Critical Minireview: The Fate of tRNACys during Oxidative Stress in Bacillus subtilis
Abstract
:1. Introduction
2. Bacillus under Oxidative Stress
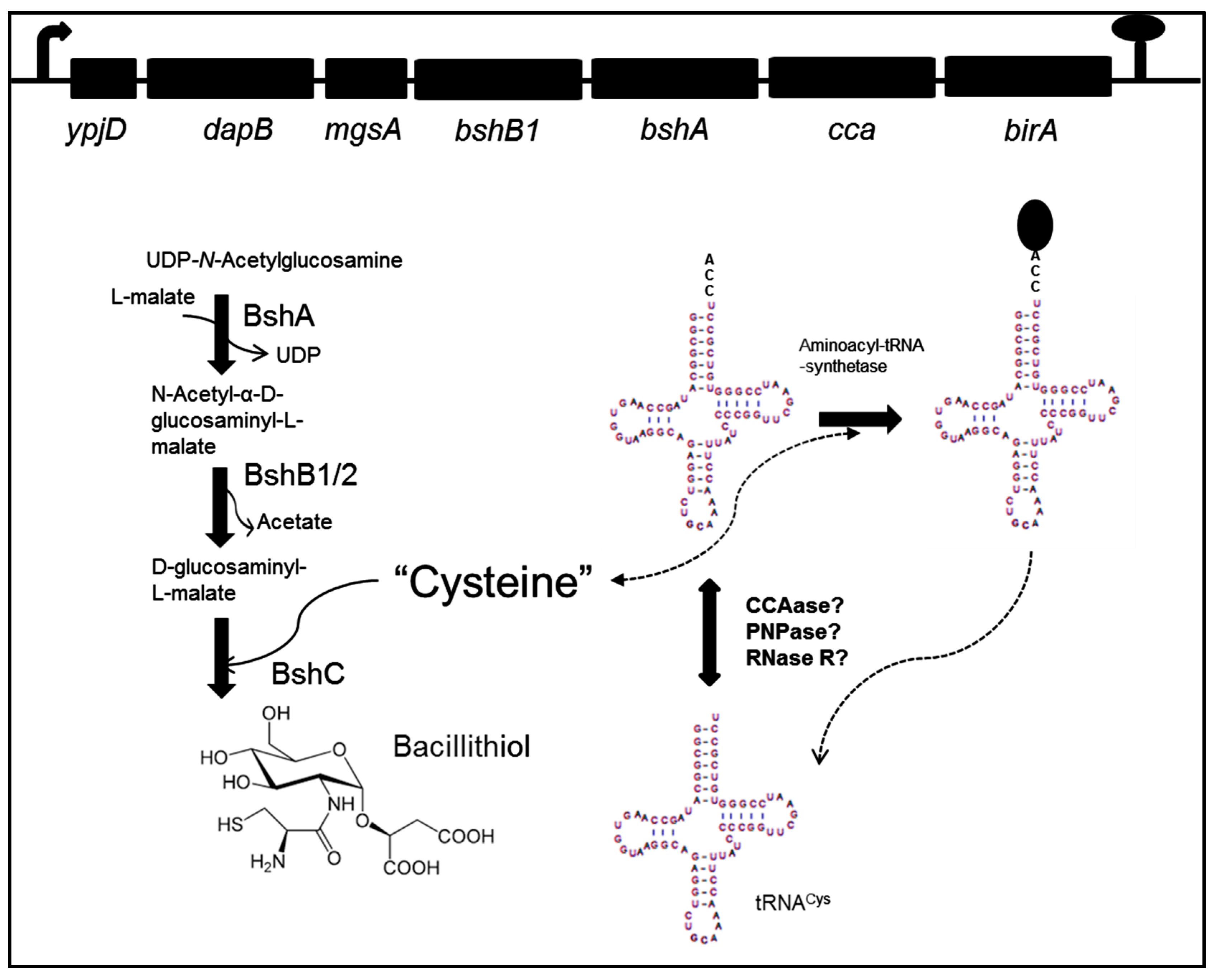
3. Bacillithiol Biosynthesis and Function
4. Role of CCA-tRNA Nucleotidyltransferase
5. A Link between Oxidative Stress, Bacillithiol, and tRNACys Maturation?
6. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Slepecky, R.A.; Hemphill, H.E. The genus Bacillus—nonmedical. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2006; pp. 530–562. [Google Scholar]
- Mols, M.; Abee, T. Primary and secondary oxidative stress in Bacillus. Environ. Microbiol. 2011, 13, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- Zuber, P. Management of oxidative stress in Bacillus. Annu. Rev. Microbiol. 2009, 63, 575–597. [Google Scholar] [CrossRef] [PubMed]
- Reder, A.; Höper, D.; Gerth, U.; Hecker, M. Contributions of Individual σB-Dependent General Stress Genes to Oxidative Stress Resistance of Bacillus subtilis. J. Bacteriol. 2012, 194, 3601–3610. [Google Scholar] [CrossRef] [PubMed]
- Herbig, A.F.; Helmann, J.D. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 2001, 41, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Hochgräfe, F.; Mostertz, J.; Albrecht, D.; Hecker, M. Fluorescence thiol modification assay: Oxidatively modified proteins in Bacillus subtilis. Mol. Microbiol. 2005, 58, 409–425. [Google Scholar] [CrossRef] [PubMed]
- Leichert, L.I.; Scharf, C.; Hecker, M. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 2003, 185, 1967–1975. [Google Scholar] [CrossRef] [PubMed]
- Narayan, M.; Welker, E.; Wedemeyer, W.J.; Scheraga, H.A. Oxidative folding of proteins. Acc. Chem. Res. 2000, 33, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Gaballa, A.; Newton, G.L.; Antelmann, H.; Parsonage, D.; Upton, H.; Rawat, M.; Claiborne, A.; Fahey, R.C.; Helmann, J.D. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc. Natl. Acad. Sci. USA 2010, 107, 6482–6486. [Google Scholar] [CrossRef] [PubMed]
- Gaballa, A.; Antelmann, H.; Hamilton, C.J.; Helmann, J.D. Regulation of Bacillus subtilis bacillithiol biosynthesis operons by Spx. Microbiology 2013, 159 Pt 10, 2025–2035. [Google Scholar] [CrossRef] [PubMed]
- Bower, S.; Perkins, J.; Yocum, R.R.; Serror, P.; Sorokin, A.; Rahaim, P.; Howitt, C.L.; Prasad, N.; Ehrlich, S.D.; Pero, J. Cloning and characterization of the Bacillus subtilis birA gene encoding a repressor of the biotin operon. J. Bacteriol. 1995, 177, 2572–2575. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Ehrlich, S.D.; Albertini, A.; Amati, G.; Andersen, K.K.; Arnaud, M.; Asai, K.; Ashikaga, S.; Aymerich, S.; Bessieres, P.; et al. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 2003, 100, 4678–4683. [Google Scholar] [CrossRef] [PubMed]
- Raynal, L.C.; Krisch, H.M.; Carpousis, A.J. The Bacillus subtilis nucleotidyltransferase is a tRNA CCA-Adding enzyme. J. Bacteriol. 1998, 180, 6276–6282. [Google Scholar] [PubMed]
- Nakano, S.; Küster-Schöck, E.; Grossman, A.D.; Zuber, P. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2003, 100, 13603–13608. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Soonsanga, S.; Helmann, J.D. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. USA 2007, 104, 8743–8748. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.A.; Sharma, S.V.; Strankman, A.W.; Duran, S.R.; Rawat, M.; Hamilton, C.J. Mechanistic studies of FosB: A divalent-metal-dependent bacillithiol-S-transferase that mediates fosfomycin resistance in Staphylococcus aureus. Biochem. J. 2013, 451, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Bernat, B.A.; Wang, Z.; Armstrong, R.N.; Helmann, J.D. FosB a Cysteine-Dependent Fosfomycin Resistance Protein under the Control of ςW, an Extracytoplasmic-Function ςfactor in Bacillus subtilis. J. Bacteriol. 2001, 183, 2380–2383. [Google Scholar] [CrossRef] [PubMed]
- Rigsby, R.E.; Fillgrove, K.L.; Beihoffer, L.A.; Armstrong, R.N. Fosfomycin resistance proteins: A nexus of glutathione transferases and epoxide hydrolases in a metalloenzyme superfamily. Methods Enzymol. 2005, 401, 367–379. [Google Scholar] [PubMed]
- Schürer, H.; Schiffer, S.; Marchfelder, A.; Mörl, M. This Is the End: Processing, Editing and Repair at the tRNA 3-Terminus. Biol. Chem. 2001, 382, 1147. [Google Scholar] [CrossRef] [PubMed]
- Neuenfeldt, A.; Just, A.; Betat, H.; Mörl, M. Evolution of tRNA nucleotidyltransferases: A small deletion generated CC-adding enzymes. Proc. Natl. Acad. Sci. USA 2008, 105, 7953–7958. [Google Scholar] [CrossRef] [PubMed]
- Mörl, M.; Marchfelder, A. The final cut: The importance of tRNA 3′-processing. EMBO Rep. 2001, 2, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Reimers, S.; Pandit, S.; Deutscher, M.P. RNA quality control: Degradation of defective transfer RNA. EMBO J. 2002, 21, 1132–1138. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E.; Whipple, J.M.; Phizicky, E.M.; Sharp, P.A. tRNAs marked with CCACCA are targeted for degradation. Science 2011, 334, 817–821. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Parker, R. Stressing out over tRNA cleavage. Cell 2009, 138, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Levitz, R.; Chapman, D.; Amitsur, M.; Green, R.; Snyder, L.; Kaufmann, G. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J. 1990, 9, 1383–1389. [Google Scholar] [PubMed]
- Fu, H.; Feng, J.; Liu, Q.; Sun, F.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009, 583, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.M.; Parker, R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 2009, 185, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.M. CCA addition to tRNA: Implications for tRNA quality control. IUBMB Life 2010, 62, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Cruz Hernandez, A.; Millan, E.S.; de Jesus Romero Gomez, S.; Antonio Cervantes Chavez, J.; Garcia Martinez, R.; Pastrana Martinez, X.; Gomez, J.L.; Jones, G.H.; Guillen, J.C. Exposure of Bacillus subtilis to mercury induces accumulation of shorter tRNA Cys species. Metallomics 2013, 5, 398–403. [Google Scholar] [CrossRef] [PubMed]
- Campos-Guillen, J.; Arvizu-Gomez, J.L.; Jones, G.H.; Olmedo-Alvarez, G. Characterization of tRNACys processing in a conditional Bacillus subtilis CCase mutant reveals the participation of RNase R in its quality control. Microbiology 2010, 156 Pt 7, 2102–2111. [Google Scholar] [CrossRef] [PubMed]
- Reuven, N.B.; Zhou, Z.; Deutscher, M.P. Functional overlap of tRNA nucleotidyltransferase, poly(A) polymerase I, and polynucleotide phosphorylase. J. Biol. Chem. 1997, 272, 33255–33259. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, H.; Kuwano, M.; Sekiguchi, M. Specific Binding of 8-Oxoguanine-Containing RNA to Polynucleotide Phosphorylase Protein. Biochemistry 2001, 40, 9977–9982. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, H.; Sekiguchi, M. Human polynucleotide phosphorylase protein in response to oxidative stress. Biochemistry 2006, 45, 6749–6755. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, Z. Human polynucleotide phosphorylase reduces oxidative RNA damage and protects HeLa cell against oxidative stress. Biochem. Biophys. Res. Commun. 2008, 372, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Jiang, Z.; Liu, M.; Gong, X.; Wu, S.; Burns, C.M.; Li, Z. Polynucleotide phosphorylase protects Escherichia coli against oxidative stress. Biochemistry 2009, 48, 2012–2020. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Deutscher, M.P. The physiological role of RNaseT can be explained by its unusual substrate specificity. J. Biol. Chem. 2002, 277, 29654–29661. [Google Scholar] [CrossRef] [PubMed]
- Dutta, T.; Malhotra, A.; Deutscher, M.P. How a CCA Sequence Protects Mature tRNAs and tRNA Precursors from Action of the Processing Enzyme RNase BN/RNase Z. J. Biol. Chem. 2013, 288, 30636–30644. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Xiao, C.; Gu, W.; Du, G.; Sun, X.; He, Q.; Zhang, G. Transfer RNAs Mediate the Rapid Adaptation of Escherichia coli to Oxidative Stress. PLoS Genet. 2015, 11, e1005302. [Google Scholar] [CrossRef] [PubMed]
- Svenningsen, S.L.; Kongstad, M.; Stenum, T.S.; Muñoz-Gómez, A.J.; Sørensen, M.A. Transfer RNA is highly unstable during early aminoacid starvation in Escherichia coli. Nucleic Acids Res. 2016. [Google Scholar] [CrossRef]
| Isotype | Ala | Gly | Pro | Thr | Val | Ser | Arg | Leu | Phe | Asn |
| With CCA | 4 | 7 | 3 | 2 | 4 | 5 | 4 | 2 | 2 | 3 |
| Without CCA | 2 | 0 | 0 | 3 | 1 | 0 | 3 | 6 | 1 | 1 |
| Isotype | Lys | Asp | Glu | His | Gln | Ile | Mel | Tyr | Cys | Trp |
| With CCA | 4 | 4 | 1 | 2 | 1 | 3 | 6 | 2 | 0 | 1 |
| Without CCA | 0 | 0 | 5 | 0 | 3 | 0 | 0 | 0 | 1 | 0 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos Guillen, J.; Jones, G.H.; Saldaña Gutiérrez, C.; Hernández-Flores, J.L.; Cruz Medina, J.A.; Valenzuela Soto, J.H.; Pacheco Hernández, S.; Romero Gómez, S.; Morales Tlalpan, V. Critical Minireview: The Fate of tRNACys during Oxidative Stress in Bacillus subtilis. Biomolecules 2017, 7, 6. https://doi.org/10.3390/biom7010006
Campos Guillen J, Jones GH, Saldaña Gutiérrez C, Hernández-Flores JL, Cruz Medina JA, Valenzuela Soto JH, Pacheco Hernández S, Romero Gómez S, Morales Tlalpan V. Critical Minireview: The Fate of tRNACys during Oxidative Stress in Bacillus subtilis. Biomolecules. 2017; 7(1):6. https://doi.org/10.3390/biom7010006
Chicago/Turabian StyleCampos Guillen, Juan, George H. Jones, Carlos Saldaña Gutiérrez, José Luis Hernández-Flores, Julio Alfonso Cruz Medina, José Humberto Valenzuela Soto, Sergio Pacheco Hernández, Sergio Romero Gómez, and Verónica Morales Tlalpan. 2017. "Critical Minireview: The Fate of tRNACys during Oxidative Stress in Bacillus subtilis" Biomolecules 7, no. 1: 6. https://doi.org/10.3390/biom7010006





