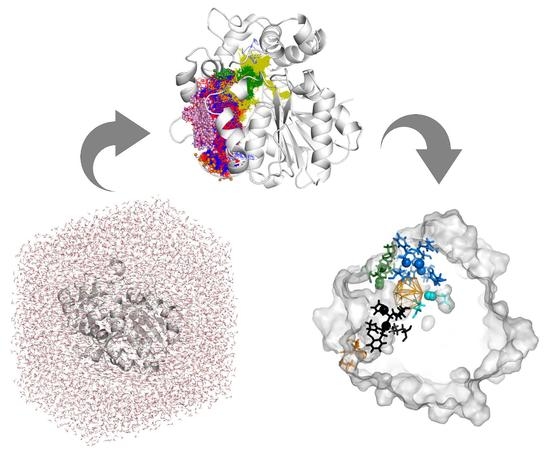
Exploring Solanum tuberosum Epoxide Hydrolase Internal Architecture by Water Molecules Tracking
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Preparation
2.2. Molecular Dynamics Simulations
2.3. Tunnel Identification
2.4. Water Tracking, Hot-Spot and Pocket Detection
2.5. Evolutionary Analysis
3. Results
3.1. Water Trajectories—Tunnels and Incidental Pathways
3.2. Cavities and Pockets
3.3. Trapped Water Molecules—Hot-Spot Identification
3.4. Evolutionary Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Biedermannová, L.; Prokop, Z.; Gora, A.; Chovancová, E.; Kovács, M.; Damborský, J.; Wade, R.C. A single mutation in a tunnel to the active site changes the mechanism and kinetics of product release in haloalkane dehalogenase LinB. J. Biol. Chem. 2012, 287, 29062–29074. [Google Scholar] [CrossRef] [PubMed]
- Kokkonen, P.; Bednar, D.; Dockalova, V.; Prokop, Z.; Damborsky, J. Conformational Changes Allow Processing of Bulky Substrates by a Haloalkane Dehalogenase with a Small and Buried Active Site. J. Biol. Chem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Prokop, Z.; Gora, A.; Brezovsky, J.; Chaloupkova, R.; Stepankova, V.; Damborsky, J. Engineering of Protein Tunnels: The Keyhole—Lock—Key Model for Catalysis by Enzymes with Buried Active Sites. Protein Eng. Handb. 2012, 3, 421–464. [Google Scholar]
- Kingsley, L.J.; Lill, M.A. Substrate tunnels in enzymes: Structure-function relationships and computational methodology. Proteins Struct. Funct. Bioinform. 2015, 83, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Marques, S.M.; Khirsariya, P.; Paruch, K.; Libichova, L.; Brezovsky, J.; Prokop, Z.; Chaloupkova, R.; Damborsky, J. Impact of the access tunnel engineering on catalysis is strictly ligand-specific. FEBS J. 2018, 285, 1456–1476. [Google Scholar] [CrossRef] [PubMed]
- Dyson, P.J.; Jessop, P.G. Solvent effects in catalysis: Rational improvements of catalysts: Via manipulation of solvent interactions. Catal. Sci. Technol. 2016, 6, 3302–3316. [Google Scholar] [CrossRef]
- Yang, L.; Dordick, J.S.; Garde, S. Hydration of enzyme in nonaqueous media is consistent with solvent dependence of its activity. Biophys. J. 2004, 87, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Stepankova, V.; Khabiri, M.; Brezovsky, J.; Pavelka, A.; Sykora, J.; Amaro, M.; Minofar, B.; Prokop, Z.; Hof, M.; Ettrich, R.; et al. Expansion of Access Tunnels and Active-Site Cavities Influence Activity of Haloalkane Dehalogenases in Organic Cosolvents. ChemBioChem 2013, 14, 890–897. [Google Scholar] [CrossRef] [PubMed]
- Persson, F.; Halle, B. Transient access to the protein interior: Simulation versus NMR. J. Am. Chem. Soc. 2013, 135, 8735–8748. [Google Scholar] [CrossRef] [PubMed]
- Gora, A.; Brezovsky, J.; Damborsky, J. Gates of enzymes. Chem. Rev. 2013, 113, 5871–5923. [Google Scholar] [CrossRef] [PubMed]
- Marques, S.; Brezovsky, J.; Damborsky, J. Role of Tunnels and Gates in Enzymatic Catalysis. Underst. Enzym. 2016, 421–463. [Google Scholar] [CrossRef]
- Raushel, F.M.; Thoden, J.B.; Holden, H.M. Enzymes with molecular tunnels. Acc. Chem. Res. 2003, 36, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Cojocaru, V.; Winn, P.J.; Wade, R.C. The Ins and Outs of Cytochrome P450s. Biochim. Biophys. Acta 2007, 1770, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Lund, L.; Shao, Q.; Gao, Y.Q.; Raushel, F.M. A combined theoretical and experimental study of the ammonia tunnel in carbamoyl phosphate synthetase. J. Am. Chem. Soc. 2009, 131, 10211–10219. [Google Scholar] [CrossRef] [PubMed]
- Magdziarz, T.; Mitusińska, K.; Gołdowska, S.; Płuciennik, A.; Stolarczyk, M.; Lugowska, M.; Góra, A. AQUA-DUCT: A ligands tracking tool. Bioinformatics 2017, 33, 2045–2046. [Google Scholar] [CrossRef] [PubMed]
- Elfström, L.T.; Widersten, M. Catalysis of potato epoxide hydrolase, StEH1. Biochem. J. 2005, 390, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Mowbray, S.L.; Elfström, L.T.; Ahlgren, K.M.; Andersson, C.E.; Widersten, M. X-ray structure of potato epoxide hydrolase sheds light on substrate specificity in plant enzymes. Protein Sci. 2006, 15, 1628–1637. [Google Scholar] [CrossRef] [PubMed]
- Thomaeus, A.; Carlsson, J.; Aqvist, J.; Widersten, M. Active site of epoxide hydrolases revisited: A noncanonical residue in potato StEH1 promotes both formation and breakdown of the alkylenzyme intermediate. Biochemistry 2007, 46, 2466–2479. [Google Scholar] [CrossRef] [PubMed]
- Thomaeus, A.; Naworyta, A.; Mowbray, S.L.; Widersten, M. Removal of distal protein-water hydrogen bonds in a plant epoxide hydrolase increases catalytic turnover but decreases thermostability. Protein Sci. 2008, 17, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T.; Bocola, M.; Wang, L.W.; Sanchis, J.; Cronin, A.; Arand, M.; Zou, J.; Archelas, A.; Bottalla, A.L.; Naworyta, A.; et al. Directed evolution of an enantioselective epoxide hydrolase: Uncovering the source of enantioselectivity at each evolutionary stage. J. Am. Chem. Soc. 2009, 131, 7334–7343. [Google Scholar] [CrossRef] [PubMed]
- Gurell, A.; Widersten, M. Modification of substrate specificity resulting in an epoxide hydrolase with shifted enantiopreference for (2,3-Epoxypropyl)benzene. ChemBioChem 2010, 11, 1422–1429. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, Å.J.; Bauer, P.; Ma, H.; Widersten, M. Obtaining optical purity for product diols in enzyme-catalyzed epoxide hydrolysis: Contributions from changes in both enantio- and regioselectivity. Biochemistry 2012, 51, 7627–7637. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, Å.J.; Bauer, P.; Dobritzsch, D.; Kamerlin, S.C.L.; Widersten, M. Epoxide hydrolysis as a model system for understanding flux through a branched reaction scheme. IUCrJ 2018, 5, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Hopmann, K.H.; Himo, F. Insights into the reaction mechanism of soluble epoxide hydrolase from theoretical active site mutants. J. Phys. Chem. B 2006, 110, 21299–21310. [Google Scholar] [CrossRef] [PubMed]
- Lonsdale, R.; Hoyle, S.; Grey, D.T.; Ridder, L.; Mulholland, A.J. Determinants of reactivity and selectivity in soluble epoxide hydrolase from quantum mechanics/molecular mechanics modeling. Biochemistry 2012, 51, 1774–1786. [Google Scholar] [CrossRef] [PubMed]
- Janfalk Carlsson, Å.; Bauer, P.; Dobritzsch, D.; Nilsson, M.; Kamerlin, S.C.L.; Widersten, M. Laboratory-Evolved Enzymes Provide Snapshots of the Development of Enantioconvergence in Enzyme-Catalyzed Epoxide Hydrolysis. ChemBioChem 2016, 17, 1693–1697. [Google Scholar] [CrossRef] [PubMed]
- Lind, M.E.S.; Himo, F. Quantum Chemical Modeling of Enantioconvergency in Soluble Epoxide Hydrolase. ACS Catal. 2016, 6, 8145–8155. [Google Scholar] [CrossRef]
- Lindberg, D.; Ahmad, S.; Widersten, M. Mutations in salt-bridging residues at the interface of the core and lid domains of epoxide hydrolase StEH1 affect regioselectivity, protein stability and hysteresis. Arch. Biochem. Biophys. 2010, 495, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Bauer, P.; Carlsson, Å.J.; Amrein, B.A.; Dobritzsch, D.; Widersten, M.; Kamerlin, S.C.L. Conformational diversity and enantioconvergence in potato epoxide hydrolase 1. Org. Biomol. Chem. 2016, 1–48. [Google Scholar] [CrossRef] [PubMed]
- Amrein, B.A.; Bauer, P.; Duarte, F.; Janfalk Carlsson, Å.; Naworyta, A.; Mowbray, S.L.; Widersten, M.; Kamerlin, S.C.L. Expanding the Catalytic Triad in Epoxide Hydrolases and Related Enzymes. ACS Catal. 2015, 5, 5702–5713. [Google Scholar] [CrossRef] [PubMed]
- Rose, P.W.; Prlić, A.; Altunkaya, A.; Bi, C.; Bradley, A.R.; Christie, C.H.; Di Costanzo, L.; Duarte, J.M.; Dutta, S.; Feng, Z.; et al. The RCSB protein data bank: Integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 2017, 45. [Google Scholar] [CrossRef]
- Anandakrishnan, R.; Aguilar, B.; Onufriev, A.V. H++ 3.0: Automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 2012, 40. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, A.; Luchko, T.; Gusarov, S.; Roe, D.R.; Simmerling, C.; Case, D.A.; Tuszynski, J. Three-dimensional molecular theory of solvation coupled with molecular dynamics in amber. J. Chem. Theory Comput. 2010, 6, 607–624. [Google Scholar] [CrossRef]
- Sindhikara, D.J.; Yoshida, N.; Hirata, F. Placevent: An algorithm for prediction of explicit solvent atom distribution-Application to HIV-1 protease and F-ATP synthase. J. Comput. Chem. 2012, 33, 1536–1543. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. Amber 14; University of San Francisco: San Francisco, CA, USA, 2014. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Chovancova, E.; Pavelka, A.; Benes, P.; Strnad, O.; Brezovsky, J.; Kozlikova, B.; Gora, A.; Sustr, V.; Klvana, M.; Medek, P.; et al. CAVER 3.0: A Tool for the Analysis of Transport Pathways in Dynamic Protein Structures. PLoS Comput. Biol. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Płuciennik, A.; Stolarczyk, M.; Bzówka, M.; Raczyńska, A.; Magdziarz, T.; Góra, A. BALCONY: An R package for MSA and functional compartments of protein variability analysis. BMC Bioinform. 2018. [Google Scholar] [CrossRef] [PubMed]
- Sander, C.; Schneider, R. Database of homology derived protein structures and the structural meaning of sequence alignment. Proteins Struct. Funct. Bioinform. 1991, 9, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Berger, V.W.; Zhou, Y. Kolmogorov–Smirnov Test: Overview. Wiley StatsRef Stat. Ref. Online 2014. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Xia, Y. Structural determinants of protein evolution are context-sensitive at the residue level. Mol. Biol. Evol. 2009, 26, 2387–2395. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, D.C.; Scherrer, M.P.; Zhou, T.; Wilke, C.O. The relationship between relative solvent accessibility and evolutionary rate in protein evolution. Genetics 2011, 188, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Horsman, G.P.; Liu, A.M.F.; Henke, E.; Bornscheuer, U.T.; Kazlauskas, R.J. Mutations in distant residues moderately increase the enantioselectivity of Pseudomonas fluorescens esterase towards methyl 3-bromo-2-methylpropanoate and ethyl 3-phenylbutyrate. Chem. Eur. J. 2003. [Google Scholar] [CrossRef] [PubMed]
- Tokuriki, N.; Tawfik, D.S. Stability effects of mutations and protein evolvability. Curr. Opin. Struct. Biol. 2009, 19, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Bednar, D.; Beerens, K.; Sebestova, E.; Bendl, J.; Khare, S.; Chaloupkova, R.; Prokop, Z.; Brezovsky, J.; Baker, D.; Damborsky, J. FireProt: Energy- and Evolution-Based Computational Design of Thermostable Multiple-Point Mutants. PLoS Comput. Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Kinoshita, M. Physicochemical origin of high correlation between thermal stability of a protein and its packing efficiency: A theoretical study for staphylococcal nuclease mutants. Biophys. Physicobiol. 2015, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Koudelakova, T.; Chaloupkova, R.; Brezovsky, J.; Prokop, Z.; Sebestova, E.; Hesseler, M.; Khabiri, M.; Plevaka, M.; Kulik, D.; Kuta Smatanova, I.; et al. Engineering enzyme stability and resistance to an organic cosolvent by modification of residues in the access tunnel. Angew. Chem. Int. Ed. 2013, 52, 1959–1963. [Google Scholar] [CrossRef] [PubMed]
- Wijma, H.J.; Floor, R.J.; Jekel, P.A.; Baker, D.; Marrink, S.J.; Janssen, D.B. Computationally designed libraries for rapid enzyme stabilization. Protein Eng. Des. Sel. 2014, 27, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Brezovsky, J.; Babkova, P.; Degtjarik, O.; Fortova, A.; Gora, A.; Iermak, I.; Rezacova, P.; Dvorak, P.; Kuta Smatanova, I.; Prokop, Z.; et al. Engineering a de Novo Transport Tunnel. ACS Catal. 2016, 6, 7597–7610. [Google Scholar] [CrossRef]
- Pavlova, M.; Klvana, M.; Prokop, Z.; Chaloupkova, R.; Banas, P.; Otyepka, M.; Wade, R.C.; Tsuda, M.; Nagata, Y.; Damborsky, J. Redesigning dehalogenase access tunnels as a strategy for degrading an anthropogenic substrate. Nat. Chem. Biol. 2009, 5, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Hendil-Forssell, P.; Martinelle, M.; Syré, P.-O. As featured in: Exploring water as building bricks in enzyme engineering. Chem. Commun. 2015, 51, 17221–17224. [Google Scholar] [CrossRef] [PubMed]
- Fink, M.J.; Syrén, P.O. Redesign of water networks for efficient biocatalysis. Curr. Opin. Chem. Biol. 2017, 37, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, K.; Góra, A.; Spruijt, R.; Mitusińska, K.; Suarez-Diez, M.; Martins dos Santos, V.; Schaap, P.J. Modulating D-amino acid oxidase (DAAO) substrate specificity through facilitated solvent access. PLoS ONE 2018, 13, e0198990. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.M.; Kang, K.; Sastry, M.; Sherman, W.; Sankaran, B.; Zwart, P.H.; Whitesides, G.M. Water-Restructuring Mutations Can Reverse the Thermodynamic Signature of Ligand Binding to Human Carbonic Anhydrase. Angew. Chem. Int. Ed. 2017. [Google Scholar] [CrossRef]
- Syrén, P.O.; Hammer, S.C.; Claasen, B.; Hauer, B. Entropy is key to the formation of pentacyclic terpenoids by enzyme-catalyzed polycyclization. Angew. Chem. Int. Ed. 2014. [Google Scholar] [CrossRef]
- Breiten, B.; Lockett, M.R.; Sherman, W.; Fujita, S.; Al-Sayah, M.; Lange, H.; Bowers, C.M.; Heroux, A.; Krilov, G.; Whitesides, G.M. Water networks contribute to enthalpy/entropy compensation in protein-ligand binding. J. Am. Chem. Soc. 2013. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.-D.; Yuan, S.; Li, L.; Chen, S.; Xu, J.-H.; Zhou, J. Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates. Proc. Natl. Acad. Sci. USA 2014, 111, 15717–15722. [Google Scholar] [CrossRef] [PubMed]







| Cavity I | Cavity II | Cavity III | |
|---|---|---|---|
| MD1 | I180, T182, Y183, R184, F189, D265*, L266, A299, H300*, F301 | H31, G32, F33, E35 *, S39, W40, H104 *, D105 *, G107, H300 *, F301 | F33, P34, F158, F168, V176, L177, I180, F191, F223, G227, F228, T229, G230, A231, V232 |
| MD2 | I180, T182, Y183, R184, F189, W210, L266, A299, H300*, F301, E305 | G32, F33, P34, E35*, H104*, D105*, H300*, F301 | F33, Y154*, F158, F168, I180, F189, F191, I200 |
| MD3 | I180, L181, T182, Y183, R184, D185, P186, A187, F189, L207, W210, F264, D265*, L266, A299, H300*, F301, E305 | - | F33, P34, Y154*, F158, F168, V176, K179, I180, F189, L197, I200, F228 |
| MD4 | I180, T182, Y183, R184, D185, F189, A203, L207, S208, W210, F264, D265*, L266, A299, H300*, F301, E305 | H31, G32, F33, E35*, S39, W40, H104*, D105*, L128, H300 *, F301, V302, S303 | F33, P34, Y154*, F158, F168, V176, L177, K179, I180, F189, F191, L197, I200, F228 |
| MD5 | K179, I180, L181, T182, Y183, R184, D185, P186, A187, A203, P204, L207, S208, W210, F264, D265 *, L266, A298, A299, H300 *, F301, V302, Q304, E305 | G32, F33, P34, E35*, H104*, D105*, H300*, F301, | F33, Y154*, F158, F168, V176, K179, I180, F189, F191, L197, I200, F228, V232 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitusińska, K.; Magdziarz, T.; Bzówka, M.; Stańczak, A.; Góra, A. Exploring Solanum tuberosum Epoxide Hydrolase Internal Architecture by Water Molecules Tracking. Biomolecules 2018, 8, 143. https://doi.org/10.3390/biom8040143
Mitusińska K, Magdziarz T, Bzówka M, Stańczak A, Góra A. Exploring Solanum tuberosum Epoxide Hydrolase Internal Architecture by Water Molecules Tracking. Biomolecules. 2018; 8(4):143. https://doi.org/10.3390/biom8040143
Chicago/Turabian StyleMitusińska, Karolina, Tomasz Magdziarz, Maria Bzówka, Agnieszka Stańczak, and Artur Góra. 2018. "Exploring Solanum tuberosum Epoxide Hydrolase Internal Architecture by Water Molecules Tracking" Biomolecules 8, no. 4: 143. https://doi.org/10.3390/biom8040143
APA StyleMitusińska, K., Magdziarz, T., Bzówka, M., Stańczak, A., & Góra, A. (2018). Exploring Solanum tuberosum Epoxide Hydrolase Internal Architecture by Water Molecules Tracking. Biomolecules, 8(4), 143. https://doi.org/10.3390/biom8040143