In Quest for Improved Drugs against Diabetes: The Added Value of X-ray Powder Diffraction Methods
Abstract
:1. Introduction
2. Overview of Insulin Crystalline Structures
2.1. First Human Insulin XRPD Studies
2.2. Characterization of Distinct Insulin Formulations Via XRPD
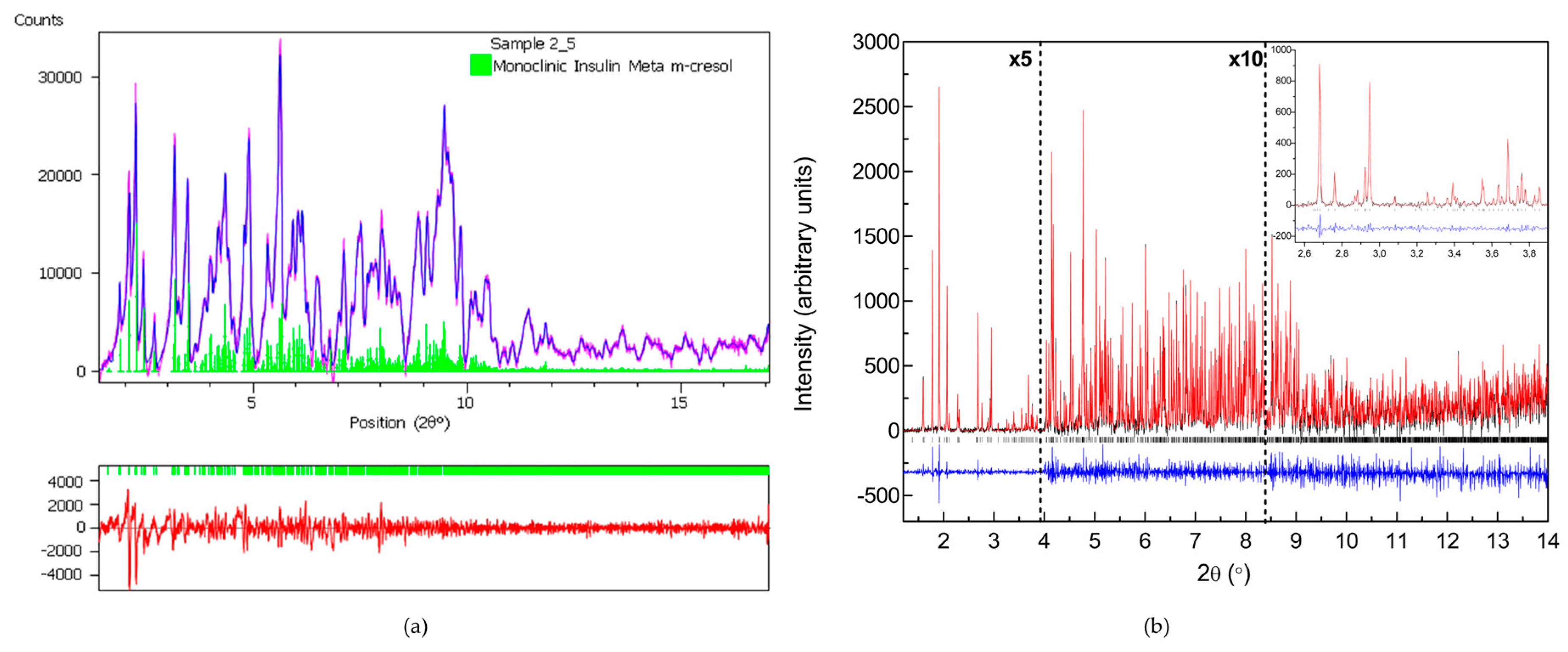
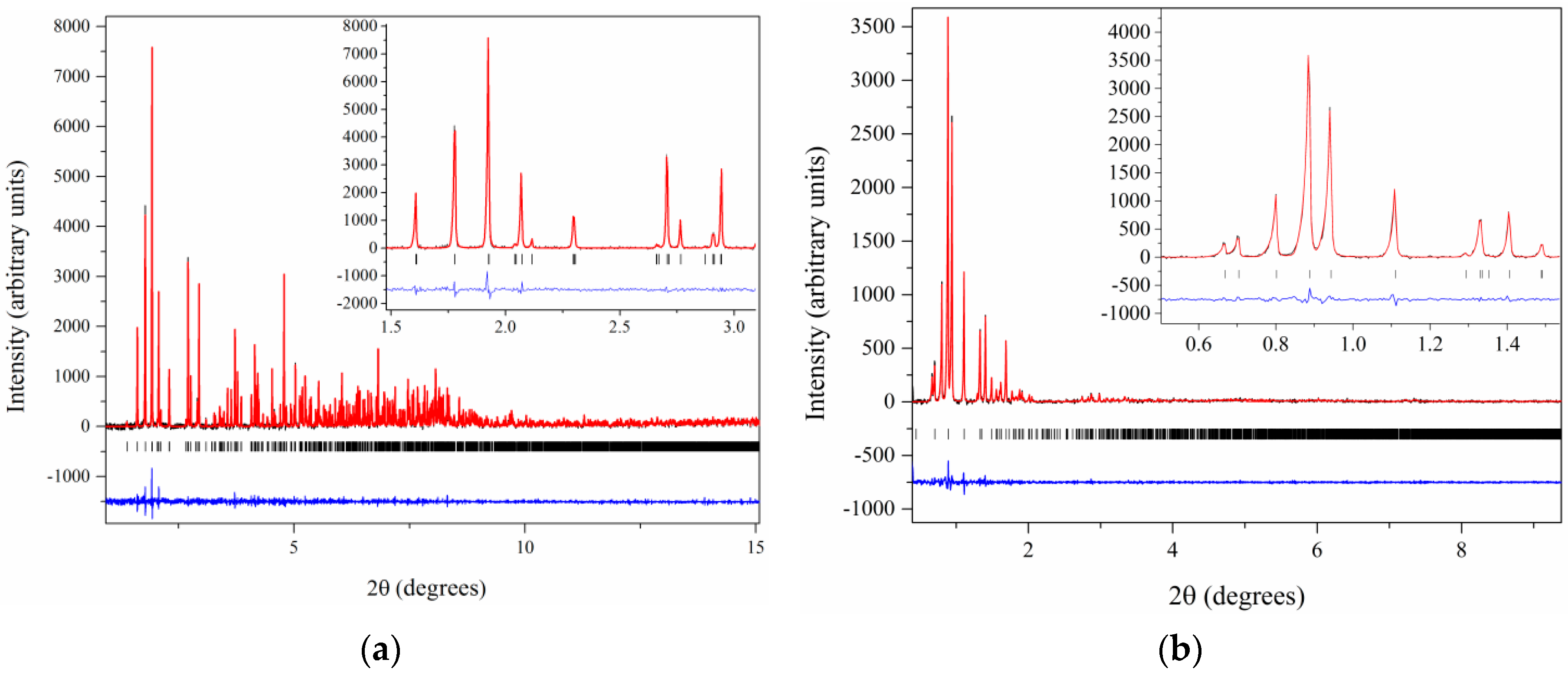
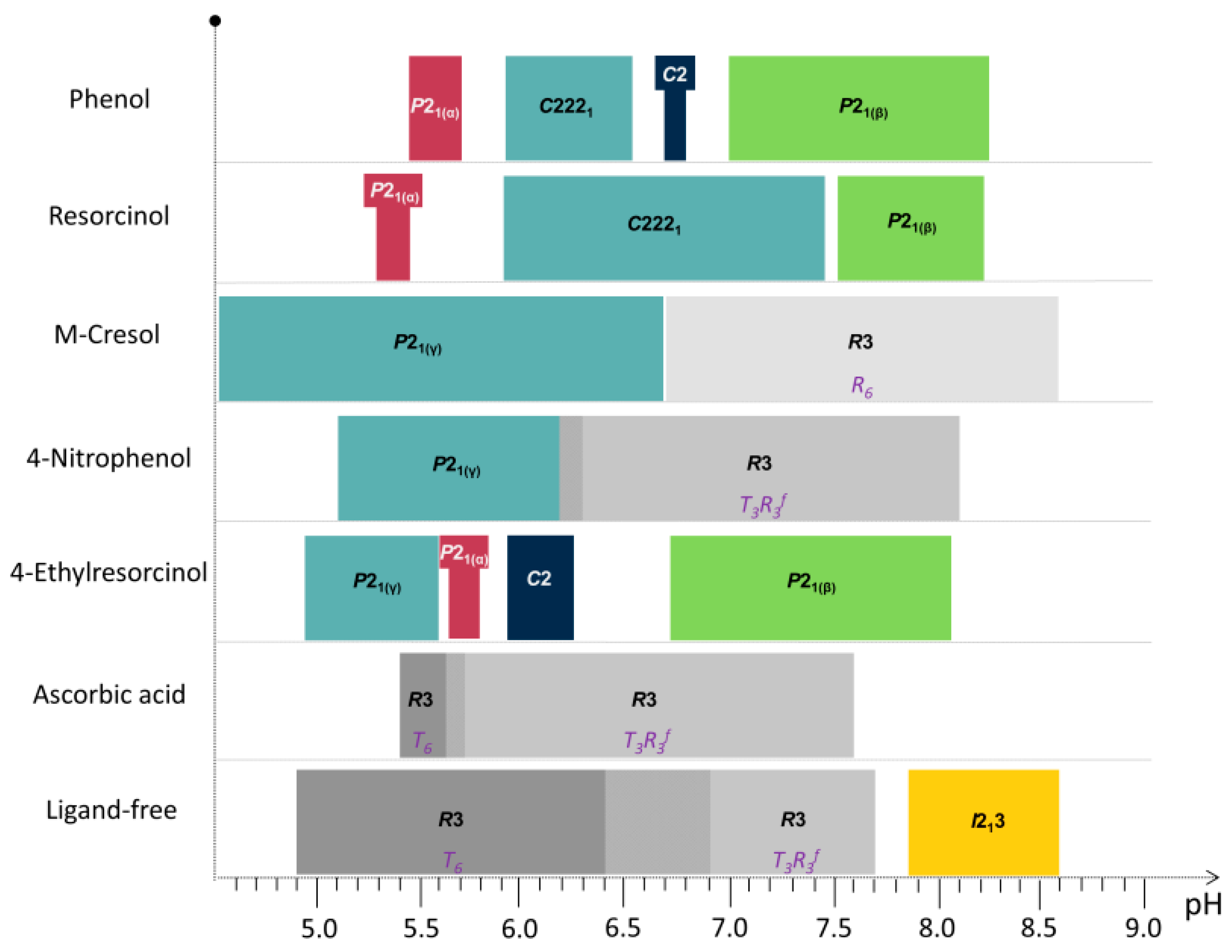
2.3. Cocrystallization of HI with Phenolic Derivatives and pH Dependence
2.4. Cocrystallization of HI with a Non-Phenolic Derivative and pH Dependence
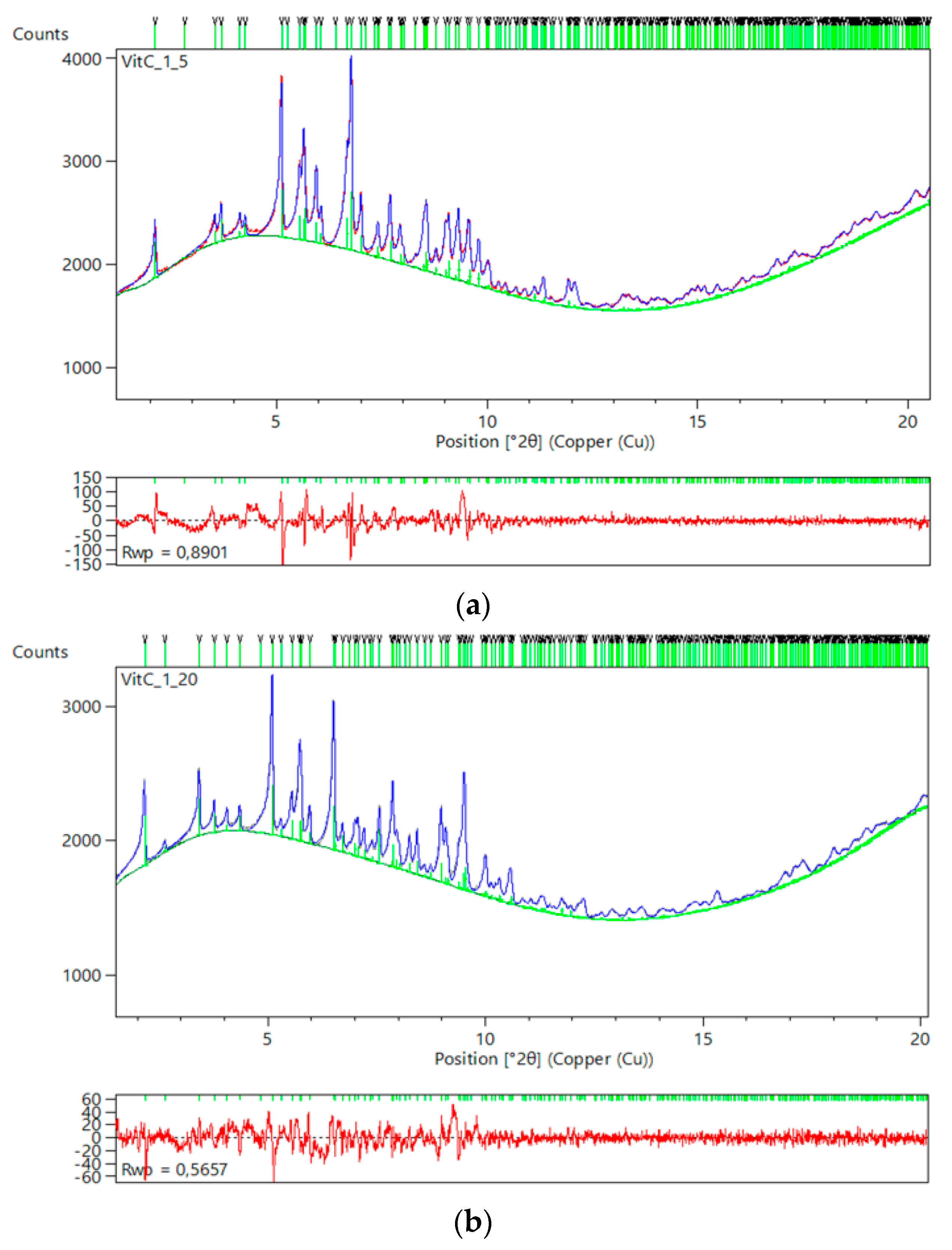
2.5. Ligand-Free Crystalline HI Studies and pH Dependence
3. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ripoll, B.C.; Leutholtz, I. Exercise and Disease Management, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2011; p. 25. Available online: http://www.crcnetbase.com/doi/10.1201/b10856-5 (accessed on 18 August 2017).
- Banting, F.G.; Best, C.H. The internal secretion of the pancreas. J. Lab. Clin. Med. 1922, 7, 251–266. [Google Scholar]
- Burn, P. Type 1 diabetes. Nat. Rev. Drug Discov. 2010, 9, 187–188. [Google Scholar] [CrossRef] [PubMed]
- Waldron-Lynch, F.; Herold, K.C. Immunomodulatory therapy to preserve pancreatic β-cell function in type 1 diabetes. Nat. Rev. Drug Discov. 2011, 10, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Danaei, G.; Finucane, M.M.; Lu, Y.; Singh, G.M.; Cowan, M.J.; Paciorek, C.J.; Lin, J.K.; Farzadfar, F.; Khang, Y.H.; Stevens, G.A.; et al. National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: Systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2·7 million participants. Lancet 2011, 378, 31–40. [Google Scholar] [CrossRef]
- Lustig, R.H.; Schmidt, L.A.; Brindis, C.D. Public health: The toxic truth about sugar. Nature 2012, 482, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.W.; D’Agostino, R.B.; Levy, D.; Belanger, A.M.; Silbershatz, H.; Kannel, W.B. Prediction of coronary heart disease using risk factor categories. Circulation 1998, 97, 1837–1847. [Google Scholar] [CrossRef] [PubMed]
- McGill, H.C., Jr.; McMahan, C.A. Pathobiological Determinants of Atherosclerosis in Youth (PDAY) Research Group. Determinants of atherosclerosis in the young. Am. J. Cardiol. 1998, 82, 30T–36T. [Google Scholar] [CrossRef]
- Grundy, S.M.; Benjamin, I.J.; Burke, G.L.; Chait, A.; Eckel, R.H.; Howard, B.V.; Mitch, W.; Smith, S.C., Jr.; Sowers, J.R. Diabetes and cardiovascular disease: a statement for healthcare professionals from the American Heart Association. Circulation 1999, 100, 1134–1146. [Google Scholar] [PubMed]
- Lipinski, B. Pathophysiology of oxidative stress in diabetes mellitus. J. Diabetes Complicat. 2001, 15, 203–210. [Google Scholar] [CrossRef]
- Williamson, R.T.; Lond, M.D. On the treatment of glycosuria and diabetes mellitus with sodium salicylate. Br. Med. J. 1901, 1, 760–762. [Google Scholar] [CrossRef] [PubMed]
- Ghiselli, A.; Laurenti, O.; DeMattia, G.; Maiani, G.; Ferro-Luzzi, A. Salicylate hydroxylation as an early marker of in vivo oxidative stress in diabetic patients. Free Radic. Biol. Med. 1992, 13, 621–626. [Google Scholar] [CrossRef]
- Adams, M.J.; Blundell, T.L.; Dodson, E.J.; Dodson, G.G.; Vijayan, M.; Baker, E.N.; Harding, M.M.; Hodgkin, D.C.; Rimmer, B.; Sheat, S. Structure of rhombohedral 2-zinc insulin crystals. Nature 1969, 224, 491–495. [Google Scholar] [CrossRef]
- Schlichtkrull, J. Chemical and Biological Studies on Insulin Crystals and Insulin Zinc Suspensions. Ph.D. Thesis, Copenhagen University, Copenhagen, Denmark, 1958. [Google Scholar]
- Kaarsholm, N.C.; Ko, H.C.; Dunn, M.F. Comparison of solution structural flexibility and zinc binding domains for insulin, proinsulin, and miniproinsulin. Biochemistry 1989, 28, 4427–4435. [Google Scholar] [CrossRef] [PubMed]
- Ciszak, E.; Beals, J.M.; Frank, B.H.; Baker, J.C.; Carter, N.D.; Smith, G.D. Role of C-terminal B-chain residues in insulin assembly: The structure of hexameric LysB28ProB29-human insulin. Structure 1995, 3, 615–622. [Google Scholar] [CrossRef]
- Norrman, M. Insulin Polymorphism. Ph.D. Thesis, Lund University, Lund, Sweden, 2007. [Google Scholar]
- Baker, E.N.; Blundell, T.L.; Cutfield, J.F.; Cutfield, S.M.; Dodson, E.J.; Dodson, G.G.; Hodgkin, D.C.; Hubbard, R.E.; Isaacs, N.W.; Reynolds, C.D.; et al. The structure of 2Zn pig insulin crystals at 1.5 Å resolution. Philos. Trans. R. Soc. Lond. B 1988, 319, 369–456. [Google Scholar] [CrossRef]
- Ciszak, E.; Smith, G.D. Crystallographic Evidence for Dual Coordination around Zinc in the T3R3 Human Insulin Hexamer. Biochemistry 1994, 33, 1512–1517. [Google Scholar] [CrossRef] [PubMed]
- Whittingham, J.L.; Chaudhurri, S.; Dodson, E.J.; Moody, P.C.E.; Dodson, G.G. X-ray Crystallographic Studies on Hexameric Insulins in the Presence of Helix-Stabilizing Agents, Thiocyanate, Methylparaben and Phenol. Biochemistry 1995, 34, 15553–15563. [Google Scholar] [CrossRef] [PubMed]
- Dunn, M.F. Zinc-ligand interactions modulate assembly and stability of the insulin hexamer—A review. Biometals 2005, 18, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Derewenda, U.; Derewenda, Z.; Dodson, E.J.; Dodson, G.G.; Reynolds, C.D.; Smith, G.D.; Sparks, C.; Swenson, D. Phenol stabilizes more helix in a new symmetrical zinc insulin hexamer. Nature 1989, 338, 594–596. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.D.; Ciszak, E. The structure of a complex of hexameric insulin and 4’-hydroxyacetanilide. Proc. Natl Acad. Sci. USA 1994, 91, 8851–8855. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.D.; Ciszak, E.; Pangborn, W.A. A novel complex of a phenolic derivative with insulin: Structural features related to the T → R transition. Protein Sci. 1996, 5, 1502–1511. [Google Scholar] [CrossRef] [PubMed]
- Hallas-Møhler, K.; Petersen, K.; Schlichtkrull, J. Crystalline and amorphous insulin–zinc compounds with prolonged action. Science 1952, 116, 394–398. [Google Scholar] [CrossRef]
- Wagner, A.; Diez, J.; Schulze-Briese, C.; Schluckebier, G. Crystal structure of Ultralente—A microcrystalline insulin suspension. Proteins 2009, 74, 1018–1027. [Google Scholar] [CrossRef] [PubMed]
- Whittingham, J.L.; Edwards, D.J.; Antson, A.A.; Clarkson, J.M.; Dodson, C.G. Interactions of Phenol and m-Cresol in the Insulin Hexamer, and Their Effect on the Association Properties of B28 Pro → Asp Insulin Analogues. Biochemistry 1998, 37, 11516–11523. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.D.; Ciszak, E.; Magrum, L.A.; Pangborn, W.A.; Blessing, R.H. R6 hexameric insulin complexed with m-cresol or resorcinol. Acta Crystallogr. D 2000, 56, 1541–1548. [Google Scholar] [CrossRef] [PubMed]
- Karavassili, F.; Giannopoulou, A.E.; Kotsiliti, E.; Knight, L.; Norrman, M.; Schluckebier, G.; Drube, L.; Fitch, A.N.; Wright, J.P.; Margiolaki, I. Structural studies of human insulin cocrystallized with phenol or resorcinol via powder diffraction. Acta Crystallogr. D 2012, 68, 1632–1641. [Google Scholar] [CrossRef] [PubMed]
- Gursky, O.; Badger, J.; Li, Y.; Caspar, D.L. Conformational changes in cubic insulin crystals in the pH range 7–11. Biophys. J. 1992, 63, 1210–1220. [Google Scholar] [CrossRef]
- McPherson, A. Crystallization of Proteins by Variation of pH or Temperature. Methods Enzymol. 1985, 114, 125–127. [Google Scholar] [PubMed]
- Farr, R.; Perryman, A.; Samudzi, C. Re-clustering the database for crystallization of macromolecules. J. Cryst. Growth 1998, 183, 653–668. [Google Scholar] [CrossRef]
- McPherson, A. Increasing the size of microcrystals by fine sampling of pH limits. J. Appl. Crystallogr. 1995, 28, 362–365. [Google Scholar] [CrossRef]
- Smith, G.D.; Pangborn, W.A.; Blessing, R.H. The structure of T6 bovine insulin. Acta Crystallogr. D 2005, 61, 1476–1482. [Google Scholar] [CrossRef] [PubMed]
- Hodgkin, D.C. Insulin molecules: The extent of our knowledge. Pure Appl. Chem. 1971, 26, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Brange, J. The new era of biotech insulin analogues. Diabetologia 1997, 40, S48–S53. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.K.; Govardhan, C.P.; Jung, C.W.; Margolin, A.L. Protein crystals for the delivery of biopharmaceuticals. Expert Opin. Biol. Ther. 2004, 4, 301–317. [Google Scholar] [CrossRef] [PubMed]
- Rietveld, H.M. A profile refinement method for nuclear and magnetic structures. J. Appl. Crystallogr. 1969, 2, 65–71. [Google Scholar] [CrossRef]
- Young, R.A.; Mackie, P.E.; Von Dreele, R.B. Application of the pattern-fitting structure-refinement method of X-ray powder diffractometer patterns. J. Appl. Crystallogr. 1977, 10, 262–269. [Google Scholar] [CrossRef]
- Alexander, L.E. Forty years of quantitative diffraction analysis. In Proceedings of Advances in X-ray Analysis; McMurdie, H.F., Barrett, C.S., Newkirk, J.B., Ruud, C.O., Eds.; Plenum Press: New York, NY, USA, 1976; Volume 20, pp. 1–13. [Google Scholar]
- Klug, H.P.; Alexander, L.E. X-ray Diffraction Procedures for Polycrystalline and Amorphous Materials, 2nd ed.; John Wiley: New York, NY, USA, 1974; pp. 271–418. [Google Scholar]
- Cheetham, A.K.; Taylor, J.C. Profile analysis of neutron powder diffraction data: Its scope, limitations and applications in solid state chemistry. J. Solid State Chem. 1977, 21, 253–275. [Google Scholar] [CrossRef]
- Harris, K.D.M.; Tremayne, M. Crystal Structure Determination from Powder Diffraction Data. Chem. Mater. 1996, 8, 2554–2570. [Google Scholar] [CrossRef]
- Von Dreele, R.B. Combined Rietveld and stereochemical restraint refinement of a protein crystal structure. J. Appl. Crystallogr. 1999, 32, 1084–1089. [Google Scholar] [CrossRef]
- Collings, I.; Watier, Y.; Giffard, M.; Dagogo, S.; Kahn, R.; Bonnete´, F.; Wright, J.P.; Fitch, A.N.; Margiolaki, I. Polymorphism of microcrystalline urate oxidase from Aspergillus flavus. Acta Crystallogr. D 2010, 66, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Das, U.; Naskar, J.; Mukherjee, A.K. Conformational analysis of an acyclic tetrapeptide: ab-initio structure determination from X-ray powder diffraction, Hirshfeld surface analysis and electronic structure. J. Pept. Sci. 2015, 21, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Liu, C.; Guo, Z. Structural Insights into Aβ42 Oligomers Using Site-directed Spin Labeling. J. Biol. Chem. 2013, 288, 18673–18683. [Google Scholar] [CrossRef] [PubMed]
- Von Dreele, R.B. Protein Crystal Structure Analysis from High-Resolution X-ray Powder-Diffraction Data. Methods Enzymol. 2003, 368, 254–267. [Google Scholar] [PubMed]
- Margiolaki, I.; Wright, J.P.; Fitch, A.N.; Fox, G.C.; Von Dreele, R.B. Synchrotron X-ray Powder Diffraction Study of Hexagonal Turkey Egg-White Lysozyme. Acta Crystallogr. D 2005, 61, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Margiolaki, I.; Wright, J.P.; Wilmanns, M.; Fitch, A.N.; Pinotsis, N. Second SH3 Domain of Ponsin Solved from Powder Diffraction. J. Am. Chem. Soc. 2007, 129, 11865–11871. [Google Scholar] [CrossRef] [PubMed]
- Margiolaki, I.; Wright, J.P. Powder crystallography on macromolecules. Acta Crystallogr. A 2008, 64, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Karavassili, F.; Margiolaki, I. Macromolecular Powder Diffraction: Ready for genuine biological problems. Protein Pept. Lett. 2016, 23, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Fili, S.; Valmas, A.; Norrman, M.; Schluckebier, G.; Beckers, D.; Degen, T.; Wright, J.; Fitch, A.; Gozzo, F.; Giannopoulou, A.E.; et al. Human insulin polymorphism upon ligand binding and pH variation: The case of 4-ethylresorcinol. IUCrJ 2015, 2, 534–544. [Google Scholar] [CrossRef] [PubMed]
- Norrman, M.; Stahl, K.; Schluckebier, G.; Al-Karadaghi, S. Characterization of insulin microcrystals using powder diffraction and multivariate data analysis. J. Appl. Crystallogr. 2006, 39, 391–400. [Google Scholar] [CrossRef]
- Norrman, M.; Schluckebier, G. Crystallographic characterization of two novel crystal forms of human insulin induced by chaotropic agents and a shift in pH. BMC Struct. Biol. 2007, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- Valmas, A.; Magiouf, K.; Fili, S.; Norrman, M.; Schluckebier, G.; Beckers, D.; Degen, T.; Wright, J.; Fitch, A.; Gozzo, F.; et al. Novel crystalline phase and first-order phase transitions of human insulin complexed with two distinct phenol derivatives. Acta Crystallogr. D 2015, 71, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Grimes, J.; Ajit, K.B.; Roy, P.; Stuart, D. The crystal structure of bluetongue virus VP7. Nature 1995, 373, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Pawley, G.S. Unit-cell refinement from powder diffraction scans. J. Appl. Crystallogr. 1981, 14, 357–361. [Google Scholar] [CrossRef]
- Smith, G.D.; Pangborn, W.A.; Blessing, R.H. Phase changes in T3R3f human insulin: Temperature or pressure induced? Acta Crystallogr. D 2001, 57, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Von Dreele, R.B.; Stephens, P.W.; Smith, G.D.; Blessing, R.H. The first protein crystal structure determined from high-resolution X-ray powder diffraction data: A variant of T3R3 human insulin-zinc complex produced by grinding. Acta Crystallogr. D 2000, 56, 1549–1553. [Google Scholar] [CrossRef] [PubMed]
- Von Dreele, R.B. Binding of N-acetylglucosamine to chicken egg lysozyme: A powder diffraction study. Acta Crystallogr. D 2001, 57, 1836–1842. [Google Scholar] [CrossRef] [PubMed]
- Von Dreele, R.B. Binding of N-acetylglucosamine oligosaccharides to hen egg-white lysozyme: A powder diffraction study. Acta Crystallogr. D 2005, 61, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Basso, S.; Fitch, A.N.; Fox, G.C.; Margiolaki, I.; Wright, J.P. High-throughput phase-diagram mapping via powder diffraction: A case study of HEWL versus pH. Acta Crystallogr. D 2005, 61, 1612–1625. [Google Scholar] [CrossRef] [PubMed]
- Krayenbühl, C.; Rosenberg, T. Crystalline protamine insulin. Rep. Steno. Mem. Hosp. Nord. Insulinlab. 1946, 1, 60–73. [Google Scholar]
- Balschmidt, P.; Hansen, F.B.; Dodson, E.J.; Dodson, G.G.; Korber, F. Structure of porcine insulin cocrystallized with clupeine Z. Acta Crystallogr. B 1991, 47, 975–986. [Google Scholar] [CrossRef] [PubMed]
- Cerenius, Y.; Stahl, K.; Svensson, L.A.; Ursby, T.; Oskarsson, A.; Albertsson, J.; Liljas, A. The crystallography beamline I711 at MAX II. J. Synchrotron Radiat. 2000, 7, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Mammen, C.B.; Ursby, T.; Cerenius, Y.; Thunnissen, M.; Als-Nielsen, J.; Larsen, S.; Liljas, A. Design of a 5-station macromolecular crystallography beamline at MAX-Lab. Acta Phys. Pol. A 2002, 101, 595–602. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Smith, G.D.; Pangborn, W.A.; Blessing, R.H. The structure of T6 human insulin at 1.0 Å resolution. Acta Crystallogr. D 2003, 59, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Margiolaki, I.; Giannopoulou, A.E.; Wright, J.P.; Knight, L.; Norrman, M.; Schluckebier, G.; Fitch, A.N.; Von Dreele, R.B. High-resolution powder X-ray data reveal the T6 hexameric form of bovine insulin. Acta Crystallogr. D 2013, 69, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Larson, A.C.; Von Dreele, R.B. General Structure Analysis System (GSAS); LAUR 86-748; Los Alamos National Laboratory: Los Alamos, NM, USA, 2004. Available online: http://11bm.xray.aps.anl.gov/documents/GSASManual.pdf (accessed on 18 August 2017).
- Toby, B.H. EXPGUI, a graphical user interface for GSAS. J. Appl. Crystallogr. 2001, 34, 210–213. [Google Scholar] [CrossRef]
- Bhat, T.N. Calculation of an OMIT map. J. Appl. Crystallogr. 1988, 21, 279–281. [Google Scholar] [CrossRef]
- Rowe, R.C.; Sheskey, P.J.; Owen, S.C. Handbook of Pharmaceuticals Excipients, 5th ed.; Pharmaceutical Press: London, UK, 2005. [Google Scholar]
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497. [Google Scholar] [CrossRef]
- Kantardjieff, K.A.; Rupp, B. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein–nucleic acid complex crystals. Protein Sci. 2003, 12, 1865–1871. [Google Scholar] [CrossRef] [PubMed]
- Karavassili, F.; Valmas, A.; Giannopoulou, E.A.; Fili, S.; Norrman, M.; Schluckebier, G; Beckers, D.; Degen, T.; Fitch, A.N.; Margiolaki, I. Novel monoclinic crystalline form of human Insulin complexed with meta-Cresol. Unpublished work. 2017. [Google Scholar]
- Degen, T.; Sadki, M.; Bron, E.; König, U.; Nénert, G. The HighScore suite. Powder Diffr. 2014, 29, S13–S18. [Google Scholar] [CrossRef]
- Rahuel-Clermont, S.; French, C.A.; Kaarsholm, N.C.; Dunn, M.F. Mechanisms of Stabilization of the Insulin Hexamer through Allosteric Ligand Interactions. Biochemistry 1997, 36, 5837–5845. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, D.; Diers, J.R.; Bocian, D.F.; Kaarsholm, N.C.; Dunn, M.F. Raman signatures of ligand binding and allosteric conformation change in hexameric insulin. Biopolymers 2001, 62, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Bloom, C.R.; Choi, W.E.; Brzovic, P.S.; Ha, J.J.; Huang, S.T.; Kaarsholm, N.C.; Dunn, M.F. Ligand binding to wild-type and E-B13Q mutant insulins: A three-state allosteric model system showing half-site reactivity. J. Mol. Biol. 1995, 245, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Rabinow, B.E. Nanosuspensions in drug delivery. Nat. Rev. Drug Discov. 2004, 3, 785–796. [Google Scholar] [CrossRef] [PubMed]
- Blair, A.S.; Hajduch, E.; Litherland, G.J.; Hundal, H.S. Regulation of glucose transport and glycogen synthesis in L6 muscle cells during oxidative stress. Evidence for cross-talk between the insulin and SAPK2/p38 mitogen-activated protein kinase signaling pathways. J. Biol. Chem. 1999, 274, 36293–36299. [Google Scholar] [CrossRef] [PubMed]
- Ceriello, A.; Motz, E. Is oxidative stress the pathogenic mechanism underlying insulin resistance, diabetes, and cardiovascular disease? The common soil hypothesis revisited. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Baynes, J.W.; Thorpe, S.R. Role of oxidative stress in diabetic complications: A new perspective on an old paradigm. Diabetes 1999, 48, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Johansen, J.S.; Harris, A.K.; Rychly, D.J.; Ergul, A. Oxidative stress and the use of antioxidants in diabetes: Linking basic science to clinical practice. Cardiovasc. Diabetol. 2005, 4, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cathcart, R.F., III. Vitamin C: The nontoxic, nonrate-limited, antioxidant free radical scavenger. Med. Hypotheses 1985, 18, 61–77. [Google Scholar] [CrossRef]
- Kodama, M.; Kodama, T.; Murakami, M.; Kodama, M. Diabetes mellitus is controlled by vitamin C treatment. In Vivo 1993, 7, 535–542. [Google Scholar] [PubMed]
- Dakhale, G.N.; Chaudhari, H.V.; Shrivastava, M. Supplementation of vitamin C reduces blood glucose and improves glycosylated hemoglobin in type 2 diabetes mellitus: A randomized, double-blind study. Adv. Pharmacol. Sci. 2011. [Google Scholar] [CrossRef] [PubMed]
- Valmas, A.; Fili, S.; Kontou, P.; Norrman, M.; Schluckebier, G.; Beckers, D.; Degen, T.; Wright, J.P.; Fitch, A.N.; Karavassili, F.; et al. Conformational responses to pH of human insulin. Unpublished work. 2017. [Google Scholar]
- Karavassili, F.; Valmas, A; Fili, S.; Kontou, P.; Spiliopoulou, M.; Bowler, M.; Beckers, D.; Degen, T.; Nenert, G; Fitch, A.N.; et al. Microcrystalline cubic human insulin at 2.7Å resolution. Unpublished work. 2017. [Google Scholar]
- Badger, J.; Harris, M.R.; Reynolds, C.D.; Evans, A.C. Structure of pig insulin dimer in the cubic crystal. Acta Crystallogr. B 1991, 47, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Caspar, D.L.D. Structure of Cubic Insulin Crystals in Glucose Solutions. Biophys. J. 1998, 74, 616–622. [Google Scholar] [CrossRef]
- Scott, D.A. Crystalline insulin. Biochem. J. 1934, 28, 1592–1602. [Google Scholar] [CrossRef] [PubMed]
- Edsall, J.T.; Felsenfeld, G.L.D.; Dewitt, S.; Goodman, D.S.; Gurd, F.R.N. The Association of Imidazole with the Ions of Zinc and Cupric Copper1a,b,c. J. Am. Chem. Soc. 1954, 76, 3054–3061. [Google Scholar] [CrossRef]
- Rotenstein, L.S.; Ran, N.; Shivers, J.P.; Yarchoan, M.; Close, K.L. Opportunities and challenges for biosimilars: What’s on the horizon in the global insulin market? Clin. Diabetes 2012, 30, 138–150. [Google Scholar] [CrossRef]
- Bernstein, J. Polymorphism in Molecular Crystals; Clarendon Press: Oxford, UK, 2002. [Google Scholar]
- Nogly, P.; James, D.; Wang, D.; White, T.A.; Zatsepin, N.; Shilova, A.; Nelson, G.; Liu, H.; Johansson, L.; Heymann, M.; et al. Lipidic cubic phase serial millisecond crystallography using synchrotron radiation. IUCrJ 2015, 2, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Coquelle, N.; Brewster, A.S.; Kapp, U.; Shilova, A.; Weinhausen, B.; Burghammer, M.; Colletier, J.P. Raster-scanning serial protein crystallography using micro- and nano-focused synchrotron beams. Acta Crystallogr. D 2015, 71, 1184–1196. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Nannenga, B.L.; Cruz, M.J.; Liu, J.; Sawtelle, S.; Calero, G.; Reyes, F.E.; Hattne, J.; Gonen, T. The collection of MicroED data for macromolecular crystallography. Nat. Protoc. 2016, 11, 895–904. [Google Scholar] [CrossRef] [PubMed]
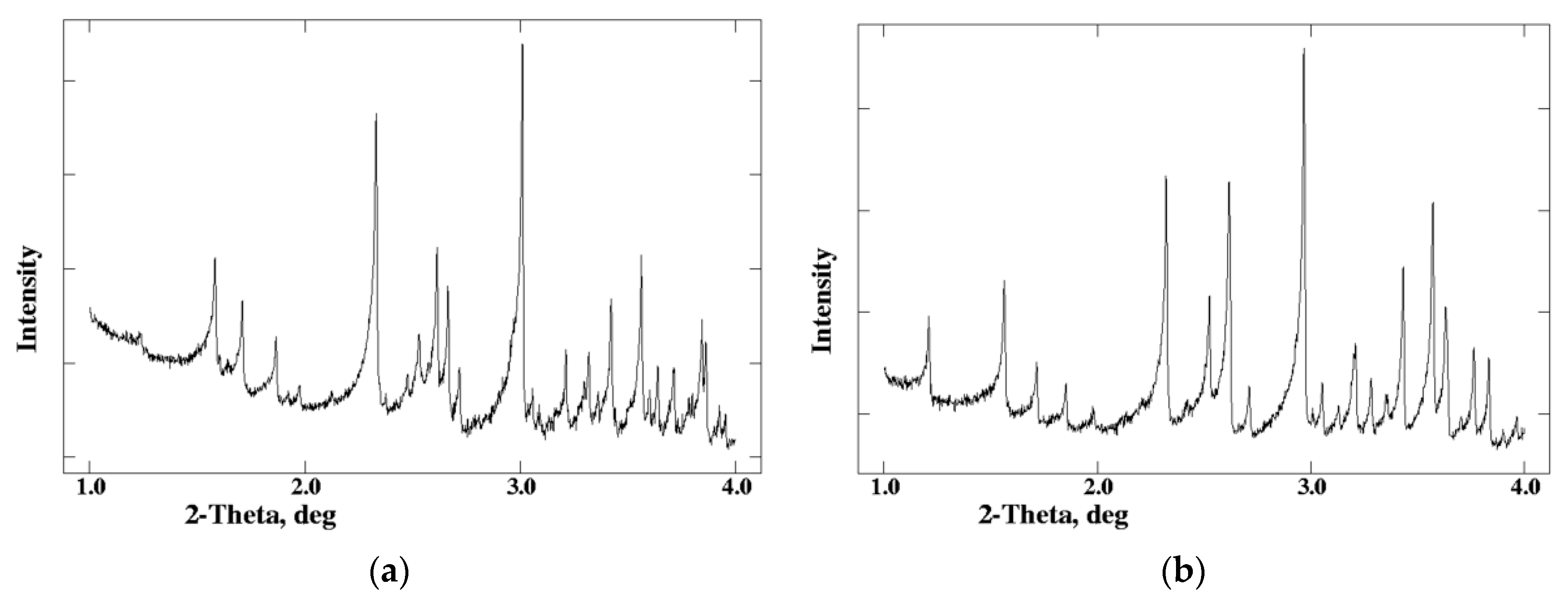
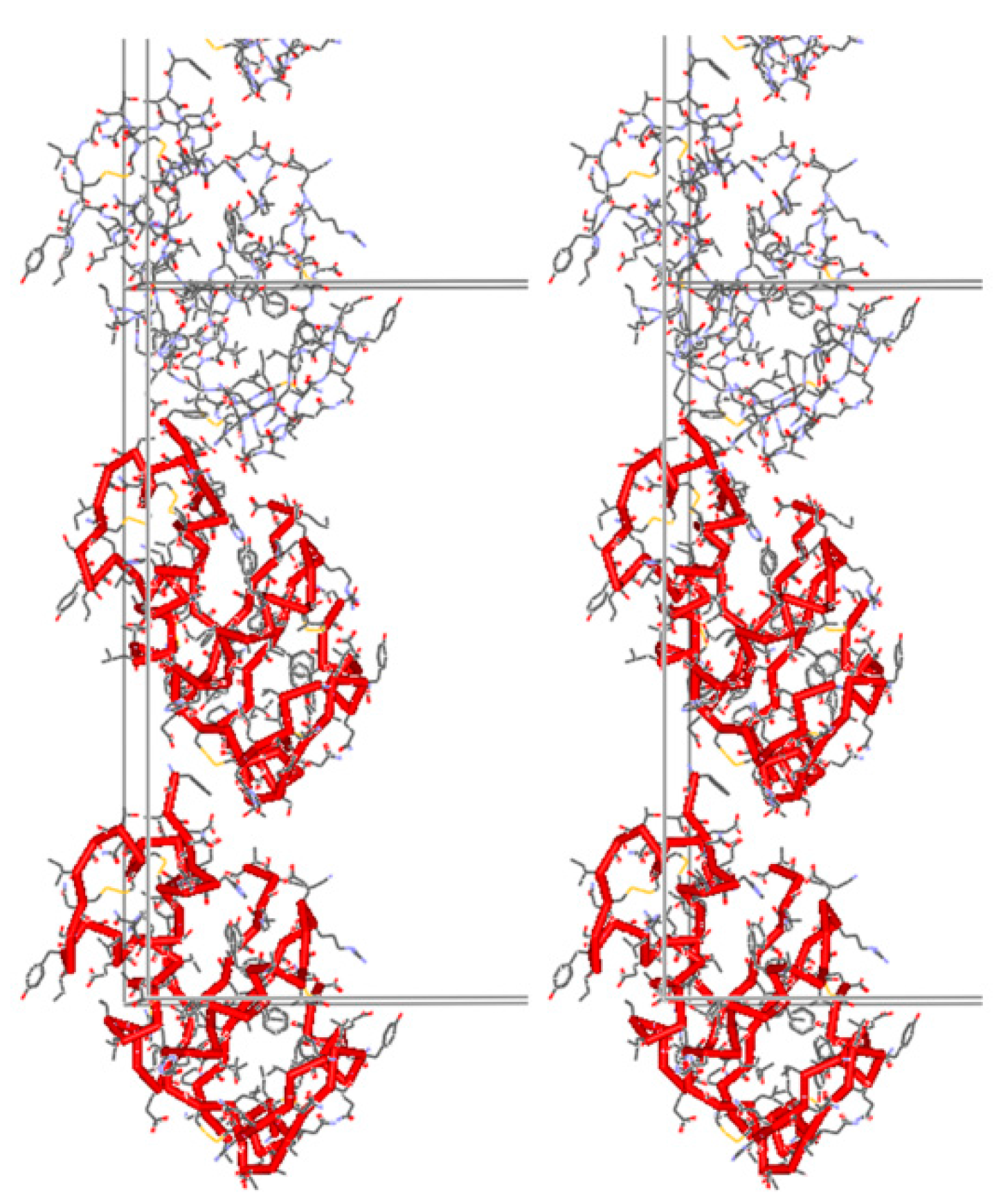

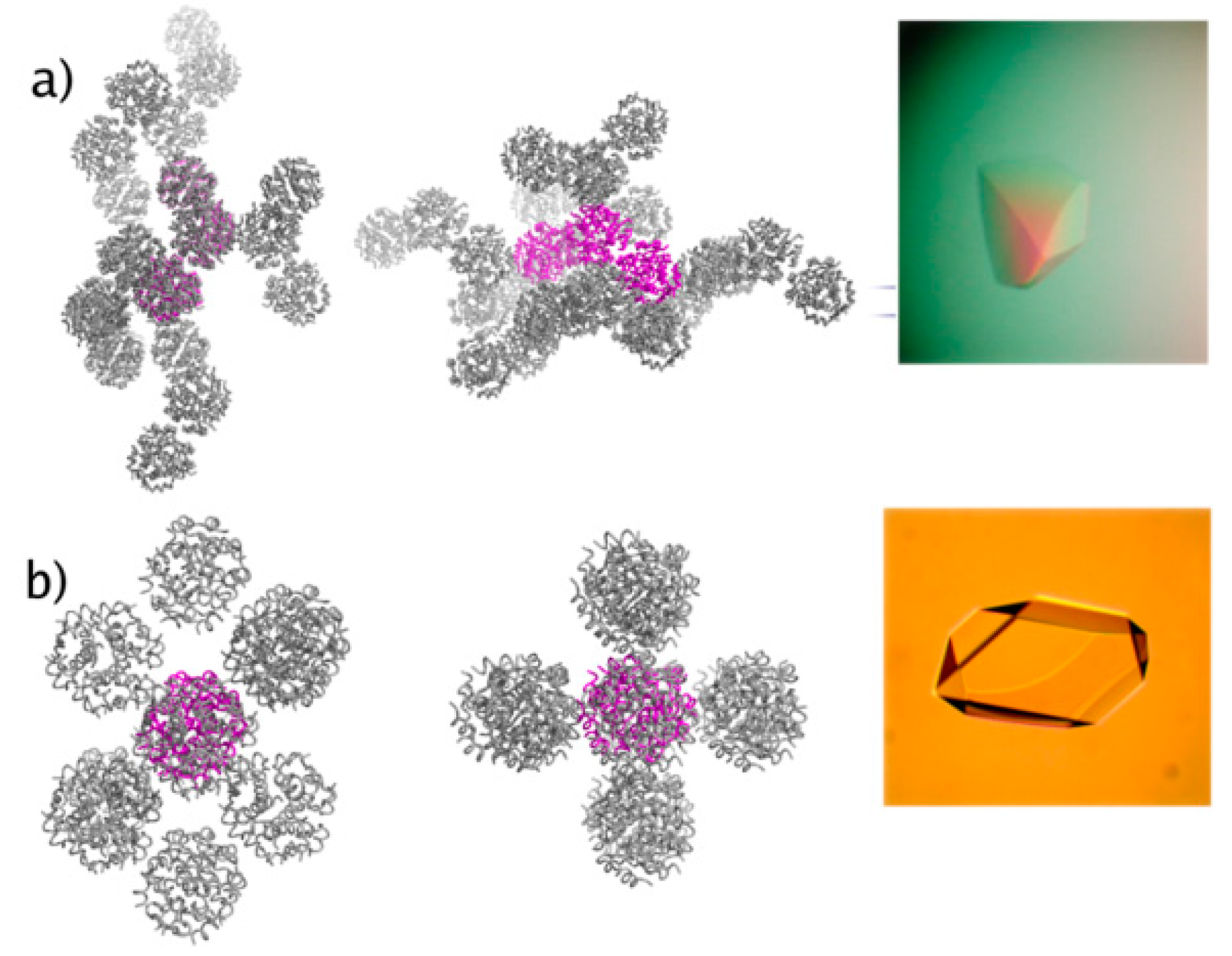
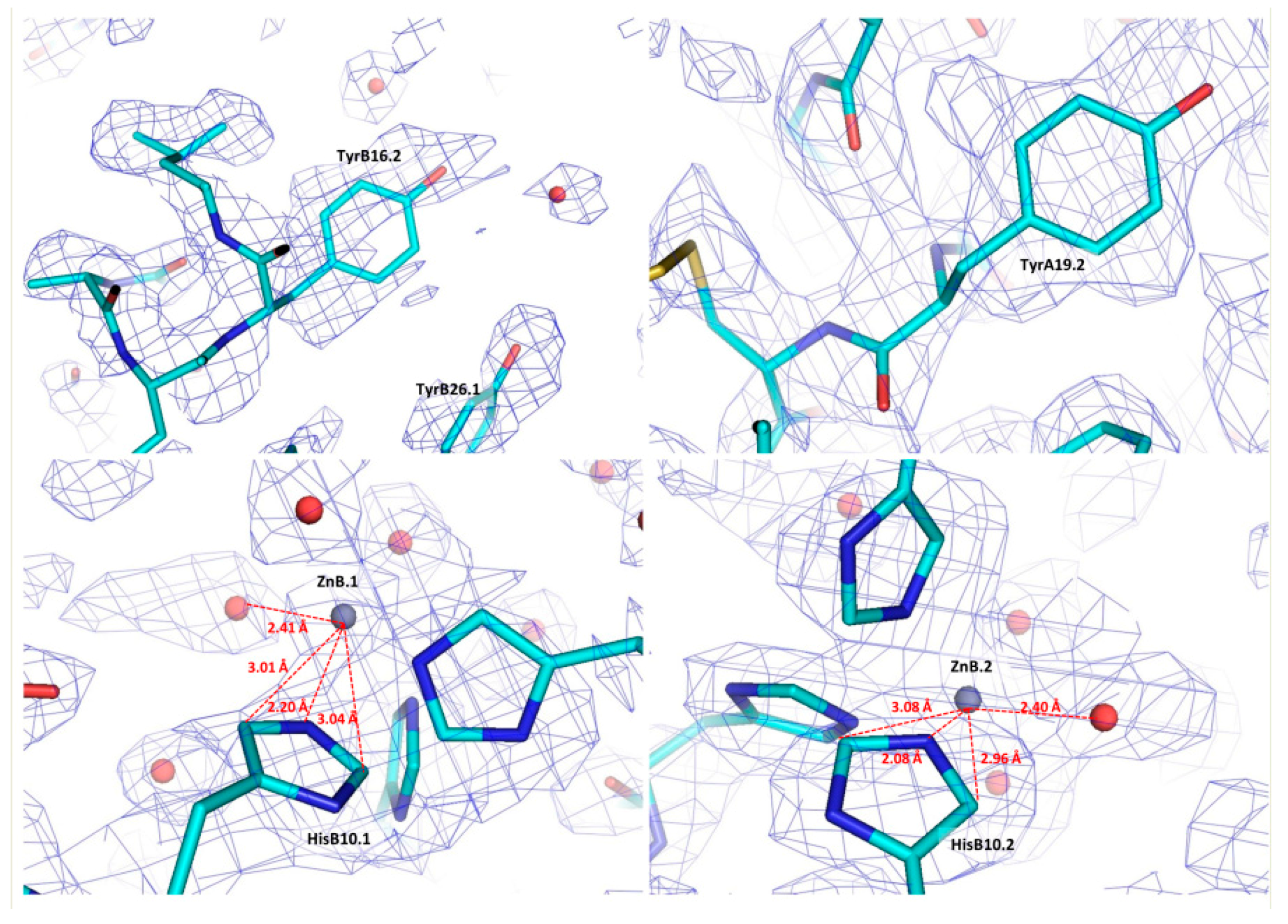



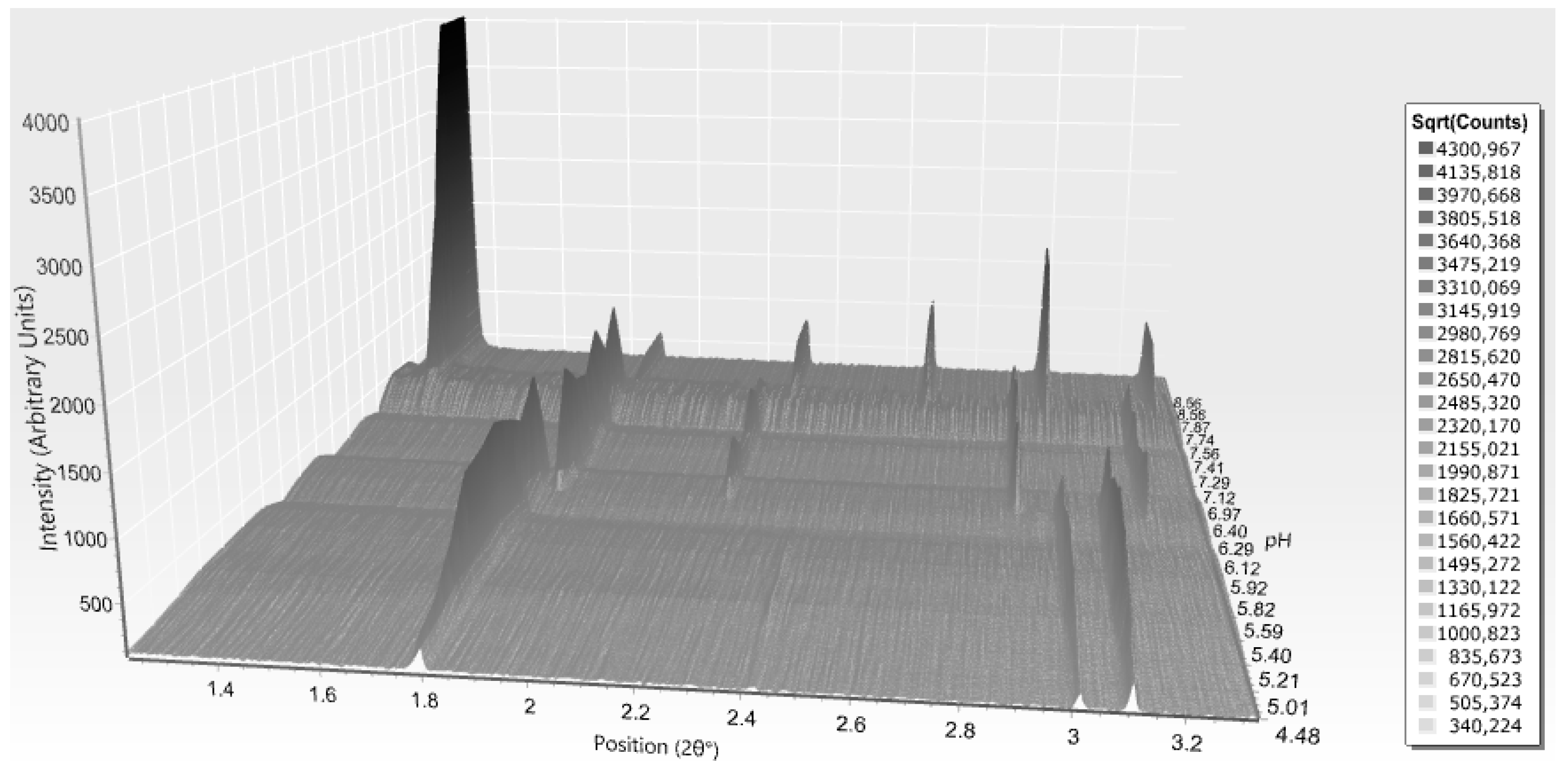
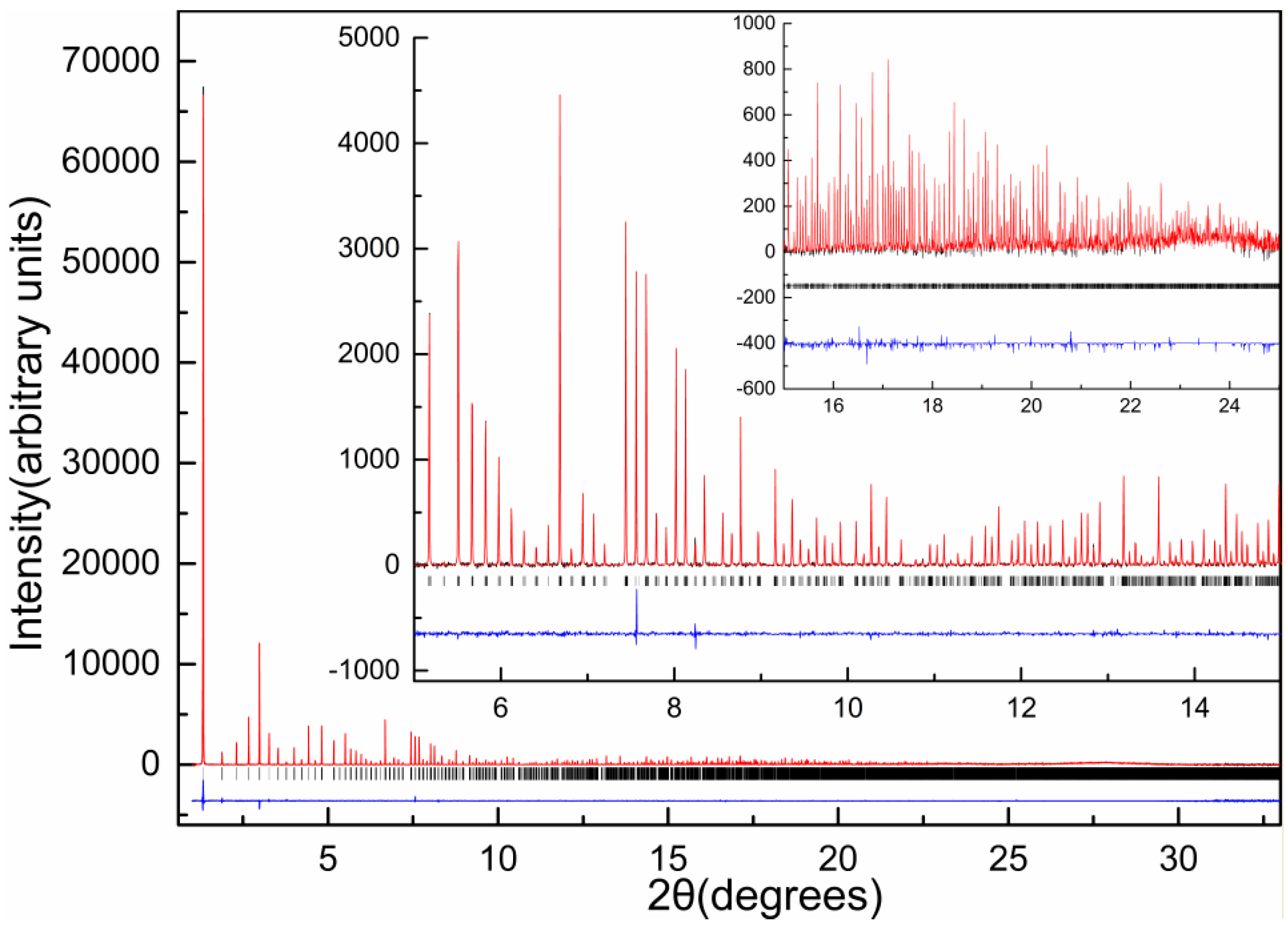

| A | B | C | D | E | F | G | H | I | J | K | Χ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Space group | P21 | C2221 | I213 | R3 | R3 | R3 | R3 | R3 | P43212 | P43212 | P43212 | C2 |
| Insulin (mg·mL−1) | 5.2 | 3.5 | 10 | 14 | 3.8 | 3.8 | 3.8 | 1.5 | 3.8 | 3.8 | 1.5 | 3.5 |
| Zn/hexamer | 2.3 | 2.3 | 2.5 | 4 | 2.2 | 2.2 | 2.2 | 3 | 3 | 3 | 2.3 | |
| Phenol derivative (mM) † | 20 1 | 25 1 | 19 3/19 4 | 65 2 | 65 2 | 65 2 | 7 3/14 4 | 7 3/14 3 | 7 3/14 3 | 25 1 | ||
| NaCl (M) | 1.0 | 1.0 | 0.02 | 0.3 | 0.12 | 0.12 | 0.12 | 1.0 | ||||
| Na acetate (M) | 0.01 | 0.01 | 0.01 | 0.01 | ||||||||
| Na citrate (M) | 0.11 | |||||||||||
| Na2HPO4 (M) | 0.48 | 0.05 | 0.04 | 0.013 | 0.013 | 0.013 | 0.05 | |||||
| Urea (M) | 1.1 | 1.1 | ||||||||||
| Tris (M) | 0.14 | |||||||||||
| Protamine | Added in isophane ratio ‡ | |||||||||||
| pH | 7.3 | 6.7 | 7.2 | 8.15 | 5.5 | 7.4 | 7.4 | 7.4 | 7.3 | 7.3 | 7.3 | 7.0 |
| Unit Cell | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal | λ (Å) †/Beamline | Trade Name | Crystal System | Space Group | Sequence Origin ‡ | B-Chain Configuration | a (Å) | b (Å) | c (Å) | β (°) | PDB Ref.§ |
| A | 0.97/911-3 | Monoclinic | P21 | H | R6 | 61.3 | 61.7 | 47.5 | 111.3 | 1EV6 [28] | |
| B | 1.00/911-2 | Orthorhombic. | C2221 | H | R6 | 58.9 | 219.4 | 223.7 | In-house database | ||
| C | 1.00/911-2 | Cubic | I213 | H | T | 78.9 | 78.9 | 78.9 | 1APH [28] | ||
| D | 0.97/911-3 | Detemir | Rhombohedral | R3 | H | R6 | 78.9 | 78.9 | 39.5 | 1EV3 [28] | |
| E | 0.97/911-3 | Rhombohedral | R3 | H | T3R3f | 80.6 | 80.6 | 37.8 | 1TRZ [19] | ||
| F | 0.969/711 | Ultralente | Rhombohedral | R3 | H | T6 | 81.3 | 81.3 | 33.7 | 1MSO [65] | |
| G | 0.969/711 | Ultratard | Rhombohedral | R3 | H | T6 | 82.5 | 82.5 | 34.0 | 4INS [18] | |
| H | 0.969/711 | Lente | B, P | ||||||||
| I | 0.969/711 | Penmix30 | Tetragonal | P43212 | H | R6 | 62.9 | 62.9 | 85.9 | In-house database | |
| J | 1.00/911-2 | Novomix30 | Tetragonal | P43212 | H B28Asp | R6 | 62.8 | 62.8 | 86.9 | In-house database | |
| K | 0.969/711 | Protaphan | Tetragonal | P43212 | P | R6 | 62.9 | 62.9 | 85.9 | 7INS [61] | |
| X | 0.97/911-2 | Monoclinic | C2 | H | Unknown | 100 | 60 | 62 | 116 | ||
| Phenol Derivative | pH Range | Space Group | Indicative Sample’s pH | a (Å) | b (Å) | c (Å) | β (°) | Resolution Range (Å) |
|---|---|---|---|---|---|---|---|---|
| phenol | ||||||||
| 5.47–5.70 | P21(α) | 5.70 | 114.682 (6) | 337.63 (2) | 49.270 (4) | 101.555 (6) | 112.2–7.5 | |
| 5.93–6.54 | C2221 | 6.14 | 60.287 (1) | 221.797 (6) | 228.812 (5) | 90 | 115–7.5 | |
| 6.70–6.75 | C2 | 6.75 | 103.0115 (5) | 61.3213 (2) | 63.5783 (4) | 117.2244 (5) | 45.9–5.3 | |
| 7.01–8.25 | P21(β) | 7.46 | 61.0920 (4) | 61.8279 (4) | 47.9302 (4) | 110.6253 (7) | 45–4.4 | |
| resorcinol | ||||||||
| 5.29–5.46 | P21(α) | 5.29 | 114.0228 (8) | 335.430 (3) | 49.211 (6) | 101.531 (8) | 112.2–7.5 | |
| 5.93–7.45 | C2221 | 6.40 | 60.5579 (7) | 220.907 (3) | 228.320 (3) | 90 | 115–7.5 | |
| 7.53–8.22 | P21(β) | 8.22 | 61.0008 (4) | 62.0040 (3) | 47.8823 (3) | 110.0465 (5) | 45–4.4 | |
| m-cresol | ||||||||
| 4.50–6.70 | P21(γ) | 87.0749 (7) | 70.1190 (5) | 48.1679 (5) | 106.7442 (8) | 46.5–6.8 | ||
| 6.70–8.60 | R3 (R6) | 8.15 | 80.0644 (6) | 80.0644 (6) | 40.8396 (3) | 90 | 40.5–3.7 | |
| 4-nitrophenol | ||||||||
| 5.1–6.3 | P21(γ) | 5.97 | 87.118 (1) | 70.9493 (9) | 48.4967 (9) | 106.653 (1) | 46.5–6.8 | |
| 6.2–8.1 | R3 (T3R3f) | 6.41 | 80.721 (1) | 80.721 (1) | 37.8039 (5) | 90 | 40.5–3.6 | |
| 4-ethylresorcinol | ||||||||
| 4.95–5.60 | P21(γ) | 5.14 | 87.132 (3) | 70.294 (2) | 48.064 (2) | 106.259 (3) | 47–6.5 | |
| 5.65–5.80 | P21(α) | 5.80 | 114.130 (7) | 336.086 (3) | 48.987 (5) | 101.935 (8) | 112–12 | |
| 5.93–6.25 | C2 | 5.97 | 103.0848 (4) | 61.6636 (2) | 63.5006 (4) | 117.417 (5) | 46–7 | |
| 6.73–8.05 | P21(β) | 6.73 | 62.8231 (7) | 62.1078 (5) | 47.8362 (6) | 111.6913 (9) | 45–6 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karavassili, F.; Valmas, A.; Fili, S.; Georgiou, C.D.; Margiolaki, I. In Quest for Improved Drugs against Diabetes: The Added Value of X-ray Powder Diffraction Methods. Biomolecules 2017, 7, 63. https://doi.org/10.3390/biom7030063
Karavassili F, Valmas A, Fili S, Georgiou CD, Margiolaki I. In Quest for Improved Drugs against Diabetes: The Added Value of X-ray Powder Diffraction Methods. Biomolecules. 2017; 7(3):63. https://doi.org/10.3390/biom7030063
Chicago/Turabian StyleKaravassili, Fotini, Alexandros Valmas, Stavroula Fili, Christos D. Georgiou, and Irene Margiolaki. 2017. "In Quest for Improved Drugs against Diabetes: The Added Value of X-ray Powder Diffraction Methods" Biomolecules 7, no. 3: 63. https://doi.org/10.3390/biom7030063






