Ectopic Neo-Formed Intracellular Membranes in Escherichia coli: A Response to Membrane Protein-Induced Stress Involving Membrane Curvature and Domains
Abstract
:1. Introduction: Membrane Domains in Bacteria
2. The Bacterium Escherichia coli as an Unexpected Model for Addressing Molecular Mechanisms Underlying Membrane Morphology and Trafficking
3. Membrane Stress Induced by Endogenous or Heterologous Membrane Protein Overexpression in Escherichia coli
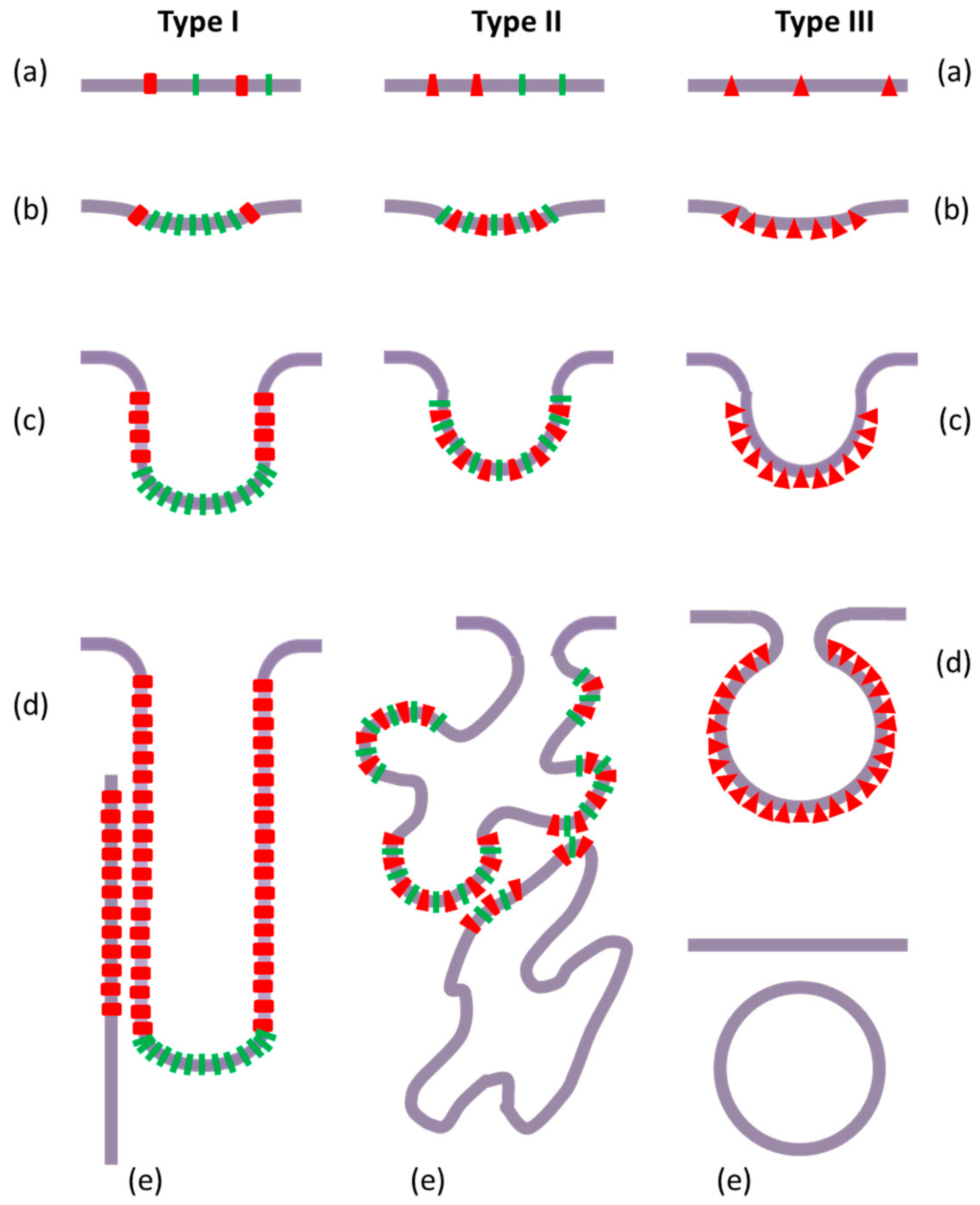
3.1. Intracellular Tubules Connected to the Cytoplasmic Membrane (“Type I” Intracellular Neo-Membranes)
3.2. Intracellular Saccules/Cisternae and Multilamellar Structures (“Type II” Intracellular Neo-Membranes)
3.3. Other Intracellular Membrane Structures (Poorly Defined)
3.4. Intracellular Vesicles (“Type III” Intracellular Neo-Membranes)
3.4.1. Viral Proteins
3.4.2. Lipid Metabolism Enzymes
3.4.3. Caveolins
3.5. Features of the Intracellular Bacterial Neo-Membranes
4. Other Cases of Membrane Stress in Different Escherichia coli Contexts
4.1. Tubule Formation Induced by Membrane Protein Mislocalization
4.2. Vesicles/Vacuoles Formation Induced by Bacterial Shape Alterations
4.2.1. Shape Mutants
4.2.2. L-Forms
5. Molecular Mechanisms
5.1. Local Protein and Lipid Heterogeneity
5.2. Local Membrane Curvature: Vesicles or Tubules
5.3. Membrane Structures More or Less Dispersed in Size, Shape, and Order
5.4. Membrane Fission or Continuity
6. Some Remaining Questions
6.1. Ectopic Neo-Formed Intracellular Membranes in Other Bacteria
6.2. Toxicity Mechanisms
6.3. Neo and Extra Lipid Synthesis
6.4. Protein–Protein Interactions
6.5. Stress-Induced Morphological Membrane Changes
Funding
Acknowledgments
Conflicts of Interest
References
- Carquin, M.; D’Auria, L.; Pollet, H.; Bongarzone, E.R.; Tyteca, D. Recent progress on lipid lateral heterogeneity in plasma membranes: From rafts to submicrometric domains. Prog. Lipid Res. 2016, 62, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaibelet, G.; Tercé, F.; Allart, S.; Lebrun, C.; Collet, X.; Jamin, N.; Orlowski, S. Fluorescent probes for detecting cholesterol-rich ordered membrane microdomains: Entangled relationships between structural analogies in the membrane and functional homologies in the cell. AIMS Biophys. 2017, 4, 121–151. [Google Scholar] [CrossRef]
- Levental, I.; Veatch, S.L. The continuing mystery of lipid rafts. J. Mol. Biol. 2016, 428, 4749–4764. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.M.; Magenau, A.; Williamson, D.; Gaus, K. The lipid raft hypothesis revisited–new insights on raft composition and function from super-resolution fluorescence microscopy. Bioessays 2012, 34, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Sevcsik, E.; Schutz, G.J. With or without rafts? Alternative views on cell membranes. Bioessays 2016, 38, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Bernardino de la Serna, J.; Schütz, G.J.; Eggeling, C.; Cebecauer, M. There is no simple model of the plasma membrane organization. Front. Cell Dev. Biol. 2016, 4, 106. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Parajo, M.F.; Cambi, A.; Torreno-Pina, J.A.; Thompson, N.; Jacobson, K. Nanoclustering as a dominant feature of plasma membrane organization. J. Cell Sci. 2014, 127, 4995–5005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honigmann, A.; Pralle, A. Compartmentalization of the cell membrane. J. Mol. Biol. 2016, 428, 4739–4748. [Google Scholar] [CrossRef] [PubMed]
- Sezgin, E.; Levental, I.; Mayor, S.; Eggeling, C. The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017, 18, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, N.C.; Masters, T.A.; Sheetz, M.P. Mechanical feedback between membrane tension and dynamics. Trends Cell Biol. 2012, 22, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Sinha, B.; Koster, D.; Ruez, R.; Gonnord, P.; Bastiani, M.; Abankwa, D.; Stan, R.V.; Butler-Browne, G.; Vedie, B.; Johannes, L.; et al. Cells respond to mechanical stress by rapid disassembly of caveolae. Cell 2011, 144, 402–413. [Google Scholar] [CrossRef] [PubMed]
- Verméglio, A.; Lavergne, J.; Rappaport, F. Connectivity of the intracytoplasmic membrane of Rhodobacter sphaeroides: A functional approach. Photosynth. Res. 2016, 127, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Furse, S.; Scott, D.J. Three-dimensional distribution of phospholipids in Gram negative bacteria. Biochemistry 2016, 55, 4742–4747. [Google Scholar] [CrossRef] [PubMed]
- Fishov, I.; Norris, V. Membrane heterogeneity created by transertion is a global regulator in bacteria. Curr. Opin. Microbiol. 2012, 15, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, S.; Nevo-Dinur, K.; Amster-Choder, O. Compartmentalization and spatiotemporal organization of macromolecules in bacteria. FEMS Microbiol. Rev. 2012, 36, 1005–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez, D. Molecular composition of functional microdomains in bacterial membranes. Chem. Phys. Lipids 2015, 192, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Hara, H.; Fishov, I.; Mileykovskaya, E.; Norris, V. The membrane: Transertion as an organizing principle in membrane heterogeneity. Front. Microbiol. 2015, 6, 572. [Google Scholar] [CrossRef] [PubMed]
- Nishibori, A.; Kusaka, J.; Hara, H.; Umeda, M.; Matsumoto, K. Phosphatidylethanolamine domains and localization of phospholipid synthases in Bacillus subtilis membranes. J. Bacteriol. 2005, 187, 2163–2174. [Google Scholar] [CrossRef] [PubMed]
- Renner, L.D.; Weibel, D.B. Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc. Natl. Acad. Sci. USA 2011, 108, 6264–6269. [Google Scholar] [CrossRef] [PubMed]
- Bramkamp, M.; Lopez, D. Exploring the existence of lipid rafts in bacteria. Microbiol. Mol. Biol. Rev. 2015, 79, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Kusaka, J.; Nishibori, A.; Hara, H. Lipid domains in bacterial membranes. Mol. Microbiol. 2006, 61, 1110–1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fishov, I.; Woldringh, C.L. Visualization of membrane domains in Escherichia coli. Mol. Microbiol. 1999, 32, 1166–1172. [Google Scholar] [CrossRef] [PubMed]
- Mileykovskaya, E.; Dowhan, W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 2000, 182, 1172–1175. [Google Scholar] [CrossRef] [PubMed]
- Vanounou, S.; Parola, A.H.; Fishov, I. Phosphatidylethanolamine and phosphatidylglycerol are segregated into different domains in bacterial membrane. A study with pyrene-labelled phospholipids. Mol. Microbiol. 2003, 49, 1067–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oswald, F.; Varadarajan, A.; Lill, H.; Peterman, E.J.G.; Bollen, Y.J.M. MreB-dependent organization of the E-coli cytoplasmic membrane controls membrane protein diffusion. Biophys. J. 2016, 110, 1139–1149. [Google Scholar] [CrossRef] [PubMed]
- Kawai, F.; Shoda, M.; Harashima, R.; Sadaie, Y.; Hara, H.; Matsumoto, K. Cardiolipin domains in Bacillus subtilis Marburg membranes. J. Bacteriol. 2004, 186, 1475–1483. [Google Scholar] [CrossRef] [PubMed]
- Mileykovskaya, E.; Dowhan, W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim. Biophys. Acta Biomembranes 2009, 1788, 2084–2091. [Google Scholar] [CrossRef] [PubMed]
- Oliver, P.M.; Crooks, J.A.; Leidl, M.; Yoon, E.J.; Saghatelian, A.; Weibel, D.B. Localization of anionic phospholipids in Escherichia coli cells. J. Bacteriol. 2014, 196, 3386–3398. [Google Scholar] [CrossRef] [PubMed]
- Saenz, J.P.; Grosser, D.; Bradley, A.S.; Lagny, T.J.; Lavrynenko, O.; Broda, M.; Simons, K. Hopanoids as functional analogues of cholesterol in bacterial membranes. Proc. Natl. Acad. Sci. USA 2015, 112, 11971–11976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saenz, J.P.; Sezgin, E.; Schwille, P.; Simons, K. Functional convergence of hopanoids and sterols in membrane ordering. Proc. Natl. Acad. Sci. USA 2012, 109, 14236–14240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzman-Flores, J.E.; Alvarez, A.F.; Poggio, S.; Gavilanes-Ruiz, M.; Georgellis, D. Isolation of detergent-resistant membranes (DRMs) from Escherichia coli. Anal. Biochem. 2017, 518, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Binenbaum, Z.; Parola, A.H.; Zaritsky, A.; Fishov, I. Transcription- and translation-dependent changes in membrane dynamics in bacteria: Testing the transertion model for domain formation. Mol. Microbiol. 1999, 32, 1173–1182. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, T.; Karczmarek, A.; Crouvoisier, M.; Bouhss, A.; Mengin-Lecreulx, D.; Den Blaauwen, T. The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol. Microbiol. 2007, 65, 1106–1121. [Google Scholar] [CrossRef] [PubMed]
- Magalon, A.; Alberge, F. Distribution and dynamics of OXPHOS complexes in the bacterial cytoplasmic membrane. Biochim. Biophys. Acta Bioenerg. 2016, 1857, 198–213. [Google Scholar] [CrossRef] [PubMed]
- Rassam, P.; Long, K.R.; Kaminska, R.; Williams, D.J.; Papadakos, G.; Baumann, C.G.; Kleanthous, C. Intermembrane crosstalk drives inner-membrane protein organization in Escherichia coli. Nat. Commun. 2018, 9, 1082. [Google Scholar] [CrossRef] [PubMed]
- Weis, R.M.; Hirai, T.; Chalah, A.; Kessel, M.; Peters, P.J.; Subramaniam, S. Electron microscopic analysis of membrane assemblies formed by the bacterial chemotaxis receptor Tsr. J. Bacteriol. 2003, 185, 3636–3643. [Google Scholar] [CrossRef] [PubMed]
- Zeev-Ben-Mordehai, T.; Vasishtan, D.; Siebert, C.A.; Whittle, C.; Grunewald, K. Extracellular vesicles: A platform for the structure determination of membrane proteins by cryo-EM. Structure 2014, 22, 1687–1692. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.A.; Sinnige, T.; Schellenberger, P.; de Keyzer, J.; Siebert, C.A.; Driessen, A.J.M.; Baldus, M.; Grunewald, K. Combined H-1-detected solid-state NMR spectroscopy and electron cryotomography to study membrane proteins across resolutions in native environments. Structure 2018, 26, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Ladizhansky, V. Applications of solid-state NMR to membrane proteins. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Weiner, J.H.; Lemire, B.D.; Elmes, M.L.; Bradley, R.D.; Scraba, D.G. Overproduction of fumarate reductase in Escherichia-coli induces a novel intracellular lipid-protein organelle. J. Bacteriol. 1984, 158, 590–596. [Google Scholar] [PubMed]
- Wilkison, W.O.; Walsh, J.P.; Corless, J.M.; Bell, R.M. Crystalline arrays of the Escherichia-coli sn-glycerol-3-phosphate acyltransferase, an integral membrane-protein. J. Biol. Chem. 1986, 261, 9951–9958. [Google Scholar] [PubMed]
- Voorhout, W.; Dekroon, T.; Leunissenbijvelt, J.; Verkleij, A.; Tommassen, J. Accumulation of LamB-LacZ hybrid proteins in intracytoplasmic membrane-like structures in Escherichia-coli-K12. J. Gen. Microbiol. 1988, 134, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Van Weeghel, R.P.; Keck, W.; Robillard, G.T. Regulated high-level expression of the mannitol permease of the phosphoenolpyruvate-dependent sugar phosphotransferase system in Escherichia-coli. Proc. Natl. Acad. Sci. USA 1990, 87, 2613–2617. [Google Scholar] [CrossRef] [PubMed]
- Lefman, J.; Zhang, P.J.; Hirai, T.; Weis, R.M.; Juliani, J.; Bliss, D.; Kessel, M.; Bos, E.; Peters, P.J.; Subramaniam, S. Three-dimensional electron microscopic imaging of membrane invaginations in Escherichia coli overproducing the chemotaxis receptor Tsr. J. Bacteriol. 2004, 186, 5052–5061. [Google Scholar] [CrossRef] [PubMed]
- Elmes, M.L.; Scraba, D.G.; Weiner, J.H. Isolation and characterization of the tubular organelles induced by fumarate reductase overproduction in Escherichia-coli. J. Gen. Microbiol. 1986, 132, 1429–1439. [Google Scholar] [CrossRef] [PubMed]
- Wilkison, W.O.; Bell, R.M.; Taylor, K.A.; Costello, M.J. Structural characterization of ordered arrays of sn-glycerol-3-phosphate acyltransferase from Escherichia-coli. J. Bacteriol. 1992, 174, 6608–6616. [Google Scholar] [CrossRef] [PubMed]
- Wilkison, W.O.; Bell, R.M. Sn-glycerol-3-phosphate acyltransferase tubule formation is dependent upon heat-shock proteins (htpR). J. Biol. Chem. 1988, 263, 14505–14510. [Google Scholar] [PubMed]
- Metzger, L.E.; Raetz, C.R.H. Purification and characterization of the lipid a disaccharide synthase (LpxB) from Escherichia coli, a peripheral membrane protein. Biochemistry 2009, 48, 11559–11571. [Google Scholar] [CrossRef] [PubMed]
- Von Meyenburg, K.; Jorgensen, B.B.; Vandeurs, B. Physiological and morphological effects of overproduction of membrane-bound ATP synthase in Escherichia-coli-K-12. EMBO J. 1984, 3, 1791–1797. [Google Scholar] [PubMed]
- Arechaga, I.; Miroux, B.; Karrasch, S.; Huijbregts, R.; de Kruijff, B.; Runswick, M.J.; Walker, J.E. Characterisation of new intracellular membranes in Escherichia coli accompanying large scale over-production of the b subunit of F1F0 ATP synthase. FEBS Lett. 2000, 482, 215–219. [Google Scholar] [CrossRef]
- Carranza, G.; Angius, F.; Ilioaia, O.; Solgadi, A.; Miroux, B.; Arechaga, I. Cardiolipin plays an essential role in the formation of intracellular membranes in Escherichia coli. Biochim. Biophys. Acta Biomembranes 2017, 1859, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
- Sobti, M.; Smits, C.; Wong, A.S.W.; Ishmukhametov, R.; Stock, D.; Sandin, S.; Stewart, A.G. Cryo-EM structures of the autoinhibited E. coli ATP synthase in three rotational states. eLife 2016, 5, e21598. [Google Scholar] [CrossRef] [PubMed]
- Montigny, C.; Penin, F.; Lethias, C.; Falson, P. Overcoming the toxicity of membrane peptide expression in bacteria by upstream insertion of Asp-Pro sequence. Biochim. Biophys. Acta Biomembr. 2004, 1660, 53–65. [Google Scholar] [CrossRef]
- Armour, G.A.; Brewer, G.J. Membrane morphogenesis from cloned fragments of bacteriophage PM2 DNA that contain the sp6.6 gene. FASEB J. 1990, 4, 1488–1493. [Google Scholar] [CrossRef] [PubMed]
- Weber, S.; Granzow, H.; Weiland, F.; Marquardt, O. Intracellular membrane proliferation in E-coli induced by foot-and-mouth disease virus 3A gene products. Virus Genes 1996, 12, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Nieboer, M.; Kingma, J.; Witholt, B. The alkane oxidation system of pseudomonas-oleovorans–induction of the alk genes in Escherichia-coli W3110(pgec47) affects membrane biogenesis and results in overexpression of alkane hydroxylase in a distinct cytoplasmic membrane subfraction. Mol. Microbiol. 1993, 8, 1039–1051. [Google Scholar] [CrossRef] [PubMed]
- Nieboer, M.; Vis, A.J.; Witholt, B. Overproduction of a foreign membrane protein in Escherichia coli stimulates and depends on phospholipid synthesis. Eur. J. Biochem. 1996, 241, 691–696. [Google Scholar] [CrossRef] [PubMed]
- Van der Brink-van der Laan, E.V.D.D.; Boots, J.W.P.; Spelbrink, R.E.J.; Kool, G.M.; Breukink, E.; Killian, J.A.; de Kruijff, B. Membrane interaction of the glycosyltransferase MurG: A special role for cardiolipin. J. Bacteriol. 2003, 185, 3773–3779. [Google Scholar] [CrossRef]
- Eriksson, H.M.; Wessman, P.; Ge, C.R.; Edwards, K.; Wieslander, A. Massive formation of intracellular membrane vesicles in Escherichia coli by a monotopic membrane-bound lipid glycosyltransferase. J. Biol. Chem. 2009, 284, 33904–33914. [Google Scholar] [CrossRef] [PubMed]
- Arioz, C.; Gotzke, H.; Lindholm, L.; Eriksson, J.; Edwards, K.; Daley, D.O.; Barth, A.; Wieslander, A. Heterologous overexpression of a monotopic glucosyltransferase (MGS) induces fatty acid remodeling in Escherichia coli membranes. Biochim. Biophys. Acta Biomembranes 2014, 1838, 1862–1870. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.R.; Gomez-Llobregat, J.; Skwark, M.J.; Ruysschaert, J.M.; Wieslander, A.; Linden, M. Membrane remodeling capacity of a vesicle-inducing glycosyltransferase. FEBS J. 2014, 281, 3667–3684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arioz, C.; Ye, W.H.; Bakali, A.; Ge, C.R.; Liebau, J.; Gotzke, H.; Barth, A.; Wieslander, A.; Maler, L. Anionic lipid binding to the foreign protein MGS provides a tight coupling between phospholipid synthesis and protein overexpression in Escherichia coli. Biochemistry 2013, 52, 5533–5544. [Google Scholar] [CrossRef] [PubMed]
- Walser, P.J.; Ariotti, N.; Howes, M.; Ferguson, C.; Webb, R.; Schwudke, D.; Leneva, N.; Cho, K.J.; Cooper, L.; Rae, J.; et al. Constitutive formation of caveolae in a bacterium. Cell 2012, 150, 752–763. [Google Scholar] [CrossRef] [PubMed]
- Ariotti, N.; Rae, J.; Leneva, N.; Ferguson, C.; Loo, D.; Okano, S.; Hill, M.M.; Walser, P.; Collins, B.M.; Parton, R.G. Molecular characterization of caveolin-induced membrane curvature. J. Biol. Chem. 2015, 290, 24875–24890. [Google Scholar] [CrossRef] [PubMed]
- Lamaze, C.; Tardif, N.; Dewulf, M.; Vassilopoulos, S.; Blouin, C.M. The caveolae dress code: Structure and signaling. Curr. Opin. Cell Biol. 2017, 47, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Parton, R.G.; del Pozo, M.A. Caveolae as plasma membrane sensors, protectors and organizers. Nat. Rev. Mol. Cell Biol. 2013, 14, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Herskovits, A.A.; Shimoni, E.; Minsky, A.; Bibi, E. Accumulation of endoplasmic membranes and novel membrane-bound ribosome-signal recognition particle receptor complexes in Escherichia coli. J. Cell Biol. 2002, 159, 403–410. [Google Scholar] [CrossRef] [PubMed]
- De Cock, H.; Meeldijk, J.; Overduin, P.; Verkleij, A.; Tommassen, J. Membrane biogenesis in Escherichia-coli–effects of a secA mutation. Biochim. Biophys. Acta 1989, 985, 313–319. [Google Scholar] [CrossRef]
- Bendezu, F.O.; de Boer, P.A.J. Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J. Bacteriol. 2008, 190, 1792–1811. [Google Scholar] [CrossRef] [PubMed]
- Onoda, T.; Enokizono, J.; Kaya, H.; Oshima, A.; Freestone, P.; Norris, V. Effects of calcium and calcium chelators on growth and morphology of Escherichia coli L-form NC-7. J. Bacteriol. 2000, 182, 1419–1422. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, T.; Hess, S.T.; Webb, W.W. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature 2003, 425, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Jimbo, T.; Sakuma, Y.; Urakami, N.; Ziherl, P.; Imai, M. Role of inverse-cone-shape lipids in temperature-controlled self-reproduction of binary vesicles. Biophys. J. 2016, 110, 1551–1562. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Flores, S. Inward multivesiculation at the basal membrane of adherent giant phospholipid vesicles. Biochim. Biophys. Acta Biomembranes 2016, 1858, 793–799. [Google Scholar] [CrossRef] [PubMed]
- Firshein, W.; Kim, P. Plasmid replication and partition in Escherichia coli: Is the cell membrane the key? Mol. Microbiol. 1997, 23, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.D.; Firshein, W. Isolation of an inner membrane-derived subfraction that supports in vitro replication of a mini-RK2 plasmid in Escherichia coli. J. Bacteriol. 2000, 182, 1757–1760. [Google Scholar] [CrossRef] [PubMed]
- Callan-Jones, A.; Sorre, B.; Bassereau, P. Curvature-driven lipid sorting in biomembranes. Cold Spring Harb. Perspect. Biol. 2011, 3, a004648. [Google Scholar] [CrossRef] [PubMed]
- Lipowsky, R. Domain-induced budding of fluid membranes. Biophys. J. 1993, 64, 1133–1138. [Google Scholar] [CrossRef] [Green Version]
- Sens, P.; Johannes, L.; Bassereau, P. Biophysical approaches to protein-induced membrane deformations in trafficking. Curr. Opin. Cell Biol. 2008, 20, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Jarsch, I.K.; Daste, F.; Gallop, J.L. Membrane curvature in cell biology: An integration of molecular mechanisms. J. Cell Biol. 2016, 214, 375–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johannes, L.; Parton, R.G.; Bassereau, P.; Mayor, S. Building endocytic pits without clathrin. Nat. Rev. Mol. Cell Biol. 2015, 16, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Kirchhausen, T. Bending membranes. Nat. Cell Biol. 2012, 14, 906–908. [Google Scholar] [CrossRef] [PubMed]
- Zimmerberg, J.; Kozlov, M.M. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 2006, 7, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Frost, A.; Perera, R.; Roux, A.; Spasov, K.; Destaing, O.; Egelman, E.H.; De Camilli, P.; Unger, V.M. Structural basis of membrane invagination by F-BAR domains. Cell 2008, 132, 807–817. [Google Scholar] [CrossRef] [PubMed]
- Busch, D.J.; Houser, J.R.; Hayden, C.C.; Sherman, M.B.; Lafer, E.M.; Stachowiak, J.C. Intrinsically disordered proteins drive membrane curvature. Nat. Commun. 2015, 6, 7875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.M.; Atefi, E.; Baumgart, T. Membrane shape instability induced by protein crowding. Biophys. J. 2016, 111, 1823–1826. [Google Scholar] [CrossRef] [PubMed]
- Stachowiak, J.C.; Hayden, C.C.; Sasaki, D.Y. Steric confinement of proteins on lipid membranes can drive curvature and tubulation. Proc. Natl. Acad. Sci. USA 2010, 107, 7781–7786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stachowiak, J.C.; Schmid, E.M.; Ryan, C.J.; Ann, H.S.; Sasaki, D.Y.; Sherman, M.B.; Geissler, P.L.; Fletcher, D.A.; Hayden, C.C. Membrane bending by protein-protein crowding. Nat. Cell Biol. 2012, 14, 944–949. [Google Scholar] [CrossRef] [PubMed]
- Kabbani, A.M.; Kelly, C.V. Nanoscale membrane budding induced by CTxB and detected via polarized localization microscopy. Biophys. J. 2017, 113, 1795–1806. [Google Scholar] [CrossRef] [PubMed]
- McMahon, H.T.; Boucrot, E. Membrane curvature at a glance. J. Cell Sci. 2015, 128, 1065–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johannes, L.; Pezeshkian, W.; Ipsen, J.H.; Shillcock, J.C. Clustering on membranes: Fluctuations and more. Trends Cell Biol. 2018, 28, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Peter, B.J.; Kent, H.M.; Mills, I.G.; Vallis, Y.; Butler, P.J.G.; Evans, P.R.; McMahon, H.T. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science 2004, 303, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Campelo, F.; McMahon, H.T.; Kozlov, M.M. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys. J. 2008, 95, 2325–2339. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, N.; Kumar, P.B.S.; Ipsen, J.H. Membrane-mediated aggregation of curvature-inducing nematogens and membrane tubulation. Biophys. J. 2013, 104, 1018–1028. [Google Scholar] [CrossRef] [PubMed]
- Sens, P.; Turner, M.S. Theoretical model for the formation of caveolae and similar membrane invaginations. Biophys. J. 2004, 86, 2049–2057. [Google Scholar] [CrossRef]
- Snead, W.T.; Hayden, C.C.; Gadok, A.K.; Zhao, C.; Lafer, E.M.; Rangamani, P.; Stachowiak, J.C. Membrane fission by protein crowding. Proc. Natl. Acad. Sci. USA 2017, 114, E3258–E3267. [Google Scholar] [CrossRef] [PubMed]
- Tian, A.; Baumgart, T. Sorting of lipids and proteins in membrane curvature gradients. Biophys. J. 2009, 96, 2676–2688. [Google Scholar] [CrossRef] [PubMed]
- Pinot, M.; Vanni, S.; Pagnotta, S.; Lacas-Gervais, S.; Payet, L.A.; Ferreira, T.; Gautier, R.; Goud, B.; Antonny, B.; Barelli, H. Polyunsaturated phospholipids facilitate membrane deformation and fission by endocytic proteins. Science 2014, 345, 693–697. [Google Scholar] [CrossRef] [PubMed]
- Gubellini, F.; Verdon, G.; Karpowich, N.K.; Luff, J.D.; Boel, G.; Gauthier, N.; Handelman, S.K.; Ades, S.E.; Hunt, J.F. Physiological response to membrane protein overexpression in E. coli. Mol. Cell. Proteom. 2011, 10, M111.007930. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Baars, L.; Ytterberg, A.J.; Klussmeier, A.; Wagner, C.S.; Nord, O.; Nygren, P.A.; van Wijk, K.J.; de Gier, J.W. Consequences of membrane protein overexpression in Escherichia coli. Mol. Cell. Proteom. 2007, 6, 1527–1550. [Google Scholar] [CrossRef] [PubMed]
- Bigay, J.; Casella, J.F.; Drin, G.; Mesmin, B.; Antonny, B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 2005, 24, 2244–2253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drin, G.; Casella, J.F.; Gautier, R.; Boehmer, T.; Schwartz, T.U.; Antonny, B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 2007, 14, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Renard, H.F.; Johannes, L.; Morsomme, P. Increasing diversity of biological membrane fission mechanisms. Trends Cell Biol. 2018, 28, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, M.M.; McMahon, H.T.; Chernomordik, L.V. Protein-driven membrane stresses in fusion and fission. Trends Biochem. Sci. 2010, 35, 699–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozlovsky, Y.; Kozlov, M.M. Membrane fission: Model for intermediate structures. Biophys. J. 2003, 85, 85–96. [Google Scholar] [CrossRef]
- Lenz, M.; Morlot, S.; Roux, A. Mechanical requirements for membrane fission: Common facts from various examples. FEBS Lett. 2009, 583, 3839–3846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobereiner, H.G.; Kas, J.; Noppl, D.; Sprenger, I.; Sackmann, E. Budding and fission of vesicles. Biophys. J. 1993, 65, 1396–1403. [Google Scholar] [CrossRef] [Green Version]
- Seigneurin-Berny, D.; King, M.S.; Sautron, E.; Moyet, L.; Catty, P.; André, F.; Rolland, N.; Kunji, E.R.; Frelet-Barrand, A. Membrane protein production in Lactococcus lactis for functional studies. Methods Mol. Biol. 2016, 1432, 79–101. [Google Scholar] [PubMed]
- Wagner, S.; Klepsch, M.M.; Schlegel, S.; Appel, A.; Draheim, R.; Tarry, M.; Hogbom, M.; van Wijk, K.J.; Slotboom, D.J.; Persson, J.O.; et al. Tuning Escherichia coli for membrane protein overexpression. Proc. Natl. Acad. Sci. USA 2008, 105, 14371–14376. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Song, J.M.; Sui, S.F.; Wang, D.N. DnaK and DnaJ facilitated the folding process and reduced inclusion body formation of magnesium transporter CorA overexpressed in Escherichia coli. Protein Expr. Purif. 2003, 32, 221–231. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Rock, C.O. Thematic review series: Glycerolipids-acyltransferases in bacterial glycerophospholipid synthesis. J. Lipid Res. 2008, 49, 1867–1874. [Google Scholar] [CrossRef] [PubMed]
- Kol, S.; Nouwen, N.; Driessen, A.J.M. Mechanisms of YidC-mediated insertion and assembly of multimeric membrane protein complexes. J. Biol. Chem. 2008, 283, 31269–31273. [Google Scholar] [CrossRef] [PubMed]
- Nagamori, S.; Smirnova, I.N.; Kaback, H.R. Role of YidC in folding of polytopic membrane proteins. J. Cell Biol. 2004, 165, 53–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoonens, M.; Miroux, B. Expression of membrane proteins at the Escherichia coli membrane for structural studies. Methods Mol. Biol. 2010, 601, 49–66. [Google Scholar] [PubMed]
- Shin, J.; Jung, Y.H.; Cho, D.H.; Park, M.; Lee, K.E.; Yang, Y.; Jeong, C.; Sung, B.H.; Sohn, J.H.; Park, J.B.; et al. Display of membrane proteins on the heterologous caveolae carved by caveolin-1 in the Escherichia coli cytoplasm. Enzyme Microb. Technol. 2015, 79–80, 55–62. [Google Scholar] [CrossRef] [PubMed]

| Type | Overexpressed Membrane Protein | References |
|---|---|---|
| I | Fumarate reductase complex (FrdABCD) | [40,45] |
| sn-Glycerol-3-phosphate acyl transferase (PlsB) | [41,46,47] | |
| LamB-LacZ fusion protein | [42] | |
| Mannitol permease (MtlA) | [43] | |
| Chemotaxis receptor (Tsr) | [44] | |
| Pseudo-phosphorylated mutant S80/Cav-1 1,a | [63,64] | |
| Truncated Cav-1(49–134) 1,b | [64] | |
| Caveolin-2 (Cav-2) 1 | [64] | |
| II | Lipid A disaccharide synthase (LpxB) | [48] |
| Lipid A disaccharide synthase (LpxB) 2 | [48] | |
| F0F1-ATP synthase | [49] | |
| F0F1-ATP synthase b subunit | [50,51] | |
| Truncated Cav-1(49–81/97–178) 1,c | [64] | |
| Nematode caveolin (Ce-Cav) 1 | [64] | |
| Caveolin-2 (Cav-2) 1 | [64] | |
| III | sp6.6 protein of PM2 bacteriophage | [54] |
| 3A protein of foot-and-mouth disease virus (FMDV) | [55] | |
| Alkane hydroxylase (AlkB) 3 | [56,57] | |
| Glycosyl transferase (MurG) | [58] | |
| Monoglycosyldiacylglycerol synthase (MGS) 4 | [59,60,61,62] | |
| Diglycosyldiacylglycerol synthase (DGS) 4 | [59] | |
| Caveolin-1 (Cav-1) 1 | [63,64] | |
| Truncated Cav-1(81–147) 1,d | [64] | |
| Caveolin-2 (Cav-2) 1 | [64] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jamin, N.; Garrigos, M.; Jaxel, C.; Frelet-Barrand, A.; Orlowski, S. Ectopic Neo-Formed Intracellular Membranes in Escherichia coli: A Response to Membrane Protein-Induced Stress Involving Membrane Curvature and Domains. Biomolecules 2018, 8, 88. https://doi.org/10.3390/biom8030088
Jamin N, Garrigos M, Jaxel C, Frelet-Barrand A, Orlowski S. Ectopic Neo-Formed Intracellular Membranes in Escherichia coli: A Response to Membrane Protein-Induced Stress Involving Membrane Curvature and Domains. Biomolecules. 2018; 8(3):88. https://doi.org/10.3390/biom8030088
Chicago/Turabian StyleJamin, Nadège, Manuel Garrigos, Christine Jaxel, Annie Frelet-Barrand, and Stéphane Orlowski. 2018. "Ectopic Neo-Formed Intracellular Membranes in Escherichia coli: A Response to Membrane Protein-Induced Stress Involving Membrane Curvature and Domains" Biomolecules 8, no. 3: 88. https://doi.org/10.3390/biom8030088





