, E.S.; Verslues, P.E. The ongoing search for the molecular basis of plant osmosensing. J. Gen. Physiol. 2015, 145, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Kalladan, R.; Lasky, J.R.; Chang, T.Z.; Sharma, S.; Juenger, T.E.; Verslues, P.E. Natural variation identifies genes affecting drought-induced abscisic acid accumulation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 11536–11541. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Guo, S.S.; Fässler, R. Integrin-mediated mechanotransduction. J. Cell Biol. 2016, 215, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Ross, T.D.; Coon, B.G.; Yun, S.; Baeyens, N.; Tanaka, K.; Ouyang, M.; Schwartz, M.A. Integrins in mechanotransduction. Curr. Opin. Cell Biol. 2013, 25, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Ruoslahti, E. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 1996, 12, 697–715. [Google Scholar] [CrossRef] [PubMed]
- Takada, Y.; Ye, X.; Simon, S. The integrins. Genome Biol. 2007, 8, 215. [Google Scholar] [CrossRef] [PubMed]
- Canut, H.; Carrasco, A.; Galaud, J.-P.; Cassan, C.; Bouyssou, H.; Vita, N.; Ferrara, P.; Pont-Lezica, R. High affinity RGD-binding sites at the plasma membrane of Arabidopsis thaliana links the cell wall. Plant J. 1998, 16, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Schindler, M.; Meiners, S.; Cheresh, D.A. RGD-dependent linkage between plant cell wall and plasma membrane: Consequences for growth. J. Cell Biol. 1989, 108, 1955–1965. [Google Scholar] [CrossRef] [PubMed]
- Lü, B.; Chen, F.; Gong, Z.H.; Xie, H.; Zhang, J.H.; Liang, J.S. Intracellular localization of integrin-like protein and its roles in osmotic stress-induced abscisic acid biosynthesis in Zea mays. Protoplasma 2007, 232, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Lü, B.; Chen, F.; Gong, Z.-H.; Xie, H.; Liang, J.-S. Integrin-like protein is involved in the osmotic stress-induced abscisic acid biosynthesis in Arabidopsis thaliana. J. Integr. Plant Biol. 2007, 49, 540–549. [Google Scholar] [CrossRef]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef]
- Zhu, J.K.; Shi, J.; Singh, U.; Wyatt, S.E.; Bressan, R.A.; Hasegawa, P.M.; Carpita, N.C. Enrichment of vitronectin-and fibronectin-like proteins in NaCl-adapted plant cells and evidence for their involvement in plasma membrane–cell wall adhesion. Plant J. 1993, 3, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Langhans, M.; Weber, W.; Babel, L.; Grunewald, M.; Meckel, T. The right motifs for plant cell adhesion: what makes an adhesive site? Protoplasma 2017, 254, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Laval, V.; Chabannes, M.; Carrière, M.; Canut, H.; Barre, A.; Rougé, P.; Pont-Lezica, R.; Galaud, J.-P. A family of Arabidopsis plasma membrane receptors presenting animal β-integrin domains. BBA-Protein Struct. Mol. 1999, 1435, 61–70. [Google Scholar] [CrossRef]
- Gouget, A.; Senchou, V.; Govers, F.; Sanson, A.; Barre, A.; Rougé, P.; Pont-Lezica, R.; Canut, H. Lectin receptor kinases participate in protein-protein interactions to mediate plasma membrane-cell wall adhesions in Arabidopsis. Plant Physiol. 2006, 140, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Kohorn, B.D.; Kohorn, S.L. The cell wall-associated kinases, WAKs, as pectin receptors. Front. Plant Sci. 2012, 3, 88. [Google Scholar] [CrossRef] [PubMed]
- Boisson-Dernier, A.; Kessler, S.A.; Grossniklaus, U. The walls have ears: The role of plant CrRLK1Ls in sensing and transducing extracellular signals. J. Exp. Bot. 2011, 62, 1581–1591. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, K.; Govers, F. Arabidopsis L-type lectin receptor kinases: Phylogeny, classification, and expression profiles. J. Exp. Bot. 2009, 60, 4383–4396. [Google Scholar] [CrossRef] [PubMed]
- Lindner, H.; Müller, L.M.; Boisson-Dernier, A.; Grossniklaus, U. CrRLK1L receptor-like kinases: Not just another brick in the wall. Curr. Opin. Plant Biol. 2012, 15, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; He, J.; Ding, H.; Liu, H.; Lü, B.; Liang, J.; Wang, L.; He, J.; Ding, H.D.; Liu, H.; et al. Overexpression of AT14A confers tolerance to drought stress-induced oxidative damage in suspension cultured cells of Arabidopsis thaliana. Protoplasma 2015, 252, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.N.; Hsieh, Y.-F.; Verslues, P.E. At14a-Like1 participates in membrane-associated mechanisms promoting growth during drought in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2015, 112, 10545–10550. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-J.; Wuriyanghan, H.; Zhang, Y.-Q.; Duan, K.-X.; Chen, H.-W.; Li, Q.-T.; Lu, X.; He, S.-J.; Ma, B.; Zhang, W.-K.; et al. An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice. Plant Physiol. 2013, 163, 1752–1765. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Yang, S.H.; Shin, K.H.; Lee, S.C.; Kim, S.H. The AtLRK10L1.2, Arabidopsis ortholog of wheat LRK10, is involved in ABA-mediated signaling and drought resistance. Plant Cell Rep. 2015, 34, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Hara, K.; Yamaguchi, Y.; Koizumi, N.; Sano, H. Osmotic stress tolerance of transgenic tobacco expressing a gene encoding a membrane-located receptor-like protein from tobacco plants. Plant Physiol. 2003, 131, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Vaid, N.; Pandey, P.; Srivastava, V.K.; Tuteja, N. Pea lectin receptor-like kinase functions in salinity adaptation without yield penalty, by alleviating osmotic and ionic stresses and upregulating stress-responsive genes. Plant Mol. Biol. 2015, 88, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Sun, M.; Luo, X.; Ding, X.; Ji, W.; Cai, H.; Bai, X.; Liu, X.; Zhu, Y. A Glycine soja ABA-responsive receptor-like cytoplasmic kinase, GsRLCK, positively controls plant tolerance to salt and drought stresses. Planta 2013, 237, 1527–1545. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Gao, Z.; Xiao, G.; Huang, R.; Zhang, H. Leucine-rich repeat receptor-like kinase FON1 regulates drought stress and seed germination by activating the expression of ABA-responsive genes in rice. Plant Mol. Biol. Report. 2014, 32, 1158–1168. [Google Scholar] [CrossRef]
- Xiong, L.; Lee, H.; Ishitani, M.; Zhu, J.-K. Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 2002, 277, 8588–8596. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Zhu, J.-K. Regulation of abscisic acid biosynthesis. Plant Physiol. 2003, 133, 29–36. [Google Scholar] [CrossRef] [PubMed]
- McAinsh, M.R.; Brownlee, C.; Hetherington, A.M. Abscisic acid-induced elevation of guard cell cytosolic Ca2+ precedes stomatal closure. Nature 1990, 343, 186–188. [Google Scholar] [CrossRef]
- Allen, G.J.; Kuchitsu, K.; Chu, S.P.; Murata, Y.; Schroeder, J.I. Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid–induced cytoplasmic calcium rises in guard cells. Plant Cell 1999, 11, 1785–1798. [Google Scholar] [CrossRef] [PubMed]
- Ranf, S.; Wünnenberg, P.; Lee, J.; Becker, D.; Dunkel, M.; Hedrich, R.; Scheel, D.; Dietrich, P. Loss of the vacuolar cation channel, AtTPC1, does not impair Ca2+ signals induced by abiotic and biotic stresses. Plant J. 2008, 53, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Knight, H.; Trewavas, A.J.; Knight, M.R. Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. 1997, 12, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Iida, K.; Nyunoya, H.; Iida, H. Determination of structural regions important for Ca2+ uptake activity in Arabidopsis MCA1 and MCA2 expressed in yeast. Plant Cell Physiol. 2011, 52, 1915–1930. [Google Scholar] [CrossRef] [PubMed]
- Kurusu, T.; Nishikawa, D.; Yamazaki, Y.; Gotoh, M.; Nakano, M.; Hamada, H.; Yamanaka, T.; Iida, K.; Nakagawa, Y.; Saji, H.; et al. Plasma membrane protein OsMCA1 is involved in regulation of hypo-osmotic shock-induced Ca2+ influx and modulates generation of reactive oxygen species in cultured rice cells. BMC Plant Biol. 2012, 12, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurusu, T.; Yamanaka, T.; Nakano, M.; Takiguchi, A.; Ogasawara, Y.; Hayashi, T.; Iida, K.; Hanamata, S.; Shinozaki, K.; Iida, H.; et al. Involvement of the putative Ca2+-permeable mechanosensitive channels, NtMCA1 and NtMCA2, in Ca2+ uptake, Ca2+-dependent cell proliferation and mechanical stress-induced gene expression in tobacco (Nicotiana tabacum) BY-2 cells. J. Plant Res. 2012, 125, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Veley, K.M.; Marshburn, S.; Clure, C.E.; Haswell, E.S. Mechanosensitive channels protect plastids from hypoosmotic stress during normal plant growth. Curr. Biol. 2012, 22, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Levina, N.; Tötemeyer, S.; Stokes, N.R.; Louis, P.; Jones, M.A.; Booth, I.R. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: Identification of genes required for MscS activity. EMBO J. 1999, 18, 1730–1737. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, E.S.; Schlegel, A.M.; Haswell, E.S. United in diversity: Mechanosensitive ion channels in plants. Annu. Rev. Plant Biol. 2015, 66, 113–137. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; et al. OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 2014, 514, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yuan, F.; Wen, Z.; Li, Y.; Wang, F.; Zhu, T.; Zhuo, W.; Jin, X.; Wang, Y.; Zhao, H.; et al. Genome-wide survey and expression analysis of the OSCA gene family in rice. BMC Plant Biol. 2015, 15, 261. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Tian, W.; Kleist, T.; He, K.; Garcia, V.; Bai, F.; Hao, Y.; Luan, S.; Li, L. DUF221 proteins are a family of osmosensitive calcium-permeable cation channels conserved across eukaryotes. Cell Res. 2014, 24, 632–635. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, H.; Shi, Y.; Song, Y.; Wang, T.; Li, Y. A maize early responsive to dehydration gene, ZmERD4, provides enhanced drought and salt tolerance in Arabidopsis. Plant Mol. Biol. Report. 2009, 27, 542. [Google Scholar] [CrossRef]
- Garapati, P.; Xue, G.-P.; Munné-Bosch, S.; Balazadeh, S. Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 2015, 168, 1122–1139. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.K.; Lindemose, S.; Masi, F.d.; Reimer, J.J.; Nielsen, M.; Perera, V.; Workman, C.T.; Turck, F.; Grant, M.R.; Mundy, J.; et al. ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio 2013, 3, 321–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Deng, Z.; Lai, J.; Zhang, Y.; Yang, C.; Yin, B.; Zhao, Q.; Zhang, L.; Li, Y.; Yang, C.; et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009, 19, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.-L.; Chen, N.-Z.; An, R.; Su, Z.; Qi, B.-S.; Ren, F.; Chen, J.; Wang, X.-C. A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol. Biol. 2007, 63, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Liang, G.; Yu, D. Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 2012, 5, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.-L.; Ba, L.-J.; Shan, W.; Kuang, J.-F.; Lu, W.-J.; Chen, J.-Y. Involvement of WRKY transcription factors in abscisic-acid-induced cold tolerance of banana fruit. J. Agric. Food Chem. 2017, 65, 3627–3635. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Avramova, Z.; Fromm, M. The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. Plant J. 2011, 66, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Sussmilch, F.C.; Brodribb, T.J.; McAdam, S.A.M. What are the evolutionary origins of stomatal responses to abscisic acid (ABA) in land plants? J. Integr. Plant Biol. 2017, 59, 240–260. [Google Scholar] [CrossRef] [PubMed]
- Brodribb, T.J.; McAdam, S.A.M. Evolution of the stomatal regulation of plant water content. Plant Physiol. 2017, 174, 639–649. [Google Scholar] [CrossRef] [PubMed]
- McAdam, S.A.M.; Brodribb, T.J. Separating active and passive influences on stomatal control of transpiration. Plant Physiol. 2014, 164, 1578–1586. [Google Scholar] [CrossRef] [PubMed]
- Jackson, G.E.; Irvine, J.; Grace, J.; Khalil, A.A.M. Abscisic acid concentrations and fluxes in droughted conifer saplings. Plant Cell Environ. 1995, 18, 13–22. [Google Scholar] [CrossRef]
- Zuccarini, P.; Ciurli, A.; Alpi, A.; Hegedüsová, K. Hydraulic and chemical mechanisms in the response of Pinus pinaster Ait. to conditions of water stress. Ekológia 2011, 30, 422. [Google Scholar] [CrossRef]
- Brodribb, T.J.; McAdam, S.A.M. Passive origins of stomatal control in vascular plants. Science 2011, 331, 582–585. [Google Scholar] [CrossRef] [PubMed]
- Zeevaart, J.A.D. Changes in the levels of abscisic acid and its metabolites in excised leaf blades of Xanthium strumarium during and after water stress. Plant Physiol. 1980, 66, 672–678. [Google Scholar] [CrossRef] [PubMed]
- González-Guzmán, M.; Apostolova, N.; Bellés, J.M.; Barrero, J.M.; Piqueras, P.; Ponce, M.R.; Micol, J.L.; Serrano, R.; Rodríguez, P.L. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 2002, 14, 1833–1846. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Hase, T.; Toyoda, T.; Shinozaki, K.; Okamoto, M. Origin and evolution of genes related to ABA metabolism and its signaling pathways. J. Plant Res. 2011, 124, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Batge, S.L.; Ross, J.J.; Reid, J.B. Abscisic acid levels in seeds of the gibberellin-deficient mutant lh-2 of pea (Pisum sativum). Physiol. Plant 1999, 105, 485–490. [Google Scholar] [CrossRef]
- Wang, T.L.; Donkin, M.E.; Martin, E.S. The physiology of a wilty pea: Abscisic acid production under water stress. J. Exp. Bot. 1984, 35, 1222–1232. [Google Scholar] [CrossRef]
- Martins, S.C.V.; McAdam, S.A.M.; Deans, R.M.; DaMatta, F.M.; Brodribb, T.J. Stomatal dynamics are limited by leaf hydraulics in ferns and conifers: Results from simultaneous measurements of liquid and vapour fluxes in leaves. Plant Cell Environ. 2016, 39, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Sweet, K.J.; Peak, D.; Mott, K.A. Stomatal heterogeneity in responses to humidity and temperature: Testing a mechanistic model. Plant Cell Environ. 2017, 40, 2771–2779. [Google Scholar] [CrossRef] [PubMed]
- Pantin, F.; Monnet, F.; Jannaud, D.; Costa, J.M.; Renaud, J.; Muller, B.; Simonneau, T.; Genty, B. The dual effect of abscisic acid on stomata. New Phytol. 2013, 197, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Wickett, N.J.; Mirarab, S.; Nguyen, N.; Warnow, T.; Carpenter, E.; Matasci, N.; Ayyampalayam, S.; Barker, M.S.; Burleigh, J.G.; Gitzendanner, M.A.; et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. USA 2014, 111, E4859–E4868. [Google Scholar] [CrossRef] [PubMed]
- Chater, C.; Kamisugi, Y.; Movahedi, M.; Fleming, A.; Cuming, A.C.; Gray, J.E.; Beerling, D.J. Regulatory mechanism controlling stomatal behavior conserved across 400 million years of land plant evolution. Curr. Biol. 2011, 21, 1025–1029. [Google Scholar] [CrossRef] [PubMed]
- Hartung, W.; Weiler, E.W.; Volk, O.H. Immunochemical evidence that abscisic acid is produced by several species of Anthocerotae and Marchantiales. Bryologist 1987, 90, 393–400. [Google Scholar] [CrossRef]
- Ruszala, E.M.; Beerling, D.J.; Franks, P.J.; Chater, C.; Casson, S.A.; Gray, J.E.; Hetherington, A.M. Land plants acquired active stomatal control early in their evolutionary history. Curr. Biol. 2011, 21, 1030–1035. [Google Scholar] [CrossRef] [PubMed]
- Hõrak, H.; Kollist, H.; Merilo, E. Fern stomatal responses to ABA and CO2 depend on species and growth conditions. Plant Physiol. 2017, 174, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Chen, G.; Wang, Y.; Huang, Y.; Marchant, B.; Yang, Q.; Dai, F.; Hills, A.; Franks, P.J.; Nevo, E.; et al. Evolutionary conservation of ABA signaling for stomatal closure in ferns. Plant Physiol. 2017, 174, 732–747. [Google Scholar] [CrossRef] [PubMed]
- Henson, I.E.; Turner, N.C. Stomatal responses to abscisic acid in three lupin species. New Phytol. 1991, 117, 529–534. [Google Scholar] [CrossRef]
- Trejo, C.L.; Davies, W.J.; Ruiz, L. Sensitivity of stomata to abscisic acid (an effect of the mesophyll). Plant Physiol. 1993, 102, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Vahisalu, T.; Kollist, H.; Wang, Y.-F.; Nishimura, N.; Chan, W.-Y.; Valerio, G.; Lamminmäki, A.; Brosché, M.; Moldau, H.; Desikan, R. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 2008, 452, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Mustilli, A.-C.; Merlot, S.; Vavasseur, A.; Fenzi, F.; Giraudat, J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 2002, 14, 3089–3099. [Google Scholar] [CrossRef] [PubMed]
- McAdam, S.A.M.; Brodribb, T.J.; Banks, J.A.; Hedrich, R.; Atallah, N.M.; Cai, C.; Geringer, M.A.; Lind, C.; Nichols, D.S.; Stachowski, K.; et al. Abscisic acid controlled sex before transpiration in vascular plants. Proc. Natl. Acad. Sci. USA 2016, 113, 12862–12867. [Google Scholar] [CrossRef] [PubMed]
- Lind, C.; Dreyer, I.; López-Sanjurjo, E.J.; von Meyer, K.; Ishizaki, K.; Kohchi, T.; Lang, D.; Zhao, Y.; Kreuzer, I.; Al-Rasheid, K.A.S.; et al. Stomatal guard cells co-opted an ancient ABA-dependent desiccation survival system to regulate stomatal closure. Curr. Biol. 2015, 25, 928–935. [Google Scholar] [CrossRef] [PubMed]
- Vesty, E.F.; Saidi, Y.; Moody, L.A.; Holloway, D.; Whitbread, A.; Needs, S.; Choudhary, A.; Burns, B.; McLeod, D.; Bradshaw, S.J.; et al. The decision to germinate is regulated by divergent molecular networks in spores and seeds. New Phytol. 2016, 211, 952–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renzaglia, K.S.; Villarreal, J.C.; Piatkowski, B.T.; Lucas, J.R.; Merced, A. Hornwort stomata: Architecture and fate shared with 400 million year old fossil plants without leaves. Plant Physiol. 2017, 174, 788–797. [Google Scholar] [CrossRef] [PubMed]
- Lucas, J.R.; Renzaglia, K.S. Structure and function of hornwort stomata. Microsc. Microanal. 2002, 8, 1090–1091. [Google Scholar] [CrossRef]
- Duckett, J.G.; Pressel, S.; P’ng, K.M.Y.; Renzaglia, K.S. Exploding a myth: The capsule dehiscence mechanism and the function of pseudostomata in Sphagnum. New Phytol. 2009, 183, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Pressel, S.; Goral, T.; Duckett, J.G. Stomatal differentiation and abnormal stomata in hornworts. J. Bryol. 2014, 36, 87–103. [Google Scholar] [CrossRef]
- Chater, C.; Caine, R.S.; Tomek, M.; Wallace, S.; Kamisugi, Y.; Cuming, A.C.; Lang, D.; MacAlister, C.A.; Casson, S.; Bergmann, D.C.; et al. Origin and function of stomata in the moss Physcomitrella patens. Nat. Plants 2016, 2, 16179. [Google Scholar] [CrossRef] [PubMed]
- Sussmilch, F.C.; Atallah, N.M.; Brodribb, T.J.; Banks, J.A.; McAdam, S.A.M. Abscisic acid (ABA) and key proteins in its perception and signaling pathways are ancient, but their roles have changed through time. Plant Signal Behav. 2017, e1365210. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Waadt, R.; Schroeder, J.I. Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 2011, 21, R346–R355. [Google Scholar] [CrossRef] [PubMed]
- Tougane, K.; Komatsu, K.; Bhyan, S.B.; Sakata, Y.; Ishizaki, K.; Yamato, K.T.; Kohchi, T.; Takezawa, D. Evolutionarily conserved regulatory mechanisms of abscisic acid signaling in land plants: Characterization of ABSCISIC ACID INSENSITIVE1-like type 2C protein phosphatase in the liverwort Marchantia polymorpha. Plant Physiol. 2010, 152, 1529–1543. [Google Scholar] [CrossRef] [PubMed]
- Takezawa, D.; Komatsu, K.; Sakata, Y. ABA in bryophytes: How a universal growth regulator in life became a plant hormone? J. Plant Res. 2011, 124, 437–453. [Google Scholar] [CrossRef] [PubMed]
- Hartung, W. The evolution of abscisic acid (ABA) and ABA function in lower plants, fungi and lichen. Funct. Plant Biol. 2010, 37, 806–812. [Google Scholar] [CrossRef]
- Tossi, V.; Cassia, R.; Bruzzone, S.; Zocchi, E.; Lamattina, L. ABA says NO to UV-B: A universal response? Trends Plant Sci. 2012, 17, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, N.; Takahata, M.; Tominaga, H. Effects of NaCl and KNO3 concentrations on the abscisic acid content of Dunaliella sp. (Chlorophyta). Hydrobiologia 1993, 267, 163–168. [Google Scholar] [CrossRef]
- Kobayashi, M.; Hirai, N.; Kurimura, Y.; Ohigashi, H.; Tsuji, Y. Abscisic acid-dependent algal morphogenesis in the unicellular green alga Haematococcus pluvialis. Plant Growth Regul. 1997, 22, 79–85. [Google Scholar] [CrossRef]
- Hirsch, R.; Hartung, W.; Gimmler, H. Abscisic acid content of algae under stress. Bot. Acta 1989, 102, 326–334. [Google Scholar] [CrossRef]
- Tietz, A.; Ruttkowski, U.; Kohler, R.; Kasprik, W. Further investigations on the occurrence and the effects of abscisic acid in algae. Biochem. Physiol. Pflanz. 1989, 184, 259–266. [Google Scholar] [CrossRef]
- Bajguz, A. Brassinosteroid enhanced the level of abscisic acid in Chlorella vulgaris subjected to short-term heat stress. J. Plant Physiol. 2009, 166, 882–886. [Google Scholar] [CrossRef] [PubMed]
- Maršálek, B.; Zahradníčková, H.; Hronková, M. Extracellular production of abscisic acid by soil algae under salt, acid or drought stress. Z. Naturforsch. C Biosci. 1992, 47, 701–704. [Google Scholar] [CrossRef]
- Hirai, N.; Yoshida, R.; Todoroki, Y.; Ohigashi, H. Biosynthesis of abscisic acid by the non-mevalonate pathway in plants, and by the mevalonate pathway in fungi. Biosci. Biotechnol. Biochem. 2000, 64, 1448–1458. [Google Scholar] [CrossRef] [PubMed]
- Inomata, M.; Hirai, N.; Yoshida, R.; Ohigashi, H. Biosynthesis of abscisic acid by the direct pathway via ionylideneethane in a fungus, Cercospora cruenta. Biosci. Biotechnol. Biochem. 2004, 68, 2571–2580. [Google Scholar] [CrossRef] [PubMed]
- Inomata, M.; Hirai, N.; Yoshida, R.; Ohigashi, H. The biosynthetic pathway to abscisic acid via ionylideneethane in the fungus Botrytis cinerea. Phytochemistry 2004, 65, 2667–2678. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Wang, Y.; Qin, S. Genomewide analysis of carotenoid cleavage dioxygenases in unicellular and filamentous cyanobacteria. Comp. Funct. Genom. 2012, 2012, 164690. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Ando, H.; Hanaoka, M.; Tanaka, K. Abscisic acid participates in the control of cell cycle initiation through heme homeostasis in the unicellular red alga Cyanidioschyzon merolae. Plant Cell Physiol. 2016, 57, 953–960. [Google Scholar] [CrossRef] [PubMed]
- Kiseleva, A.A.; Tarachovskaya, E.R.; Shishova, M.F. Biosynthesis of phytohormones in algae. Russ. J. Plant Physiol. 2012, 59, 595–610. [Google Scholar] [CrossRef]
- Burbidge, A.; Grieve, T.M.; Jackson, A.; Thompson, A.; McCarty, D.R.; Taylor, I.B. Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. Plant J. 1999, 17, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Bang, S.W.; Park, S.-H.; Jeong, J.S.; Kim, Y.S.; Jung, H.; Ha, S.-H.; Kim, J.-K. Characterization of the stress-inducible OsNCED3 promoter in different transgenic rice organs and over three homozygous generations. Planta 2013, 237, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Schwartz, S.H.; Zeevaart, J.A.D.; McCarty, D.R. Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. USA 1997, 94, 12235–12240. [Google Scholar] [CrossRef] [PubMed]
- Sederias, J.; Colman, B. The interaction of light and low temperature on breaking the dormancy of Chara vulgaris oospores. Aquat. Bot. 2007, 87, 229–234. [Google Scholar] [CrossRef]
- Salerno, G.L. Measurement of enzymes related to sucrose metabolism in permeabilized Chlorella vulgaris cells. Physiol. Plant 1985, 64, 259–264. [Google Scholar] [CrossRef]
- Ullrich, W.R.; Kunz, G. Effect of abscisic acid on nitrate uptake, respiration and photosynthesis in green algae. Plant Sci. Lett. 1984, 37, 9–14. [Google Scholar] [CrossRef]
- Holzinger, A.; Kaplan, F.; Blaas, K.; Zechmann, B.; Komsic-Buchmann, K.; Becker, B. Transcriptomics of desiccation tolerance in the streptophyte green alga Klebsormidium reveal a land plant-like defense reaction. PLoS ONE 2014, 9, e110630. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, S.R.; Kamisugi, Y.; Trinh, C.H.; Schmutz, J.; Jenkins, J.W.; Grimwood, J.; Muchero, W.; Tuskan, G.A.; Rensing, S.A.; Lang, D.; et al. Genetic analysis of Physcomitrella patens identifies ABSCISIC ACID NON-RESPONSIVE, a regulator of ABA responses unique to basal land plants and required for desiccation tolerance. Plant Cell 2016, 28, 1310–1327. [Google Scholar] [CrossRef] [PubMed]
- Hellwege, E.M.; Dietz, K.-J.; Volk, O.H.; Hartung, W. Abscisic acid and the induction of desiccation tolerance in the extremely xerophilic liverwort Exormotheca holstii. Planta 1994, 194, 525–531. [Google Scholar] [CrossRef]
- Cuming, A.C.; Cho, S.H.; Kamisugi, Y.; Graham, H.; Quatrano, R.S. Microarray analysis of transcriptional responses to abscisic acid and osmotic, salt, and drought stress in the moss, Physcomitrella patens. New Phytol. 2007, 176, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Khandelwal, A.; Cho, S.H.; Marella, H.; Sakata, Y.; Perroud, P.-F.; Pan, A.; Quatrano, R.S. Role of ABA and ABI3 in desiccation tolerance. Science 2010, 327, 546. [Google Scholar] [CrossRef] [PubMed]
- Kamisugi, Y.; Cuming, A.C. The evolution of the abscisic acid-response in land plants: Comparative analysis of Group 1 LEA gene expression in moss and cereals. Plant Mol. Biol. 2005, 59, 723–737. [Google Scholar] [CrossRef] [PubMed]
- Shinde, S.; Nurul Islam, M.; Ng, C.K.Y. Dehydration stress-induced oscillations in LEA protein transcripts involves abscisic acid in the moss, Physcomitrella patens. New Phytol. 2012, 195, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, T.L.; Bewley, J.D. Characterization of protein synthetic changes in a desiccation-tolerant fern, Polypodium virginianum. Comparison of the effects of drying, rehydration and abscisic acid. J. Exp. Bot. 1993, 44, 921–928. [Google Scholar] [CrossRef]
- Rinne, P.; Welling, A.; Kaikuranta, P. Onset of freezing tolerance in birch (Betula pubescens Ehrh.) involves LEA proteins and osmoregulation and is impaired in an ABA-deficient genotype. Plant Cell Environ. 1998, 21, 601–611. [Google Scholar] [CrossRef]
- Wang, X.; Chen, S.; Zhang, H.; Shi, L.; Cao, F.; Guo, L.; Xie, Y.; Wang, T.; Yan, X.; Dai, S. Desiccation tolerance mechanism in resurrection fern-ally Selaginella tamariscina revealed by physiological and proteomic analysis. J. Proteome Res. 2010, 9, 6561–6577. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.-Z.; Dunstan, D.I. Characterization of cDNAs representing five abscisic acid-responsive genes associated with somatic embryogenesis in Picea glauca, and their responses to abscisic acid stereostructure. Planta 1997, 203, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Meurs, C.; Basra, A.S.; Karssen, C.M.; van Loon, L.C. Role of abscisic acid in the induction of desiccation tolerance in developing seeds of Arabidopsis thaliana. Plant Physiol. 1992, 98, 1484–1493. [Google Scholar] [CrossRef] [PubMed]
- Mishra, G.; Zhang, W.; Deng, F.; Zhao, J.; Wang, X. A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 2006, 312, 264–266. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Zhu, J.-K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [PubMed]
- Creelman, R.A.; Mason, H.S.; Bensen, R.J.; Boyer, J.S.; Mullet, J.E. Water deficit and abscisic acid cause differential inhibition of shoot versus root growth in soybean seedlings: Analysis of growth, sugar accumulation, and gene expression. Plant Physiol. 1990, 92, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Saab, I.N.; Sharp, R.E.; Pritchard, J.; Voetberg, G.S. Increased endogenous abscisic acid maintains primary root growth and inhibits shoot growth of maize seedlings at low water potentials. Plant Physiol. 1990, 93, 1329–1336. [Google Scholar] [CrossRef] [PubMed]
- Graeber, K.A.I.; Nakabayashi, K.; Miatton, E.; Leubner-Metzger, G.; Soppe, W.J.J. Molecular mechanisms of seed dormancy. Plant Cell Environ. 2012, 35, 1769–1786. [Google Scholar] [CrossRef] [PubMed]
- Ram, H.Y.M.; Jaiswal, V.S. Induction of male flowers on female plants of Cannabis sativa by gibberellins and its inhibition by abscisic acid. Planta 1972, 105, 263–266. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, M.; Atsmon, D.; Galun, E. Sexual differentiation in cucumber: The effects of abscisic acid and other growth regulators on various sex genotypes. Plant Cell Physiol. 1977, 18, 261–269. [Google Scholar] [CrossRef]
- Rudich, J.; Halevy, A.H. Involvement of abscisic acid in the regulation of sex expression in the cucumber. Plant Cell Physiol. 1974, 15, 635–642. [Google Scholar] [CrossRef]
- Fujii, H.; Verslues, P.E.; Zhu, J.-K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Molina, L.; Mongrand, S.; Chua, N.-H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 2001, 98, 4782–4787. [Google Scholar] [CrossRef] [PubMed]
- Kane, M.E.; Albert, L.S. Abscisic acid induces aerial leaf morphology and vasculature in submerged Hippuris vulgaris L. Aquat. Bot. 1987, 28, 81–88. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef] [PubMed]
- Nystedt, B.; Street, N.R.; Wetterbom, A.; Zuccolo, A.; Lin, Y.-C.; Scofield, D.G.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The Norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, B.-C.; Joseph, L.M.; Deng, W.-T.; Liu, L.; Li, Q.-B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Vallabhaneni, R.; Bradbury, L.M.T.; Wurtzel, E.T. The carotenoid dioxygenase gene family in maize, sorghum, and rice. Arch. Biochem. Biophys. 2010, 504, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Wan, H.; Wu, Z.; Wang, R.; Ruan, M.; Ye, Q.; Li, Z.; Zhou, G.; Yao, Z.; Yang, Y. A comprehensive analysis of carotenoid cleavage dioxygenases genes in Solanum lycopersicum. Plant Mol. Biol. Report. 2016, 34, 512–523. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sussmilch, F.C.; McAdam, S.A.M. Surviving a Dry Future: Abscisic Acid (ABA)-Mediated Plant Mechanisms for Conserving Water under Low Humidity. Plants 2017, 6, 54. https://doi.org/10.3390/plants6040054
Sussmilch FC, McAdam SAM. Surviving a Dry Future: Abscisic Acid (ABA)-Mediated Plant Mechanisms for Conserving Water under Low Humidity. Plants. 2017; 6(4):54. https://doi.org/10.3390/plants6040054
Chicago/Turabian StyleSussmilch, Frances C., and Scott A. M. McAdam. 2017. "Surviving a Dry Future: Abscisic Acid (ABA)-Mediated Plant Mechanisms for Conserving Water under Low Humidity" Plants 6, no. 4: 54. https://doi.org/10.3390/plants6040054






