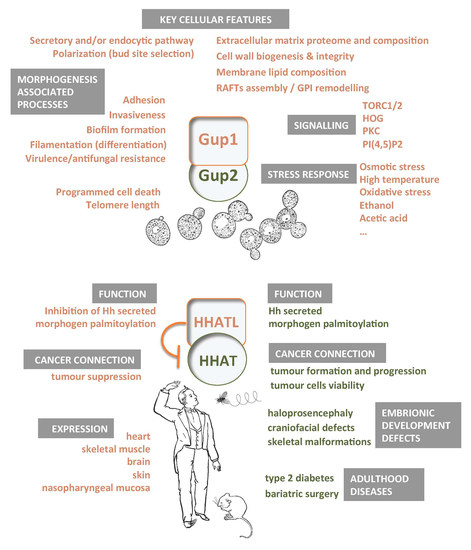
Yeast Gup1(2) Proteins Are Homologues of the Hedgehog Morphogens Acyltransferases HHAT(L): Facts and Implications
Abstract
:1. Introduction
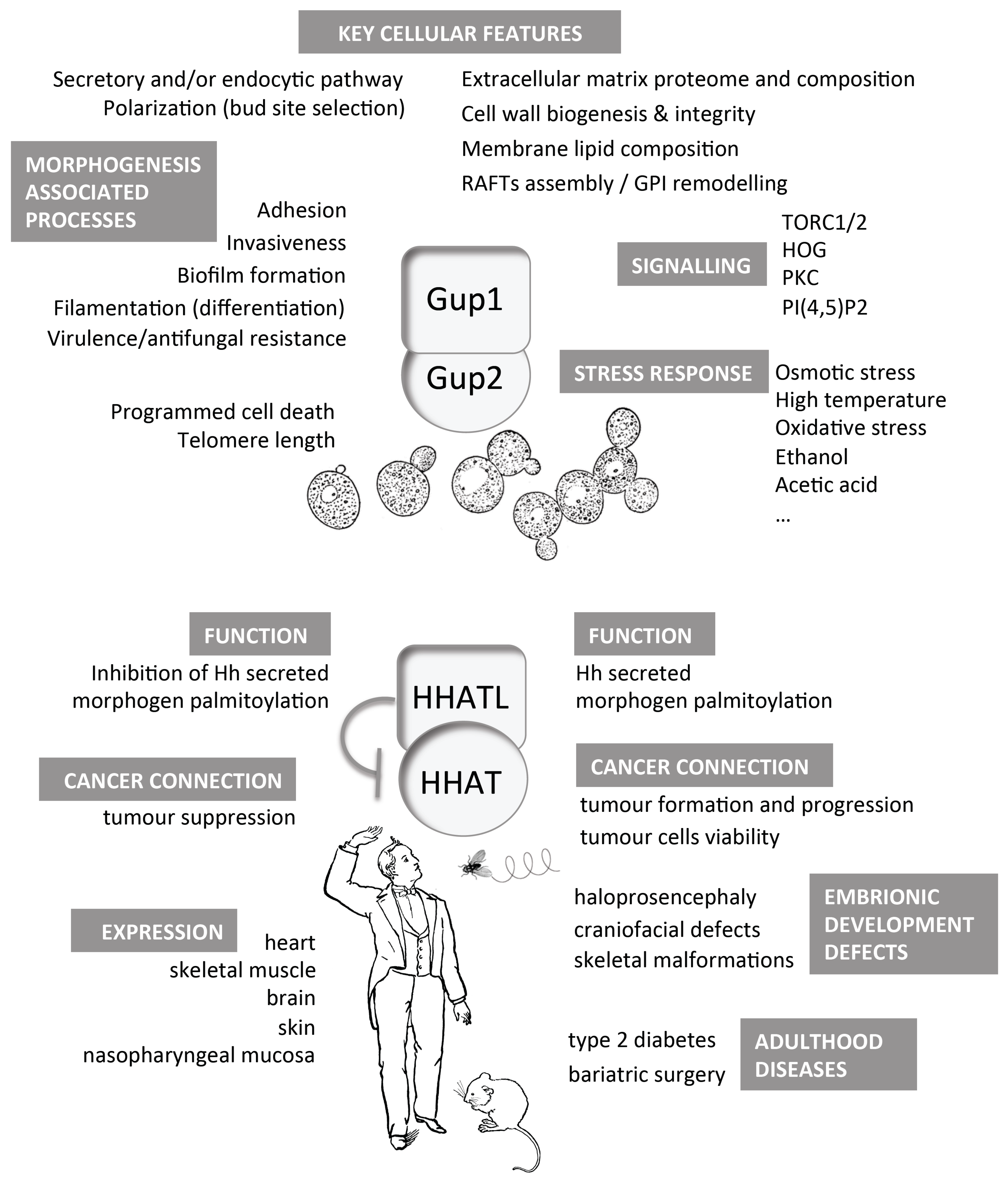
2. Yeast Gup1 and Gup2 Proteins
2.1. Gup1 and Gup2 Are Members of the MBOAT Superfamily
2.2. Gup1 and Gup2 Subcellular Localisation
2.3. GUP1 and GUP2 Expression in Yeast
3. Yeast Phenotypes Associated with the Deletion of GUP1
3.1. Phenotypes Emerging from Genome-Wide Yeast Screenings
3.2. Cell Wall Integrity and Biogenesis
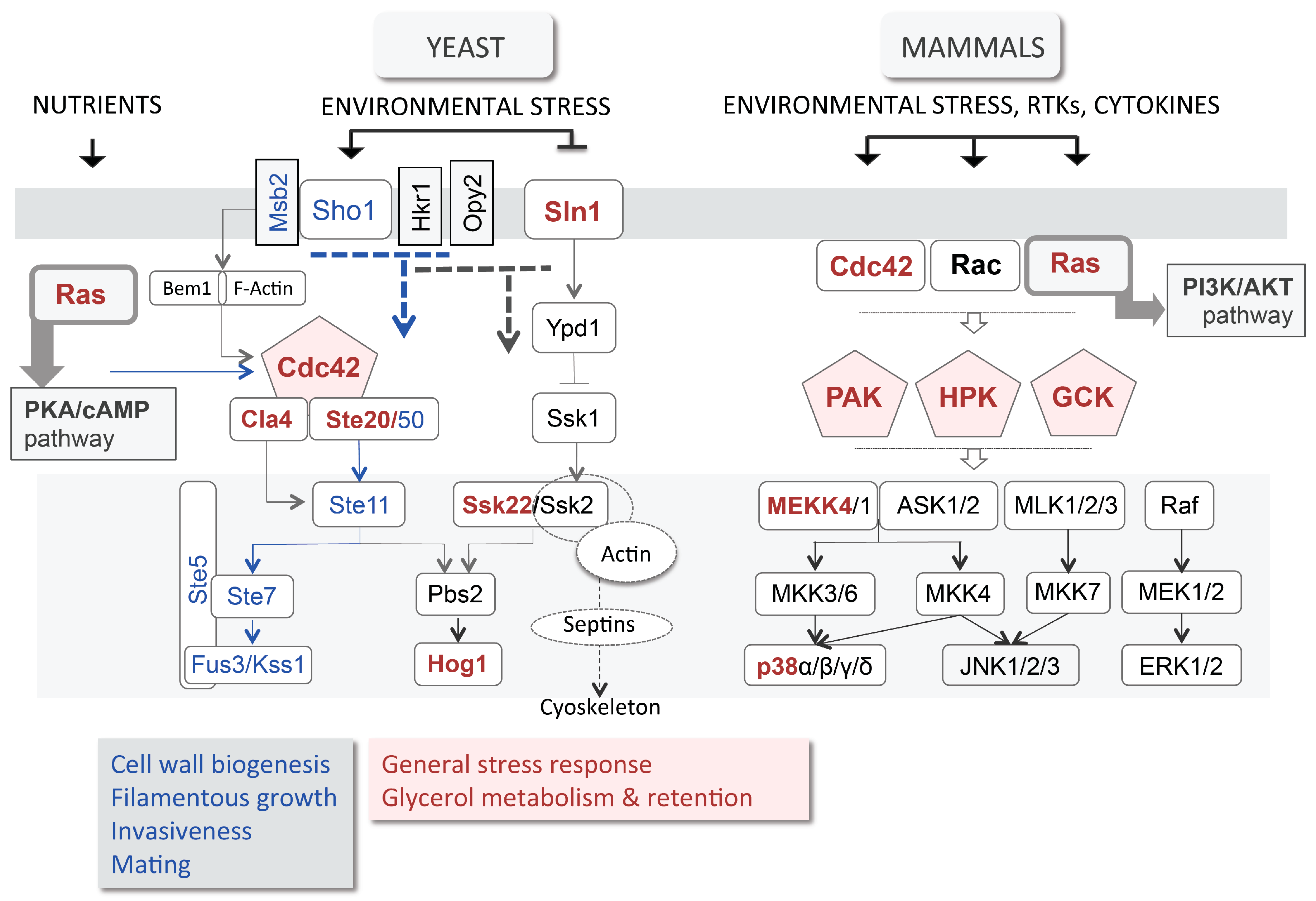
3.3. High Osmolarity Glycerol (HOG) Pathway
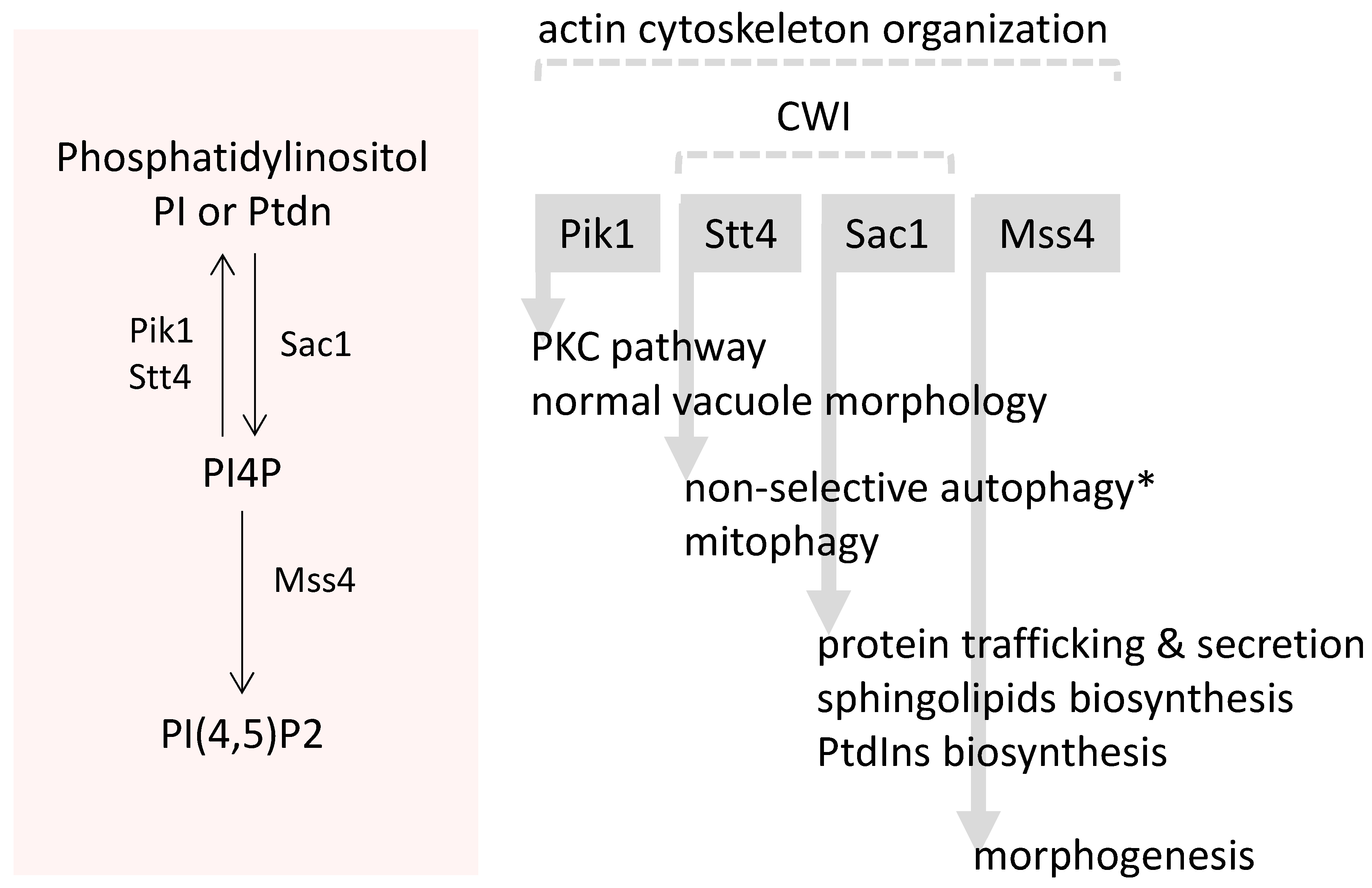
3.4. Plasma Membrane Composition and Associated Signalling
3.5. Cell Death
3.6. Differentiation and Morphology
4. Proteins Interacting with Yeast Gup1 and Gup2
5. Gup1/2 Homologues from High Eukaryotes
5.1. Hedgehog Pathway
5.2. HHAT(L) Proteins Expression and Associated Pathologies
6. Similarity Analysis of the GUP/HHAT(L) Amino Acid Sequences
7. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Nomenclature
References
- Verduyckt, M.; Vignaud, H.; Bynens, T.; van den Brande, J.; Franssens, V.; Cullin, C.; Winderickx, J. Yeast as a model for Alzheimer’s disease: Latest studies and advanced strategies. Meth. Mol. Biol. 2016, 1303, 197–215. [Google Scholar]
- Brückner, S.; Mösch, H.U. Choosing the right lifestyle: Adhesion and development in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2012, 36, 25–58. [Google Scholar] [CrossRef] [PubMed]
- Čáp, M.; Štepánek, L.; Harant, K.; Váchová, L.; Palková, Z. Cell differentiation within a yeast colony: Metabolic and regulatory parallels with a tumor-affected organism. Mol. Cell 2012, 46, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Váchová, L.; Palková, Z. Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia. J. Cell Biol. 2005, 169, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Váchová, L.; Palková, Z. Caspases in yeast apoptosis-like death: Facts and artefacts. FEMS Yeast Res. 2007, 7, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Turrà, D.; Nordzieke, D.; Vitale, S.; El Ghalid, M.; di Pietro, A. Hyphal chemotropism in fungal pathogenicity. Semin. Cell Dev. Biol. 2016, 57, 69–75. [Google Scholar] [CrossRef] [PubMed]
- St’ovíček, V.; Váchová, L.; Kuthan, M.; Palková, Z. General factors important for the formation of structured biofilm-like yeast colonies. Fungal Genet. Biol. 2010, 47, 1012–1022. [Google Scholar] [CrossRef] [PubMed]
- Váchová, L.; Kucerová, H.; Devaux, F.; Ulehlová, M.; Palková, Z. Metabolic diversification of cells during the development of yeast colonies. Environ. Microbiol. 2009, 11, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Zupan, J.; Raspor, P. Invasive growth of Saccharomyces cerevisiae depends on environmental triggers: A quantitative model. Yeast 2010, 27, 217–228. [Google Scholar] [PubMed]
- Faria-Oliveira, F.; Carvalho, J.; Belmiro, C.L.; Ramalho, G.; Pavão, M.; Lucas, C.; Ferreira, C. Elemental biochemical analysis of the polysaccharides in the extracellular matrix of the yeast Saccharomyces cerevisiae. J. Basic Microbiol. 2015, 55, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Faria-Oliveira, F.; Carvalho, J.; Ferreira, C.; Hernáez, M.L.; Gil, C.; Lucas, C. Quantitative differential proteomics of yeast extracellular matrix: There is more to it than meets the eye. BMC Microbiol. 2015, 15, 271. [Google Scholar] [CrossRef] [PubMed]
- Palková, Z.; Váchová, L. Ammonia signaling in yeast colony formation. Int. Rev. Cytol. 2003, 225, 229–272. [Google Scholar] [PubMed]
- Chen, H.; Fujita, M.; Feng, Q.; Clardy, J.; Fink, G.R. Tyrosol is a quorum sensing molecule in Candida albicans. Proc. Nat. Acad. Sci. USA 2004, 101, 5048–5052. [Google Scholar] [CrossRef] [PubMed]
- Sprague, G.F.; Winans, S.C. Eukaryotes learn how to count: Quorum sensing by yeast. Genes Dev. 2006, 20, 1045–1049. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, I.; Kornberg, T.B. Hedgehog and its circuitous journey from producing to target cells. Semin. Cell Dev. Biol. 2014, 33, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, T.; Nagai, Y.; Ohya, Y. Molecular mechanism of VDE-initiated intein homing in yeast nuclear genome. Adv. Biophys. 2004, 38, 215–232. [Google Scholar] [CrossRef]
- Chong, S.; Shao, Y.; Paulus, H.; Benner, J.; Perler, F.B.; Xu, M.Q. Protein splicing involving the Saccharomyces cerevisiae VMA intein. The steps in the splicing pathway, side reactions leading to protein cleavage, and establishment of an in vitro splicing system. J. Biol. Chem. 1997, 271, 22159–22168. [Google Scholar]
- McGuire, C.; Cotter, K.; Stransky, L.; Forgac, M. Regulation of V-ATPase assembly and function of V-ATPases in tumor cell invasiveness. Biochim. Biophys. Acta 2016, 1857, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Hölst, B.; Lunde, C.; Lages, F.; Oliveira, R.; Lucas, C.; Kielland-Brandt, M.C. GUP1 and its close homologue GUP2, encoding multimembrane-spanning proteins involved in active glycerol uptake in Saccharomyces cerevisiae. Mol. Microbiol. 2000, 37, 108–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamoun, Z.; Mann, R.K.; Nellen, D.; von Kessler, D.P.; Bellotto, M.; Beachy, P.A.; Basler, K. Skinny Hedgehog, an acyltransferase required for palmitoylation and activity of the Hedgehog signal. Science 2001, 293, 2080–2084. [Google Scholar] [CrossRef] [PubMed]
- Buglino, J.A.; Resh, M.D. Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. J. Biol. Chem. 2008, 283, 22076–22088. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Kita, Y.; Niikura, T. Mammalian Gup1, a homolog of Saccharomyces cerevisiae glycerol uptake/transporter 1, acts as a negative regulator for N-terminal palmitoylation of Sonic hedgehog. FEBS J. 2008, 275, 318–331. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, K. A superfamily of membrane-bound O-acyltransferases with implications for Wnt signaling. Trends Biochem. Sci. 2000, 26, 111–112. [Google Scholar] [CrossRef]
- Neves, L.; Oliveira, R.; Lucas, C. Yeast orthologues associated with glycerol transport and metabolism. FEMS Yeast Res. 2004, 5, 51–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neves, L.; Lages, F.; Lucas, C. New insights on glycerol transport in Saccharomyces cerevisiae. FEBS Lett. 2004, 565, 160–162. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.; van Voorst, F.; Martins, A.; Neves, L.; Oliveira, R.; Kielland-Brandt, M.; Lucas, C.; Brandt, A. A member of the sugar transporter family, Stl1p is the glycerol/H+ symporter in Saccharomyces cerevisiae. Mol. Biol. Cell 2005, 16, 2068–2076. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.O.; Kim, S.W.; Han, S.O. Engineering of glycerol utilization pathway for ethanol production by Saccharomyces cerevisiae. Bioresour. Technol. 2010, 101, 4157–4161. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.; Silva, S.; van Voorst, F.; Aguiar, C.; Kielland-Brandt, M.C.; Brandt, A.; Lucas, C. Absence of Gup1p in Saccharomyces cerevisiae results in defective cell wall composition, assembly, stability and morphology. FEMS Yeast Res. 2006, 6, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
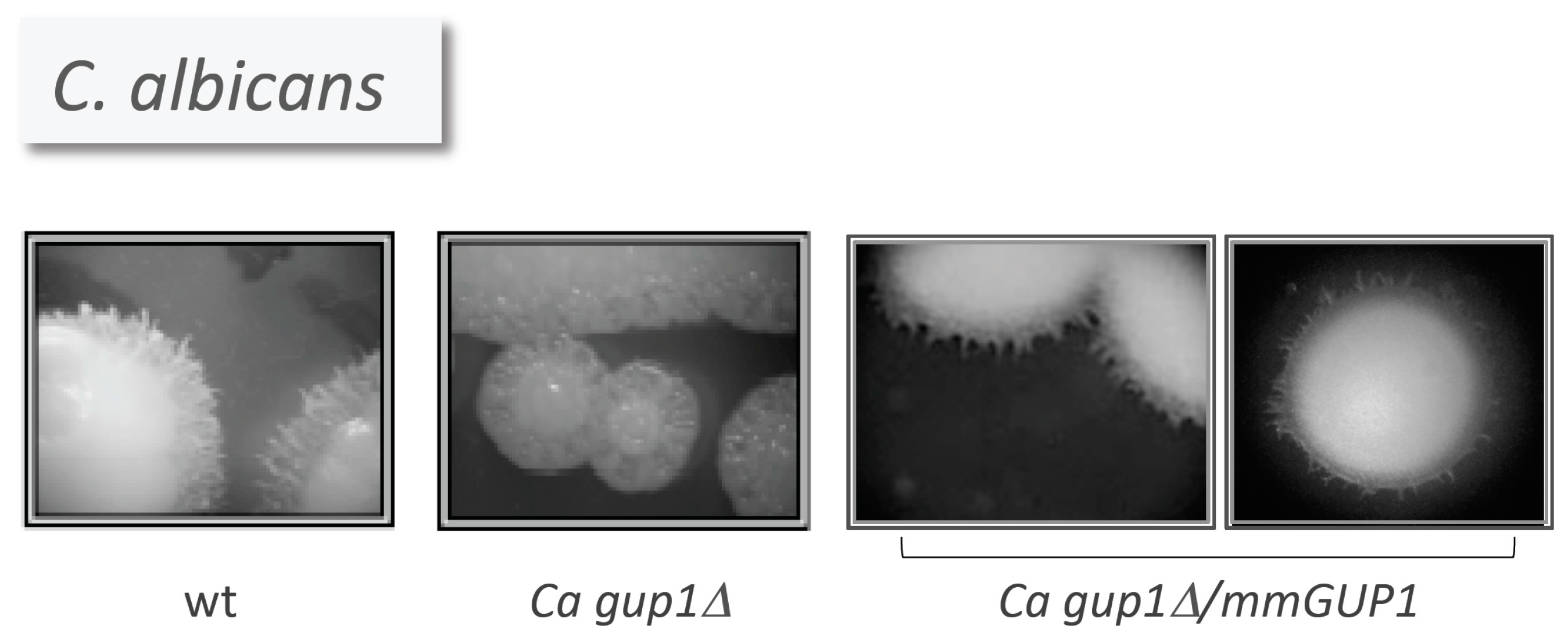
- Ferreira, C.; Silva, S.; Faria-Oliveira, F.; Pinho, E.; Henriques, M.; Lucas, C. Candida albicans virulence and drug-resistance requires the O-acyltransferase Gup1p. BMC Microbiol. 2010, 10, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Micchelli, C.A.; The, I.; Selva, E.; Mogila, V.; Perrimon, N. Rasp, a putative transmembrane acyltransferase, is required for Hedgehog signaling. Development 2002, 129, 843–851. [Google Scholar] [PubMed]
- Lee, J.D.; Treisman, J.E. Sightless has homology to transmembrane acyltransferases and is required to generate active Hedgehog protein. Curr. Biol. 2001, 11, 1147–1152. [Google Scholar] [CrossRef]
- Amanai, K.; Jiang, J. Distinct roles of Central Missing and Dispatched in sending the Hedgehog signal. Development 2001, 128, 5119–5127. [Google Scholar] [PubMed]
- NCBI—National Center for Biotechnological Information. Available online: http://www.ncbi.nlm.nih.gov/ (assessed on 28 October 2016).
- Soejima, H.; Kawamoto, S.; Akai, J.; Miyoshi, O.; Arai, Y.; Morohka, T.; Matsuo, S.; Niikawa, N.; Kimura, A.; Okubo, K.; et al. Isolation of novel heart-specific genes using the Body Map database. Genomics 2001, 74, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Q.; Peng, H.; Jiang, H.Y.; Hu, H.; Zhang, J.H.; Yao, K.T.; Zhao, T. Cloning of human KIAA1173 gene and biological characterization of transfected 6–10B cells. Di Yi Jun Yi Da Xue Xue Bao 2005, 25, 1216–1220. [Google Scholar] [PubMed]
- Zhang, S.Q.; Peng, H.; Song, L.Y.; Li, X.M.; Jiang, H.Y.; Yao, K.T.; Zhao, T. Detection of KIAA1173 gene expression in nasopharyngeal carcinoma tissues and cell lines on tissue microarray. Ai Zheng 2005, 24, 1322–1326. [Google Scholar] [PubMed]
- Ferreira, C.; Lucas, C. The yeast O-acyltransferase Gup1p interferes in lipid metabolism with direct consequences on the sphingolipid-sterol-ordered domains integrity/assembly. Biochim. Biophys. Acta 2008, 1778, 2648–2653. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.Y.; Sun, J.; Chang, T.-Y. Membrane-bound O-acyltransferases (MBOATs). Front. Biol. 2011, 6, 177–182. [Google Scholar]
- Bleve, G.; di Sansebastiano, G.P.; Grieco, F. Over-expression of functional Saccharomyces cerevisiae GUP1, induces proliferation of intracellular membranes containing ER and Golgi resident proteins. Biochim. Biophys. Acta 2011, 1808, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Buglino, J.A.; Resh, M.D. Identification of conserved regions and residues within Hedgehog acyltransferase critical for palmitoylation of Sonic Hedgehog. PLoS ONE 2010, 5, e11195. [Google Scholar] [CrossRef] [PubMed]
- Bleve, G.; Zacheo, G.; Cappello, M.S.; Dellaglio, F.; Grieco, F. Subcellular localization and functional expression of the glycerol uptake protein 1 (GUP1) of Saccharomyces cerevisiae tagged with green fluorescent protein. Biochem. J. 2005, 390, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Konitsiotis, A.D.; Jovanović, B.; Ciepla, P.; Spitaler, M.; Lanyon-Hogg, T.; Tate, E.W.; Magee, A.I. Topological analysis of Hedgehog acyltransferase, a multipalmitoylated transmembrane protein. J. Biol. Chem. 2015, 290, 3293–3307. [Google Scholar] [CrossRef] [PubMed]
- Transmembrane Helix Prediction. Available online: http://www.cbs.dtu.dk/services/TMHMM/ (assessed on 15 July 2016).
- CSS-Palm. Prediction of Palmitoylation Site. Available online: http://csspalm.biocuckoo.org (assessed on 15 July 2016).
- Tulha, J.; Lucas, C.; CBMA-Centre of Molecular and Environmental Biology, University of Minho, Braga, Portugal. Unpublished work. 2016.
- YEASTRACT, Yeast Search for Transcriptional Regulators and Consensus Tracking. Available online: http://www.yeastract.com/ (assessed on 15 July 2016).
- Oliveira, R.; Lucas, C. Expression studies of GUP1 and GUP2, genes involved in glycerol active transport in Saccharomyces cerevisiae, using semi-quantitative RT-PCR. Curr. Genet. 2004, 46, 140–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Posas, F.; Chambers, J.R.; Heyman, J.A.; Hoeffler, J.P.; de Nadal, E.; Ariño, J. The transcriptional response of yeast to saline stress. J. Biol. Chem. 2000, 275, 17249–17255. [Google Scholar] [CrossRef] [PubMed]
- Rep, M.; Krantz, M.; Thevelein, J.M.; Hohmann, S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 2000, 275, 8290–8300. [Google Scholar] [CrossRef] [PubMed]
- Yale, J.; Bohnert, H.J. Transcript expression in Saccharomyces cerevisiae at high salinity. J. Biol. Chem. 2001, 276, 15996–16007. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 300–372. [Google Scholar] [CrossRef] [PubMed]
- Henry, S.A.; Kohlwein, S.D.; Carman, G.M. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 2012, 190, 317–349. [Google Scholar] [CrossRef] [PubMed]
- Larsson, C.; Påhlman, I.-L.; Ansell, R.; Rigoulet, M.; Adler, L.; Gustafsson, L. The importance of the glycerol 3-phosphate shuttle during aerobic growth of Saccharomyces cerevisiae. Yeast 1988, 14, 347–357. [Google Scholar] [CrossRef]
- Siderius, M.; van Wuytswinkel, O.; Reijenga, K.A.; Kelders, M.; Mager, W.H. The control of intracellular glycerol in Saccharomyces cerevisiae influences osmotic stress response and resistance to increased temperature. Mol. Microbiol. 2000, 36, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Oelkers, P.; Tinkelenberg, A.; Erdeniz, N.; Cromley, D.; Billheimer, T.; Sturley, S.L. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J. Biol. Chem. 2000, 275, 15609–15612. [Google Scholar] [CrossRef] [PubMed]
- Dardalhon, M.; Lin, W.; Nicolas, A.; Averbeck, D. Specific transcriptional responses induced by 8-methoxypsoralen and UVA in yeast. FEMS Yeast Res. 2007, 7, 866–878. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Snyder, M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 2001, 12, 2147–2170. [Google Scholar] [CrossRef] [PubMed]
- Casamayor, A.; Snyder, M. Bud-site selection and cell polarity in budding yeast. Curr. Opin. Microbiol. 2002, 5, 179–186. [Google Scholar] [CrossRef]
- Bonangelino, C.J.; Chavez, E.M.; Bonifacino, J.S. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 2002, 13, 2486–2501. [Google Scholar] [CrossRef] [PubMed]
- Reiner, S.; Micolod, D.; Zellnig, G.; Schneiter, R. A genomewide screen reveals a role of mitochondria in anaerobic uptake of sterols in yeast. Mol. Biol. Cell 2006, 17, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Askree, S.H.; Yehuda, T.; Smolikov, S.; Gurevich, R.; Hawk, J.; Coker, C.; Krauskopf, A.; Kupiec, M.; McEachern, M.J. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA 2004, 101, 8658–8663. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, S.C.; Sá-Correia, I. Genome-wide identification of genes required for yeast growth under imatinib stress: Vacuolar H+-ATPase function is an important target of this anticancer drug. OMICS 2009, 13, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Hu, B.; Arno, M.J.; Panaretou, B. Genomic screening in vivo reveals the role played by vacuolar H+ ATPase and cytosolic acidification in sensitivity to DNA-damaging agents such as cisplatin. Mol. Pharmacol. 2007, 71, 2416–2425. [Google Scholar] [CrossRef] [PubMed]
- Tucker, C.L.; Fields, S. Quantitative genome-wide analysis of yeast deletion strain sensitivities to oxidative and chemical stress. Comp. Funct. Genom. 2004, 5, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Viladevall, L.; Serrano, R.; Ruiz, A.; Domenech, G.; Giraldo, J.; Barceló, A.; Ariño, J. Characterization of the calcium-mediated response to alkaline stress in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 43614–43624. [Google Scholar] [CrossRef] [PubMed]
- Darvishi, E.; Omidi, M.; Bushehri, A.A.; Golshani, A.; Smith, M.L. Thymol antifungal mode of action involves telomerase inhibition. Med. Mycol. 2013, 51, 826–834. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Lin, W.; Ma, X.; Lu, Q.; Ma, X.; Bian, G.; Jiang, L. The protein kinase Hal5p is the high-copy suppressor of lithium-sensitive mutations of genes involved in the sporulation and meiosis as well as the ergosterol biosynthesis in Saccharomyces cerevisiae. Genomics 2010, 95, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Arita, A.; Ellen, T.P.; Liu, X.; Bai, J.; Rooney, J.P.; Kurtz, A.D.; Klein, C.B.; Dai, W.; Begley, T.J.; et al. A genome-wide screen in Saccharomyces cerevisiae reveals pathways affected by arsenic toxicity. Genomics 2009, 94, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Rossouw, D.; Bauer, F.F. Comparing the transcriptomes of wine yeast strains: Toward understanding the interaction between environment and transcriptome during fermentation. Appl. Microbiol. Biotechnol. 2009, 84, 937–954. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Liu, L.Z. Quantitative transcription dynamic analysis reveals candidate genes and key regulators for ethanol tolerance in Saccharomyces cerevisiae. BMC Microbiol. 2010, 10, 169. [Google Scholar] [CrossRef] [PubMed]
- Lesage, G.; Shapiro, J.; Specht, C.A.; Sdicu, A.M.; Ménard, P.; Hussein, S.; Tong, A.H.; Boone, C.; Bussey, H. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 2005, 6, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Nobel, H.; Ruiz, C.; Martin, H.; Morris, W.; Brul, S.; Molina, M.; Klis, F.M. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology 2000, 46, 2121–2132. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C. Identification and characterisation of the glycerol/H+ symporter in Saccharomyces cerevisiaie and the involvement of related genes in the cell wall integrity. Ph.D. Thesis, University of Minho, Braga, Portugal, September 2005. Available online: http://repositorium.sdum.uminho.pt/ (assessed on 28 October 2016). [Google Scholar]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [PubMed]
- Lockshon, D.; Olsen, C.P.; Brett, C.L.; Chertov, A.; Merz, A.J.; Lorenz, D.A.; van Gilst, M.R.; Kennedy, B.K. Rho signaling participates in membrane fluidity homeostasis. PLoS ONE 2012, 7, e45049. [Google Scholar] [CrossRef] [PubMed]
- Audhya, A.; Loewith, R.; Parsons, A.B.; Gao, L.; Tabuchi, M.; Zhou, H.; Boone, C.; Hall, M.N.; Emr, S.D. Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 2004, 23, 3747–3757. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Lai, Y.; Jiang, Y. The TOR complex 1 is a direct target of Rho1 GTPase. Mol. Cell 2012, 45, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Perez, P.; Rincón, S.A. Rho GTPases: Regulation of cell polarity and growth in yeasts. Biochem. J. 2010, 426, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Hall, M.N. Target of Rapamycin (TOR) in nutrient signaling and growth control. Genetics 2011, 189, 1177–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacinto, E.; Lorberg, A. TOR regulation of AGC kinases in yeast and mammals. Biochem. J. 2008, 410, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Kuranda, K.; Leberre, V.; Sokol, S.; Palamarczyk, G.; François, J. Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol. Microbiol. 2006, 61, 1147–1166. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, J.S.; Kuruvilla, F.G.; Tong, J.K.; Shamji, A.F.; Schreiber, S.L. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 14866–14870. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, M.C.; Heitman, J. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J. Biol. Chem. 1995, 270, 27531–27537. [Google Scholar] [PubMed]
- Kunz, J.; Loeschmann, A.; Deuter-Reinhard, M.; Hall, M.N. FAP1, a homologue of human transcription factor NF-X1, competes with rapamycin for binding to FKBP12 in yeast. Mol. Microbiol. 2000, 37, 1480–9143. [Google Scholar] [CrossRef] [PubMed]
- Simamora, P.; Alvarez, J.M.; Yalkowsky, S.H. Solubilization of rapamycin. Int. J. Pharm. 2001, 213, 25–29. [Google Scholar] [CrossRef]
- Cameron, A.M.; Nucifora, F.C., Jr.; Fung, E.T.; Livingston, D.J.; Aldape, R.A.; Ross, C.A.; Snyder, S.H. FKBP12 binds the inositol 1,4,5-trisphosphate receptor at leucine-proline (1400–1401) and anchors calcineurin to this FK506-like domain. J. Biol. Chem. 1997, 272, 27582–27588. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, H.; Saltsman, K.; Gasch, A.P.; Li, H.X.; Ogawa, N.; Botstein, D.; Brown, P.O.; Cyert, M.S. Genome-wide analysis of gene expression regulated by the Calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 31079–31088. [Google Scholar] [CrossRef] [PubMed]
- Odorizzi, G.; Babst, M.; Emr, S.D. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biol. Sci. 2000, 25, 229–235. [Google Scholar] [CrossRef]
- Staschke, K.A.; Dey, S.; Zaborske, J.M.; Palam, L.R.; McClintick, J.N.; Pan, T.; Edenberg, H.J.; Wek, R.C. Integration of general amino acid control and target of rapamycin (TOR) regulatory pathways in nitrogen assimilation in yeast. J. Biol. Chem. 2010, 285, 16893–16911. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, L.; Aranda, C.; González, A. TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J. Bacteriol. 2001, 183, 2331–2334. [Google Scholar] [CrossRef] [PubMed]
- Schuldiner, M.; Collins, S.R.; Thompson, N.J.; Denic, V.; Bhamidipati, A.; Punna, T.; Ihmels, J.; Andrews, B.; Boone, C.; Greenblatt, J.F.; et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 2005, 123, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Burston, H.E.; Maldonado-Báez, L.; Davey, M.; Montpetit, B.; Schluter, C.; Wendland, B.; Conibear, E. Regulators of yeast endocytosis identified by systematic quantitative analysis. J. Cell Biol. 2009, 185, 1097–1110. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.G. Spontaneous cell polarization: Feedback control of Cdc42 GTPase breaks cellular symmetry. Bioessays 2015, 37, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Raitt, D.C.; Posas, F.; Saito, H. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 2000, 19, 4623–4631. [Google Scholar] [CrossRef] [PubMed]
- Styles, E.; Youn, J.Y.; Mattiazzi Usaj, M.; Andrews, B. Functional genomics in the study of yeast cell polarity: Moving in the right direction. Philos. Trans. R. Soc. B 2016, 368, 20130118. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, S. An integrated view on a eukaryotic osmoregulation system. Curr. Genet. 2015, 61, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Gordon, M.D.; Nusse, R. Wnt signaling: Multiple pathways, multiple receptors, and multiple transcription factors. J. Biol. Chem. 2006, 281, 22429–22433. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, A.; Nebreda, A.R. Mechanisms and functions of p38 MAPK signalling. Biochem. J. 2010, 429, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, J.D.; Bibbs, L.; Ulevitch, R.J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 1994, 265, 808–811. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; McLaughlin, M.M.; McDonnell, P.C.; Lee, J.C.; Livi, G.P.; Young, P.R. Human mitogen-activated protein kinase CSBP1, but not CSBP2, complements a hog1 deletion in yeast. J. Biol. Chem. 1995, 270, 29043–29046. [Google Scholar] [PubMed]
- Takekawa, M.; Posas, F.; Saito, H. A human homolog of the yeast Ssk2/Ssk22 MAP kinase kinase kinases, MTK1, mediates stress-induced activation of the p38 and JNK pathways. EMBO J. 1997, 16, 4973–4982. [Google Scholar] [CrossRef] [PubMed]
- Bikkavilli, R.K.; Feigin, M.E.; Malbon, C.C. p38 mitogen-activated protein kinase regulates canonical Wnt–β-catenin signaling by inactivation of GSK3β. J. Cell Sci. 2008, 121, 3598–3607. [Google Scholar] [CrossRef] [PubMed]
- Gallo, K.A.; Johnson, G.L. Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat. Rev. 2002, 3, 663–672. [Google Scholar] [CrossRef] [PubMed]
- The Wnt Homepage. Available online: http://web.stanford.edu/group/nusselab/cgi-bin/wnt/ (assessed on 8 August 2016).
- Tatebayashi, H.; Yamamoto, K.; Nagoya, M.; Takayama, T.; Nishimura, A.; Sakurai, M.; Momma, T.; Saito, H. Osmosensing and scaffolding functions of the oligomeric four-transmembrane domain osmosensor Sho1. Nat. Commun. 2015, 6, 6975. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, A.; Yamamoto, K.; Oyama, M.; Kozuka-Hata, H.; Saito, H.; Tatebayashi, K. Scaffold protein Ahk1, which associates with Hkr1, Sho1, Ste11, and Pbs2, inhibits cross talk signaling from the Hkr1 osmosensor to the Kss1 mitogen-activated protein kinase. Mol. Cell Biol. 2016, 36, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Peña, J.M.; García, R.; Nombela, C.; Arroyo, J. The high-osmolarity glycerol (HOG) and cell wall integrity (CWI) signalling pathways interplay: A yeast dialogue between MAPK routes. Yeast 2010, 27, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Winkler, A.; Arkind, C.; Mattison, C.P.; Burkholder, A.; Knoche, K.; Ota, I. Heat stress activates the yeast high-osmolarity glycerol mitogen-activated protein kinase pathway, and protein tyrosine phosphatases are essential under heat stress. Eukaryot. Cell 2002, 1, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Monge, R.; Real, E.; Wojda, I.; Bebelman, J.P.; Mager, W.H.; Siderius, M. Hyperosmotic stress response and regulation of cell wall integrity in Saccharomyces cerevisiae share common functional aspects. Mol. Microbiol. 2001, 41, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Bermejo, C.; Rodríguez, E.; García, R.; Rodríguez-Peña, J.M.; de la Concepción, M.L.R.; Rivas, C.; Arias, P.; Nombela, C.; Posas, F.; Arroyo, J. The sequential activation of the yeast HOG and SLT2 pathways is required for cell survival to cell wall stress. Mol. Biol. Cell 2008, 19, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Faria-Oliveira, F. First molecular and biochemical characterization of the extracelular matrix of Saccharomyces cerevisiae. Ph.D. Thesis, University of Minho, Braga, Portugal, September 2013. Available online: http://repositorium.sdum.uminho.pt/ (assessed on 28 October 2016). [Google Scholar]
- García, R.; Bermejo, C.; Grau, C.; Pérez, R.; Rodríguez-Peña, J.M.; Francois, J.; Nombela, C.; Arroyo, J. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 2004, 279, 15183–15195. [Google Scholar] [CrossRef] [PubMed]
- Bosson, R.; Jaquenoud, M.; Conzelmann, A. GUP1 of Saccharomyces cerevisiae encodes an O-acyltransferase involved in remodeling of the GPI anchor. Mol. Biol. Cell 2006, 17, 2736–2645. [Google Scholar] [CrossRef] [PubMed]
- Friant, S.; Lombardi, R.; Schmelze, T.; Hall, M.N.; Riezman, H. Sphingolipid base signalling via PKh kinases is required for endocytosis in yeast. EMBO J. 2001, 20, 6783–6792. [Google Scholar] [CrossRef] [PubMed]
- Roelants, F.M.; Torrance, P.D.; Bezman, N.; Thorner, J. Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell 2002, 13, 3005–3028. [Google Scholar] [CrossRef] [PubMed]
- Roelants, F.M.; Torrance, P.D.; Thorner, J. Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pck1 and Sch9. Nat. Rev. Microbiol. 2004, 150, 3289–3304. [Google Scholar]
- Friant, S.; Zanolari, B.; Riezman, H. Increased protein kinase or decreased PP2A activity bypasses sphigoid base requirement in endocytosis. EMBO J. 2000, 19, 2834–2844. [Google Scholar] [CrossRef] [PubMed]
- Dupré, S.; Haguenauer-Tsapis, R. Raft partitioning of the yeast uracil permease during trafficking along the endocytic pathway. Traffic 2003, 4, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Neubauer, H.A.; Pitson, S.M. Roles, regulation and inhibitors of sphingosine kinase 2. FEBS J. 2013, 280, 5317–5336. [Google Scholar] [CrossRef] [PubMed]
- Roelants, F.M.; Breslow, D.K.; Muir, A.; Weissman, J.S.; Thornera, J. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2011, 108, 19222–19227. [Google Scholar] [CrossRef] [PubMed]
- Muir, A.; Roelants, F.M.; Timmons, G.; Leskoske, K.L.; Thorner, J. Down-regulation of TORC2-Ypk1 signaling promotes MAPK-independent survival under hyperosmotic stress. eLife 2015, 4, e09336. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Reiter, W.; Dohnal, I.; Gregori, C.; Beese-Sims, S.; Kuchler, K.; Ammerer, G.; Levin, D.E. MAPK Hog1 closes the S. cerevisiae glycerol channel Fps1 by phosphorylating and displacing its positive regulators. Genes Dev. 2013, 27, 2590–2601. [Google Scholar] [CrossRef] [PubMed]
- Gomes, K.N.; Freitas, S.M.; Pais, T.M.; Fietto, J.L.; Totola, A.H.; Arantes, R.M.; Martins, A.; Lucas, C.; Schuller, D.; Casal, M.; et al. Deficiency of Pkc1 activity affects glycerol metabolism in Saccharomyces cerevisiae. FEMS Yeast Res. 2005, 5, 767–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Meer, G. Lipid traffic in animal cells. Annu. Rev. Cell Biol. 1989, 5, 247–275. [Google Scholar] [CrossRef] [PubMed]
- Bagnat, M.; Keranen, S.; Shevchenko, A.; Simons, K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 3254–3259. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, K.; Mouritsen, O.G.; Anderson, R.G.W. Lipid rafts: At a crossroad between cell biology and physics. Nat. Cell Biol. 2007, 9, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Mollinedo, F. Lipid raft involvement in yeast cell growth and death. Front. Oncol. 2012, 2, 140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spira, F.; Mueller, N.S.; Beck, G.; von Olshausen, P.; Beig, J.; Wedlich-Söldner, R. Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat. Cell Biol. 2012, 14, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Bagnat, M.; Simons, K. Lipid rafts in protein sorting and cell polarity in budding yeast Saccharomyces cerevisiae. Biol. Chem. 2002, 383, 1475–1480. [Google Scholar] [CrossRef] [PubMed]
- Bagnat, M.; Chang, A.; Simons, K. Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol. Biol. Cell 2001, 12, 4129–4138. [Google Scholar] [CrossRef] [PubMed]
- Malinska, K.; Malinsky, J.; Opekarova, M.; Tanner, W. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell 2003, 14, 4427–4436. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, Y.; Suzuki, N.; Kataoka, T. The effect of posttranslational modifications on the interaction of Ras2 with adenylyl cyclase. Science 1993, 259, 683–686. [Google Scholar] [CrossRef] [PubMed]
- Berchtold, D.; Walther, T. TORC2 Plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol. Biol. Cell 2009, 20, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Prinz, W.A. ATP-binding cassette (ABC) transporters mediate nonvesicular, raft-modulated sterol movement from the plasma membrane to the endoplasmic reticulum. J. Biol. Chem. 2004, 279, 45226–45234. [Google Scholar] [CrossRef] [PubMed]
- Odorizzi, G.; Cowles, C.R.; Emr, S.D. The AP-3 complex: A coat of many colours. Trends Cell Biol. 1998, 8, 282–288. [Google Scholar] [CrossRef]
- Bosson, R.; Guillas, I.; Vionnet, C.; Roubaty, C.; Conzelmann, A. Incorporation of ceramides into Saccharomyces cerevisiae glycosylphosphatidylinositol-anchored proteins can be monitored in vitro. Euk. Cell 2009, 8, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Watanabe, R.; Jaensch, N.; Romanova-Michaelides, M.; Satoh, T.; Kato, M.; Riezman, H.; Yamaguchi, Y.; Maeda, Y.; Kinoshita, T. Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI. J. Cell Biol. 2011, 194, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Sipos, G.; Reggiori, F.; Vionnet, C.; Conzelmann, A. Alternative lipid remodelling pathways for glycosylphosphatidylinositol membrane anchors in Saccharomyces cerevisiae. EMBO J. 1997, 16, 3494–3505. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Umemura, M.; Yoko-o, T.; Jigami, Y. PER1 is required for GPI-phospholipase A2 activity and involved in lipid remodeling of GPI-anchored proteins. Mol. Biol. Cell 2006, 17, 5253–5264. [Google Scholar] [CrossRef] [PubMed]
- Umemura, M.; Fujita, M.; Yoko-o, T.; Fukamizu, A.; Jigami, Y. Saccharomyces cerevisiae CWH43 is involved in the remodelling of the lipid moiety of GPI anchors to ceramides. Mol. Biol. Cell 2007, 18, 4304–4316. [Google Scholar] [CrossRef] [PubMed]
- Yoko-o, T.; Ichikawa, D.; Miyagishi, Y.; Kato, A.; Umemura, M.; Takase, K.; Ra, M.; Ikeda, K.; Taguchi, R.; Jigami, Y. Determination and physiological roles of the glycosylphosphatidylinositol lipid remodelling pathway in yeast. Mol. Microbiol. 2013, 88, 140–155. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Gutierrez, D.; Eisenberg, T.; Büttner, S.; Meisinger, C.; Kroemer, G.; Madeo, F. Apoptosis in yeast: Triggers, pathways, subroutines. Cell Death Differ. 2010, 17, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Palková, Z.; Váchová, L. Yeast cell differentiation: Lessons from pathogenic and non-pathogenic yeasts. Semin. Cell Dev. Biol. 2016, 57, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Laun, P.; Pichova, A.; Madeo, F.; Fuchs, J.; Ellinger, A.; Kohlwein, S.; Dawes, I.; Fröhlich, K.U.; Breitenbach, M. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol. Microbiol. 2001, 39, 1166–1173. [Google Scholar] [CrossRef] [PubMed]
- Fabrizio, P.; Pletcher, S.D.; Minois, N.; Vaupel, J.W.; Longo, V.D. Chronological aging-independent replicative life span regulation by Msn2/Msn4 and Sod2 in Saccharomyces cerevisiae. FEBS Lett. 2004, 557, 136–142. [Google Scholar] [CrossRef]
- Herker, E.; Jungwirth, H.; Lehmann, K.A.; Maldener, C.; Fröhlich, K.U.; Wissing, S.; Büttner, S.; Fehr, M.; Sigrist, S.; Madeo, F. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 2004, 164, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Skinner, C.; Castello, P.R.; Kato, M.; Easlon, E.; Xie, L.; Li, T.; Lu, S.P.; Wang, C.; Tsang, F.; et al. Identification of potential calorie restriction-mimicking yeast mutants with increased mitochondrial respiratory chain and nitric oxide levels. J. Aging Res. 2011, 2011, 673185. [Google Scholar] [CrossRef] [PubMed]
- Tulha, J.; Faria-Oliveira, F.; Costa, C.; Lucas, C.; Ferreira, C. Saccharomyces cerevisiae Gup1p is required for a programmed cell death process. BMC Microbiol. 2012, 12, 80. [Google Scholar] [CrossRef] [PubMed]
- Fabrizio, P.; Longo, V.D. Chronological aging-induced apoptosis in yeast. Biochim. Biophys. Acta 2008, 1783, 1280–1285. [Google Scholar] [CrossRef] [PubMed]
- Guaragnella, N.; Antonacci, L.; Passarella, S.; Marra, E.; Giannattasio, S. Achievements and perspectives in yeast acetic acid-induced programmed cell death pathways. Biochem. Soc. Trans. 2011, 39, 1538–1543. [Google Scholar] [CrossRef] [PubMed]
- Shintani, T.; Klionsky, D.J. Autophagy in health and disease: A double-edged sword. Science 2004, 306, 990–995. [Google Scholar] [CrossRef] [PubMed]
- Granek, J.A.; Magwene, P.M. Environmental and genetic determinants of colony morphology in yeast. PLoS Genet. 2010, 6, e1000823. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eggimann, P.; Pittet, D. Candida colonization index and subsequent infection in critically ill surgical patients: 20 years later. Intensive Care Med. 2014, 40, 1429–1448. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.C.; Joosten, L.A.; Kullberg, B.J.; Netea, M.G. Interplay between Candida albicans and the mammalian innate host defense. Infect. Immun. 2012, 80, 1304–1313. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Johnson, A.D. Candida albicans biofilms and human disease. Annu. Rev. Microbiol. 2015, 69, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Kayingo, G.; Martins, A.; Andrie, R.; Neves, L.; Lucas, C.; Wong, B. A permease encoded by STL1 is required for active glycerol uptake by Candida albicans. Microbiology 2009, 155, 1547–1557. [Google Scholar] [CrossRef] [PubMed]
- Armada, R. Human, mouse, fly and yeast GUP1 orthologues in Candida albicans. Master’s Thesis, University of Minho, Braga, Portugal, September 2011. Available online: http://repositorium.sdum.uminho.pt/ (assessed on 28 October 2016). [Google Scholar]
- Pemán, J.; Cantón, E.; Espinel-Ingroff, A. Antifungal drug resistance mechanisms. Expert Rev. Anti Infect. Ther. 2009, 7, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Sanglard, D.; Ischer, F.; Parkinson, T.; Falconer, D.; Bille, J. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agentes. Antimicrob. Agents Chemother. 2003, 47, 2040–2412. [Google Scholar] [CrossRef]
- Reynolds, T.B.; Fink, G.R. Bakers’ yeast, a model for fungal biofilm formation. Science 2001, 291, 878–881. [Google Scholar] [CrossRef] [PubMed]
- Bojsen, R.K.; Andersen, K.S.; Regenberg, B. Saccharomyces cerevisiae—A model to uncover molecular mechanisms for yeast biofilm biology. FEMS Immunol. Med. Microbiol. 2012, 65, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Faria-Oliveira, F.; Carvalho, J.; Belmiro, C.L.; Martinez-Gomariz, M.; Hernaez, M.L.; Pavão, M.; Gil, C.; Lucas, C.; Ferreira, C. Methodologies to generate, extract, purify and fractionate yeast ECM for analytical use in proteomics and glycomics. BMC Microbiol. 2014, 14, 244. [Google Scholar] [CrossRef] [PubMed]
- Alvaro, C.G.; O’Donnell, A.F.; Prosser, D.C.; Augustine, A.A.; Goldman, A.; Brodsky, J.L.; Cyert, M.S.; Wendland, B.; Thorner, J. Specific α-arrestins negatively regulate Saccharomyces cerevisiae pheromone response by down-modulating the G-protein-coupled receptor Ste2. Mol. Cell Biol. 2014, 34, 2660–2681. [Google Scholar] [CrossRef] [PubMed]
- Van Zeebroeck, G.; Kimpe, M.; Vandormael, P.; Thevelein, J.M. A split-ubiquitin two-hybrid screen for proteins physically interacting with the yeast amino acid transceptor Gap1 and ammonium transceptor Mep2. PLoS ONE 2011, 6, e24275. [Google Scholar] [CrossRef] [PubMed]
- Butcher, R.A.; Schreiber, S.L. A small molecule suppressor of FK506 that targets the mitochondria and modulates ionic balance in Saccharomyces cerevisiae. Chem. Biol. 2003, 10, 521–531. [Google Scholar] [CrossRef]
- Andrade, S.L.; Einsle, O. The Amt/Mep/Rh family of ammonium transport proteins. Mol. Membr. Biol. 2007, 24, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Marini, A.-M.; Soussi-Boudekou, S.; Vissers, S.; André, B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell Biol. 1997, 17, 4282–4293. [Google Scholar] [CrossRef] [PubMed]
- Boeckstaens, M.; André, B.; Marini, A.M. The yeast ammonium transport protein Mep2 and its postivie regulator, the Npr1 kinase, play an importante role in normal and pesudohyphal growth on various nitrogen media through retrieval of excreted ammonium. Mol. Microbiol. 2007, 64, 534–546. [Google Scholar] [CrossRef] [PubMed]
- Boeckstaens, M.; André, B.; Marini, A.M. Distinct transport mechanisms in yeast ammonium transport/sensor proteins of the Mep/Amt/Rh family and impact on filamentation. J. Biol. Chem. 2008, 283, 21362–21370. [Google Scholar] [CrossRef] [PubMed]
- Neuhäuser, B.; Dunkel, N.; Satheesh, S.V.; Morschhäuser, J. Role of the Npr1 kinase in ammonium transport and signaling by the ammonium permease Mep2 in Candida albicans. Euk. Cell 2011, 10, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Powers, R.W., III; Kaeberlein, M.; Caldwell, S.D.; Kennedy, B.K.; Fields, S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006, 20, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Thevelein, J.M.; Geladé, R.; Holsbeeks, I.; Lagatie, O.; Popova, Y.; Rolland, F.; Stolz, F.; van de Velde, S.; van Dijck, P.; Vandormael, P.; et al. Nutrient sensing systems for rapid activation of the protein kinase A pathway in yeast. Biochem. Soc. Trans. 2005, 33, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.P.; Lo, R.S.; Ben-Hur, A.; Desmarais, C.; Stagljar, I.; Noble, W.S.; Fields, S. Large-scale identification of yeast integral membrane protein interactions. Proc. Natl. Acad. Sci. USA 2005, 102, 12123–12128. [Google Scholar] [CrossRef] [PubMed]
- Ptacek, J.; Devgan, G.; Michaud, G.; Zhu, H.; Zhu, X.; Fasolo, J.; Guo, H.; Jona, G.; Breitkreutz, A.; Sopko, R.; et al. Global analysis of protein phosphorylation in yeast. Nature 2005, 438, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Irie, K.; Gerber, A.P. Distinct roles for Khd1p in the localization and expression of bud-localized mRNAs in yeast. RNA 2008, 14, 2333–2347. [Google Scholar] [CrossRef] [PubMed]
- Schlecht, U.; Miranda, M.; Suresh, S.; Davis, R.W.; St Onge, R.P. Multiplex assay for condition-dependent changes in protein-protein interactions. Proc. Natl. Acad. Sci. USA 2012, 109, 9213–9218. [Google Scholar] [CrossRef] [PubMed]
- Batisse, J.; Batisse, C.; Budd, A.; Boettcher, B.; Hurt, E. Purification of nuclear poly(A)-binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J. Biol. Chem. 2009, 284, 34911–34917. [Google Scholar] [CrossRef] [PubMed]
- Snider, J.; Hanif, A.; Lee, M.E.; Jin, K.; Yu, A.R.; Graham, C.; Chuk, M.; Damjanovic, D.; Wierzbicka, M.; Tang, P.; et al. Mapping the functional yeast ABC transporter interactome. Nat. Chem. Biol. 2013, 9, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.J.; He, F.; Spatrick, P.; Li, C.; Jacobson, A. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc. Natl. Acad. Sci. USA 2007, 104, 20872–20877. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.C.; Magee, A.I. Acyltransferases for secreted signalling proteins. Mol. Membr. Biol. 2009, 26, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Masumoto, N.; Lanyon-Hogg, T.; Rodgers, U.R.; Konitsiotis, A.D.; Magee, A.I.; Tate, E.W. Membrane bound O-acyltransferases and their inhibitors. Biochem. Soc. Trans. 2015, 43, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Matevossian, A.; Resh, M.D. Membrane topology of hedgehog acyltransferase. J. Biol. Chem. 2014, 290, 2235–2243. [Google Scholar] [CrossRef] [PubMed]
- Cases, S.; Smith, S.J.; Zheng, Y.W.; Myers, H.M.; Lear, S.R.; Sande, E.; Novak, S.; Collins, C.; Welch, C.B.; Lusis, A.J.; et al. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA 1998, 95, 13018–13023. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Lee, C.Y.; Chang, E.T.; Cruz, J.C.; Levesque, M.C.; Chang, T.Y. Recombinant acyl-CoA:cholesterol acyltransferase-1 (ACAT-1) purified to essential homogeneity utilizes cholesterol in mixed micelles or in vesicles in a highly cooperative manner. J. Biol. Chem. 1998, 273, 35132–35141. [Google Scholar] [CrossRef] [PubMed]
- Stahl, U.; Stalberg, K.; Stymne, S.; Ronne, H. A family of eukaryotic lysophospholipid acyltransferases with broad specificity. FEBS Lett. 2008, 582, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.L.; Monetti, M.; Burri, B.J.; Farese, R.V., Jr. The triacylglycerol synthesis enzyme DGAT1 also catalyzes the synthesis of diacylglycerols, waxes, and retinyl esters. J. Lipid Res. 2005, 46, 1502–1511. [Google Scholar] [CrossRef] [PubMed]
- Echelard, Y.; Epstein, D.J.; St-Jacques, B.; Shen, L.; Mohler, J.; McMahon, J.A.; McMahon, A.P. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 1993, 75, 1417–1430. [Google Scholar] [CrossRef]
- Ma, C.; Zhou, Y.; Beachy, P.A.; Moses, K. The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell 1993, 75, 927–938. [Google Scholar] [CrossRef]
- Heemskerk, J.; DiNardo, S. Drosophila hedgehog acts as a morphogen in cellular patterning. Cell 1994, 76, 449–460. [Google Scholar] [CrossRef]
- Ericson, J.; Muhr, J.; Placzek, M.; Lints, T.; Jessell, T.M.; Edlund, T. Sonic hedgehog induces the differentiation of ventral forebrain neurons: A common signal for ventral patterning within the neural tube. Cell 1995, 81, 747–756. [Google Scholar] [CrossRef]
- Nybakken, K.; Perrimon, N. Hedgehog signal transduction: Recent findings. Curr. Opin. Genet. Dev. 2002, 12, 503–511. [Google Scholar] [CrossRef]
- Gritli-Linde, A.; Lewis, P.; McMahon, A.P.; Linde, A. The whereabouts of a morphogen: Direct evidence for short- and graded long-range activity of hedgehog signaling peptides. Dev. Biol. 2001, 236, 364–386. [Google Scholar] [CrossRef] [PubMed]
- Stamataki, D.; Ulloa, F.; Tsoni, S.V.; Mynett, A.; Briscoe, J. A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes Dev. 2005, 19, 626–641. [Google Scholar] [CrossRef] [PubMed]
- Fuccillo, M.; Joyner, A.L.; Fishell, G. Morphogen to mitogen: The multiple roles of hedgehog signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006, 7, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Panaková, D.; Sprong, H.; Marois, E.; Thiele, C.; Eaton, S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature 2005, 435, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Callejo, A.; Culi, J.; Guerrero, I. Patched, the receptor of Hedgehog, is a lipoprotein receptor. Proc. Natl. Acad. Sci. USA 2008, 105, 912–917. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Goetz, J.A.; Suber, L.M.; Scott, W.J.; Schreiner, C.M.; Robbins, D.J. A freely diffusible form of Sonic hedgehog mediates long-range signalling. Nature 2001, 411, 716–720. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.H.; Li, Y.J.; Kawakami, T.; Xu, M.; Chuang, P.T. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 2004, 18, 641–659. [Google Scholar] [CrossRef] [PubMed]
- Goetz, J.A.; Singh, S.; Suber, L.M.; Kull, F.J.; Robbins, D.J. A highly conserved amino-terminal region of sonic hedgehog is required for the formation of its freely diffusible multimeric form. J. Biol. Chem. 2006, 281, 4087–4093. [Google Scholar] [CrossRef] [PubMed]
- Steinhauer, J.; Treisman, J.E. Lipid-modified morphogens: Functions of fats. Curr. Opin. Genet. Dev. 2009, 19, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Burke, R.; Nellen, D.; Bellotto, M.; Hafen, E.; Senti, K.A.; Dickson, B.J.; Basler, K. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell 1999, 99, 803–815. [Google Scholar] [CrossRef]
- Katanaev, V.L.; Solis, G.P.; Hausmann, G.; Buestorf, S.; Katanayeva, N.; Schrock, Y.; Stuermer, C.A.; Basler, K. Reggie-1/Flotillin-2 promotes secretion of the long-range signalling forms of Wingless and Hedgehog in Drosophila. EMBO J. 2008, 27, 509–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palm, W.; Swierczynska, M.M.; Kumari, V.; Ehrhart-Bornstein, M.; Bornstein, S.R.; Eaton, S. Secretion and signaling activities of lipoprotein-associated hedgehog and non-sterol-modified hedgehog in flies and mammals. PLoS Biol. 2013, 11, e1001505. [Google Scholar] [CrossRef] [PubMed]
- Liegeois, S.; Benedetto, A.; Garnier, J.M.; Schwab, Y.; Labouesse, M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J. Cell Biol. 2006, 173, 949–961. [Google Scholar] [CrossRef] [PubMed]
- Vyas, N.; Walvekar, A.; Tate, D.; Lakshmanan, V.; Bansal, D.; Lo Cicero, A.; Raposo, G.; Palakodeti, D.; Dhawan, J. Vertebrate Hedgehog is secreted on two types of extracellular vesicles with different signaling properties. Nat. Sci. Rep. 2014, 4, 7357. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.L.; Nakayasu, E.S.; Joffe, L.S.; Guimarães, A.J.; Sobreira, T.J.P.; Nosanchuk, J.D.; Cordero, R.J.B.; Frases, S.; Casadevall, A.; Almeida, I.C.; et al. Biogenesis of extracellular vesicles in yeast. Many questions and few answers. Commun. Integr. Biol. 2010, 3, 533–535. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.L.; Nakayasu, E.S.; Joffe, L.S.; Guimarães, A.J.; Sobreira, T.J.P.; Nosanchuk, J.D.; Cordero, R.J.B.; Frases, S.; Casadevall, A.; Almeida, I.C.; et al. Characterization of yeast extracellular vesicles: Evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS ONE 2010, 5, e11113. [Google Scholar] [CrossRef] [PubMed]
- Gil-Bona, A.; Llama-Palacios, A.; Parra, C.M.; Vivanco, F.; Nombela, C.; Monteoliva, L.; Gil, C. Proteomics unravels extracellular vesicles as carriers of classical cytoplasmic proteins in Candida albicans. J. Prot. Res. 2015, 14, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Gil-Bona, A.; Monteoliva, L.; Gil, C. Global proteomic profiling of the secretome of Candida albicans ecm33 cell wall mutant reveals the involvement of Ecm33 in Sap2 secretion. J. Prot. Res. 2015, 14, 4270–4281. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ding, Q.; Yen, C.J.; Xia, W.; Izzo, J.G.; Lang, J.Y.; Li, C.W.; Hsu, J.L.; Miller, S.A.; Wang, X.; et al. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer Cell 2012, 21, 374–387. [Google Scholar] [CrossRef] [PubMed]
- Stecca, B.; Mas, C.; Clement, V.; Zbinden, M.; Correa, R.; Piguet, V.; Beermann, F.; Ruiz, I.; Altaba, A. Melanomas require Hedgehog-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc. Natl. Acad. Sci. USA 2007, 104, 5895–5900. [Google Scholar] [CrossRef] [PubMed]
- Hemmings, B.A.; Restuccia, D.F. PI3K-PKB/AKT pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011189. [Google Scholar] [CrossRef] [PubMed]
- Seto, M.; Ohta, M.; Asaoka, Y.; Ikenoue, T.; Tada, M.; Miyabayashi, K.; Mohri, D.; Tanaka, Y.; Ijichi, H.; Tateishi, K.; et al. Regulation of the hedgehog signaling by the mitogen-activated protein kinase cascade in gastric cancer. Mol. Carcinog. 2009, 48, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Nolan-Stevaux, O.; Lau, J.; Truitt, M.L.; Chu, G.C.; Hebrok, M.; Fernández-Zapico, M.E.; Hanahan, D. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 2009, 23, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.D.; Kraus, P.; Gaiano, N.; Nery, S.; Kohtz, J.; Fishell, G.; Loomis, C.A.; Treisman, J.E. An acylatable residue of Hedgehog is differentially required in Drosophila and mouse limb development. Dev. Biol. 2001, 233, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Pepinsky, R.B.; Zeng, C.; Wen, D.; Rayhorn, P.; Baker, D.P.; Williams, K.P.; Bixler, S.A.; Ambrose, C.M.; Garber, E.A.; Miatkowski, K.; et al. Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem. 1998, 273, 14037–14045. [Google Scholar] [CrossRef] [PubMed]
- Callier, P.; Calvel, P.; Matevossian, A.; Makrythanasis, P.; Bernard, P.; Kurosaka, H.; Vannier, A.; Thauvin-Robinet, C.; Borel, C.; Mazaud-Guittot, S.; et al. Loss of function mutation in the palmitoyltransferase HHAT leads to syndromic 46,XY disorder of sex development by impeding Hedgehog protein palmitoylation and signaling. PLoS Genet. 2014, 10, e1004340. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.K.; Porter, J.A.; Young, K.E.; Beachy, P.A. Teratogen-mediated inhibition of target tissue response to Shh signaling. Science 1998, 280, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- Hahn, H.; Christiansen, J.; Wicking, C.; Zaphiropoulos, P.G.; Chidambaram, A.; Gerrard, B.; Vorechovsky, I.; Bale, A.E.; Toftgard, R.; Dean, M.; et al. A mammalian patched homolog is expressed in target tissues of sonic hedgehog and maps to a region associated with developmental abnormalities. J. Biol. Chem. 1996, 271, 12125–12128. [Google Scholar] [CrossRef] [PubMed]
- Incardona, J.P.; Roelink, H. The role of cholesterol in Shh signaling and teratogen-induced holoprosencephaly. Cell Mol. Life Sci. 2000, 57, 1709–1719. [Google Scholar] [CrossRef] [PubMed]
- The Human Protein Atlas. Available online: http://www.proteinatlas.org/ (assessed on 17 August 2016).
- Karhadkar, S.S.; Bova, G.S.; Abdallah, N.; Dhara, S.; Gardner, D.; Maitra, A.; Isaacs, J.T.; Berman, D.M.; Beachy, P.A. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature 2004, 431, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, G.; Dhara, S.; Fendrich, V.; Bedja, D.; Beaty, R.; Mullendore, M.; Karikari, C.; Alvarez, H.; Iacobuzio-Donahue, C.; Jimeno, A.; et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: A new paradigm for combination therapy in solid cancers. Cancer Res. 2007, 67, 2187–2196. [Google Scholar] [CrossRef] [PubMed]
- Yauch, R.L.; Gould, S.E.; Scales, S.J.; Tang, T.; Tian, H.; Ahn, C.P.; Marshall, D.; Fu, L.; Januario, T.; Kallop, D.; et al. A paracrine requirement for hedgehog signalling in cancer. Nature 2008, 455, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Callahan, C.A.; DuPree, K.J.; Darbonne, W.C.; Ahn, C.P.; Scales, S.J.; de Sauvage, F.J. Hedgehog signaling is restricted to the stromal compartment during pancreatic carcinogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 4254–4259. [Google Scholar] [CrossRef] [PubMed]
- Jacob, L.; Lum, L. Deconstructing the Hedgehog pathway in development and disease. Science 2007, 318, 66–68. [Google Scholar] [CrossRef] [PubMed]
- Konitsiotis, A.D.; Chang, S.C.; Jovanović, B.; Ciepla, P.; Masumoto, N.; Palmer, C.P.; Tate, E.W.; Couchman, J.R.; Magee, A.I. Attenuation of hedgehog acyltransferase-catalyzed Sonic Hedgehog palmitoylation causes reduced signaling, proliferation and invasiveness of human carcinoma cells. PLoS ONE 2014, 9, e89899. [Google Scholar] [CrossRef] [PubMed]
- Matevossian, A.; Resh, M.D. Hedgehog acyltransferase as a target in estrogen receptor positive, HER2 amplified, and tamoxifen resistant breast cancer cells. Mol. Cancer 2015, 14, 72. [Google Scholar] [CrossRef] [PubMed]
- Petrova, E.; Matevossian, A.; Resh, M.D. Hedgehog acyltransferase as a target in pancreatic ductal adenocarcinoma. Oncogene 2015, 34, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Q.; Tian, X.; Luo, Y.W.; Zhong, D.Q.; Huang, Z.M.; Zhang, X.B. Expression, clinical and pathological significance of KIAA1173 gene in skin squamous cell carcinoma. Zhonghua Yi Xue Za Zhi 2010, 90, 1243–1246. [Google Scholar] [PubMed]
- Berisha, S.Z.; Serre, D.; Schauer, P.; Kashyap, S.R.; Smith, J.D. Changes in whole blood gene expression in obese subjects with type 2 diabetes following bariatric surgery: A pilot study. PLoS ONE 2011, 6, e16729. [Google Scholar] [CrossRef] [PubMed]
- Al-Goblan, A.; Al-Alfi, M.; Khan, M.Z. Mechanism linking diabetes mellitus and obesity. Diab. Metabol. Synd. Obes. Targ. Ther. 2014, 7, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; MacLennan, N.K.; Horvath, S.; Liao, J.C.; Dipple, K.M. Glycerol kinase deficiency alters expression of genes involved in lipid metabolism, carbohydrate metabolism, and insulin signaling. Eur. J. Hum. Genet. 2007, 15, 646–657. [Google Scholar] [CrossRef] [PubMed]
- Maeda, N.; Funahashi, T.; Shimomura, I. Metabolic impact of adipose and hepatic glycerol channels aquaporin 7 and aquaporin 9. Nat. Clin. Pract. Endocrinol. Metab. 2008, 4, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. Available online: www.ebi.ac.uk/Tools/msa/clustalo/ (assessed on 8 July 2016). [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.A. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 1998, 52, 81–104. [Google Scholar] [CrossRef] [PubMed]
- Palková, Z.; Janderová, B.; Gabriel, J.; Zikánová, B.; Pospísek, M.; Forstová, J. Ammonia mediates communication between yeast colonies. Nature 1997, 390, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Palková, Z.; Forstová, J. Yeast colonies synchronise their growth and development. J. Cell Sci. 2000, 113, 1923–1928. [Google Scholar] [PubMed]

 ), glucosamine (✩) and ethanolamine-P (☐). Dark thick scrawls represent peptide chains. PI: phosphatidylinositol; IPC: inositol phosphoceramide. Types B, C and D were defined from bands obtained using TLC [136].
), glucosamine (✩) and ethanolamine-P (☐). Dark thick scrawls represent peptide chains. PI: phosphatidylinositol; IPC: inositol phosphoceramide. Types B, C and D were defined from bands obtained using TLC [136].
 ), glucosamine (✩) and ethanolamine-P (☐). Dark thick scrawls represent peptide chains. PI: phosphatidylinositol; IPC: inositol phosphoceramide. Types B, C and D were defined from bands obtained using TLC [136].
), glucosamine (✩) and ethanolamine-P (☐). Dark thick scrawls represent peptide chains. PI: phosphatidylinositol; IPC: inositol phosphoceramide. Types B, C and D were defined from bands obtained using TLC [136].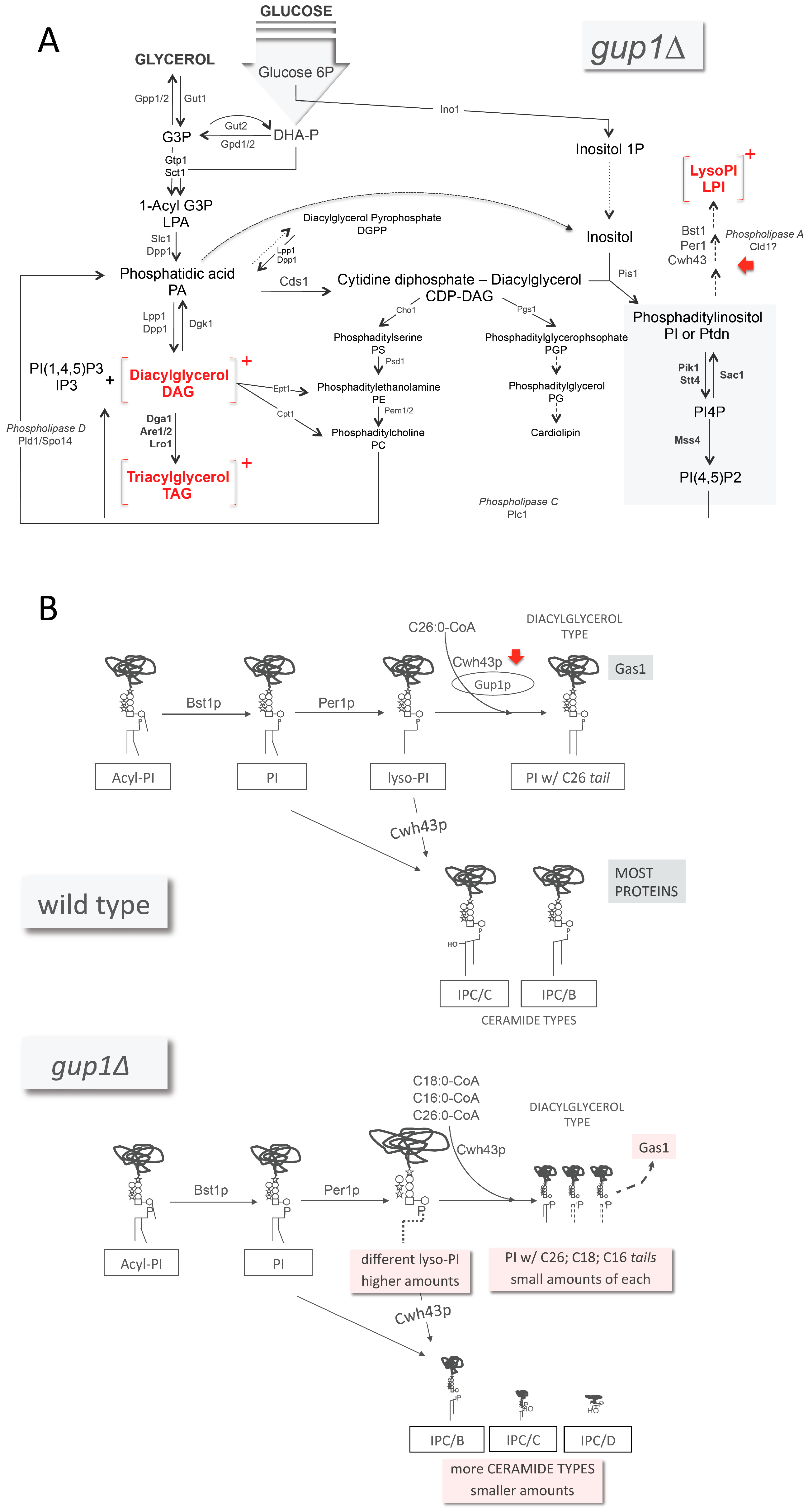
| Aliases | Organism | Key References | |
|---|---|---|---|
| Gup1 | Gup2 | Saccharomyces cerevisiae | [19,28] |
| Gup1 | - | Candida albicans | [29] |
| - | RASP | Drosophila melanogaster | [30] |
| Skinny Hedgehog | [20] | ||
| Sightless | [31] | ||
| Central Missing | [32] | ||
| HHATL | HHAT | Mus musculus | [22] |
| HHATL | HHAT | Homo sapiens | [33] |
| c3orf3 | [34] | ||
| KIAA117 | [35,36] | ||
| MBOAT3 * | [33] | ||
| MSTP002 # | [33] | ||
| OACT3 * | [33] | ||
| Predicted Function in GUP1 and GUP2 Transcription Regulation | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GUP1 | GUP2 | Identical Regulation | Opposite Regulation | ||||||||
| Activ. | Inhib. | Both | Activ. | Inhib. | Both | Activ. | Inhib. | Both | Activ. GUP1 + Inhib. GUP2 | Inhib. GUP1 + Activ. GUP2 | |
| Ash1 | Abf1 | Rap1 | Ecm22 | Sfp1 | Ace2 | Aft1 | Tec1 | Mig3 | Rap1 | Ash1 | none |
| Met4 | Cac2 | Spt23 | Gat4 | Snf2 | Ash1 | Cbf1 | |||||
| Tec1 | Cup9 | Yrm1 | Gcr2 | Snf6 | Bas1 | Cin5 | |||||
| Gal4 | Gis1 | Sok2 | Fhl1 | Plm2 | |||||||
| Hir3 | Gln3 | Spt10 | Gcn4 | Pho4 | |||||||
| Mig3 | Gzf3 | Spt2 | Gcr2 | Rap1 | |||||||
| Sfp1 | Hap5 | Ste12 | Mig3 | Rfx1 | |||||||
| Sum1 | Hda1 | Sut1 | Msn4 | Sko1 | |||||||
| Uls1 | Hms1 | Swi3 | Rif2 | Xbp1 | |||||||
| Isw2 | Taf14 | Set2 | Yap6 | ||||||||
| Mbp1 | Tec1 | Swi5 | Msn2 | ||||||||
| Mcm1 | Tup1 | Tbf1 | Swi4 | ||||||||
| Mga1 | Uls1 | ||||||||||
| Mig1 | Uc2 | ||||||||||
| Mot3 | Urc2 | ||||||||||
| Nrg1 | Xbp1 | ||||||||||
| Rgm1 | Yhp1 | ||||||||||
| Rox1 | Yox1 | ||||||||||
| YR015C | |||||||||||
| Partner | Assay | Type of Study | Key Reference | Assigned Function | |
|---|---|---|---|---|---|
| Gup1 | Fet3 | PCA | HTP | [174] | Iron-O2-oxidoreductase; multicopper oxidase that oxidizes ferrous (Fe2+) to ferric iron (Fe3+) for subsequent cellular uptake by transmembrane permease Ftr1. |
| Frk1 | BA | HTP | [175] | Protein kinase of unknown cellular role; interacts with rRNA transcription and ribosome biogenesis factors, and the long chain fatty acyl-CoA synthetase Faa3p. | |
| Hek2 | AC | HTP | [176] | RNA-binding protein involved in asymmetric localization of ASH1 mRNA; represses translation of ASH1 mRNA, an effect reversed by Yck1-dependent phosphorylation; regulates telomere position effect and length. | |
| Mep2 | 2H/Co-L | Single study | [165] | NH4+ permease; regulation of pseudohyphal growth; expression regulated by nitrogen catabolite repression (NCR). | |
| Msc7 | PCA | HTP | [177] | Cytoplasmic protein of unknown function. | |
| Nab2 | AC | HTP | [178] | Nuclear poly(A)-binding protein; required for nuclear mRNA export and tail length control. | |
| Pil1 | Co-IP | Single study | [45] | Eisosome core component; detected in phosphorylated state in mitochondria; phosphorylated upon Pkc1 hyperactivation in a Slt2p MAPK-dependent fashion. | |
| Por1 | Co-IP/Co-L | Single study | [45] | Mitochondrial voltage-dependent anion channel (VDAC); required for maintenance of mitochondrial osmotic stability and membrane permeability; couples the glutathione pools of the intermembrane space and the cytosol. | |
| Sat4 | BA | HTP | [175] | Ser/Thr protein kinase involved in salt tolerance; functions in regulation of Trk1-Trk2 potassium transporter. | |
| Vtc4 | PCA | HTP | [174] | Vacuolar membrane polyP polymerase; subunit of the vacuolar transporter chaperone (VTC) complex; regulates membrane trafficking; role in non-autophagic vacuolar fusion. | |
| YHL042W | PCA | HTP | [174] | Putative protein of unknown function. | |
| Gup2 | Aqy1 | PCA | HTP | [174] | Spore-specific aquaporin. |
| Aus1 | PCA | HTP | [179] | Plasma membrane sterol transporter of the ATP-binding cassette family; required, along with Pdr11, for uptake of exogenous sterols and their incorporation into the plasma membrane. | |
| Hsp30 | PCA | HTP | [174] | Negative regulator of the H(+)-ATPase Pma1; stress-responsive; induced by heat shock, ethanol, weak organic acid, glucose limitation, and entry into stationary phase. | |
| Ifa38 | PCA | HTP | [174] | Microsomal beta-keto-reductase; mutants exhibit reduced VLCFA synthesis, accumulate high levels of dihydrosphingosine, phytosphingosine and medium-chain ceramides. | |
| Nam7 | AC | HTP | [180] | ATP-dependent RNA helicase. | |
| Pdr10 | PCA | HTP | [179] | ATP-binding cassette (ABC) transporter; multidrug transporter involved in the pleiotropic drug resistance network; regulated by Pdr1p and Pdr3p. | |
| Pho88 | PCA | HTP | [174] | Probable membrane protein involved in phosphate transport; role in the maturation of secretory proteins. | |
| Sss1 | PCA | HTP | [174] | Subunit of the Sec61 translocation complex (Sec61-Sss1-Sbh1); this complex forms a channel for passage of secretory proteins through the ER membrane. | |
| Ste2 | PCA | HTP | [174] | Receptor for α-factor pheromone; interacts with both pheromone and a heterotrimeric G-protein to initiate the signaling response that leads to mating. |
| C. albicans SC5314 Gup1 | S. cerevisiae S288c Gup1 | S. cerevisiae S288c Gup2 | D. melanogaster Gup2/RASP | M. musculus Gup1/HHATL | H. sapiens Gup1/HHATL | M. musculus Gup2/HHAT | H. sapiens Gup2/HHAT | |
|---|---|---|---|---|---|---|---|---|
| C. albicans SC5314 Gup1 | 100.00 | 57.64 | 47.02 | 16.63 | 19.15 | 18.45 | 18.49 | 18.77 |
| S. cerevisiae S288c Gup1 | 57.64 | 100.00 | 56.61 | 17.84 | 20.70 | 20.45 | 19.30 | 19.80 |
| S. cerevisiae S288c Gup2 | 47.02 | 56.61 | 100.00 | 17.80 | 20.92 | 20.20 | 20.00 | 21.07 |
| D. melanogaster Gup2/RASP | 16.63 | 17.84 | 17.80 | 100.00 | 21.62 | 21.14 | 25.94 | 26.58 |
| M. musculus Gup1/HHATL | 19.15 | 20.70 | 20.92 | 21.62 | 100.00 | 90.07 | 28.16 | 27.92 |
| H. sapiens Gup1/HHATL | 18.45 | 20.45 | 20.20 | 21.14 | 90.07 | 100.00 | 28.33 | 27.86 |
| M. musculus Gup2/HHAT | 18.49 | 19.30 | 20.00 | 25.94 | 28.16 | 28.33 | 100.00 | 82.91 |
| H. sapiens Gup2/HHAT | 18.77 | 19.80 | 21.07 | 26.58 | 27.92 | 27.86 | 82.91 | 100.00 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lucas, C.; Ferreira, C.; Cazzanelli, G.; Franco-Duarte, R.; Tulha, J. Yeast Gup1(2) Proteins Are Homologues of the Hedgehog Morphogens Acyltransferases HHAT(L): Facts and Implications. J. Dev. Biol. 2016, 4, 33. https://doi.org/10.3390/jdb4040033
Lucas C, Ferreira C, Cazzanelli G, Franco-Duarte R, Tulha J. Yeast Gup1(2) Proteins Are Homologues of the Hedgehog Morphogens Acyltransferases HHAT(L): Facts and Implications. Journal of Developmental Biology. 2016; 4(4):33. https://doi.org/10.3390/jdb4040033
Chicago/Turabian StyleLucas, Cândida, Célia Ferreira, Giulia Cazzanelli, Ricardo Franco-Duarte, and Joana Tulha. 2016. "Yeast Gup1(2) Proteins Are Homologues of the Hedgehog Morphogens Acyltransferases HHAT(L): Facts and Implications" Journal of Developmental Biology 4, no. 4: 33. https://doi.org/10.3390/jdb4040033