Functional Consequences of Differential O-glycosylation of MUC1, MUC4, and MUC16 (Downstream Effects on Signaling)
Abstract
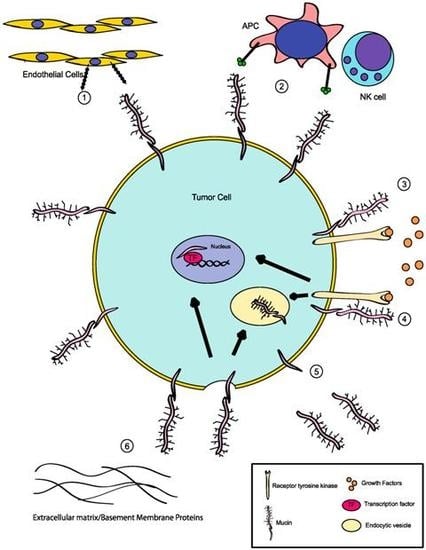
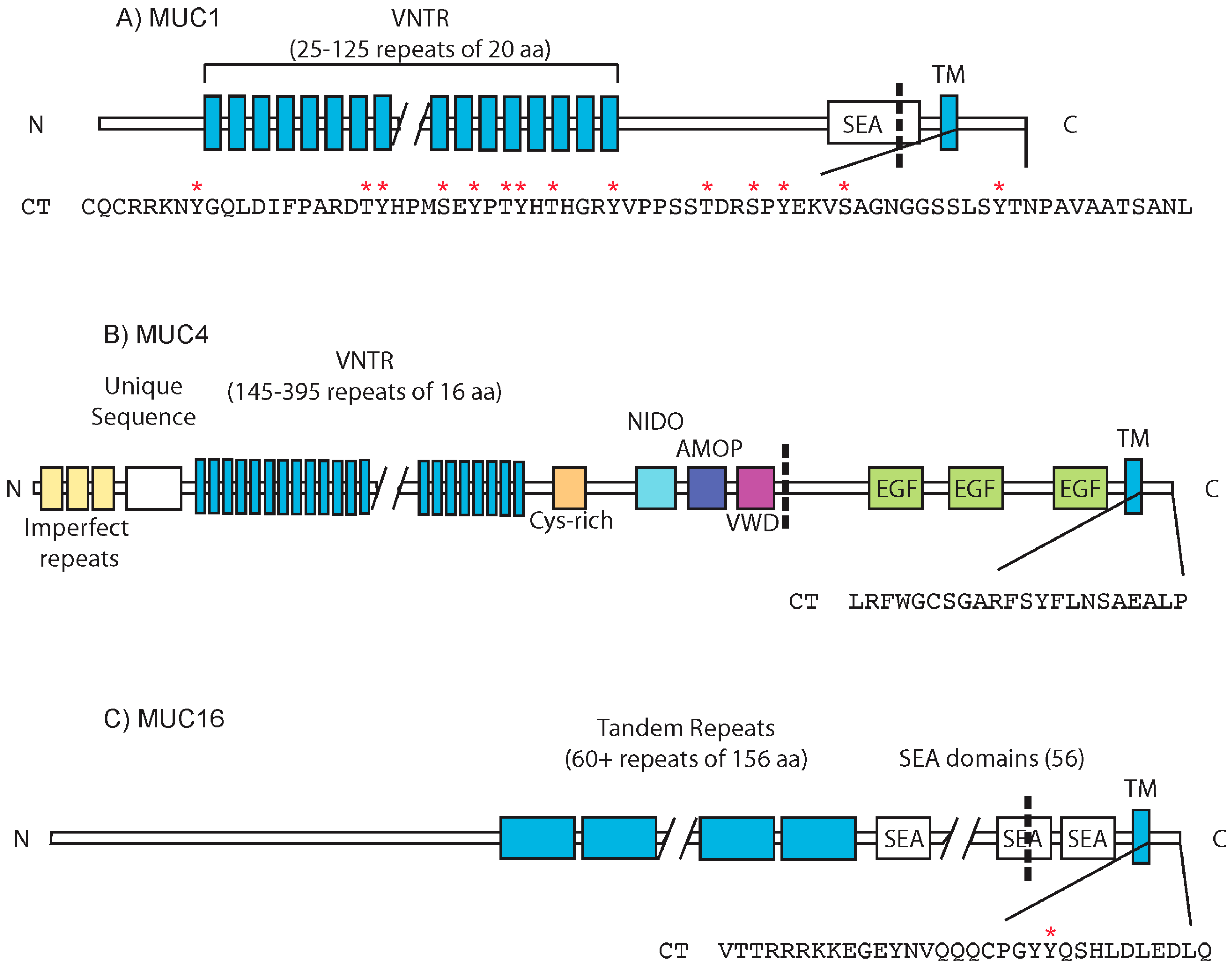
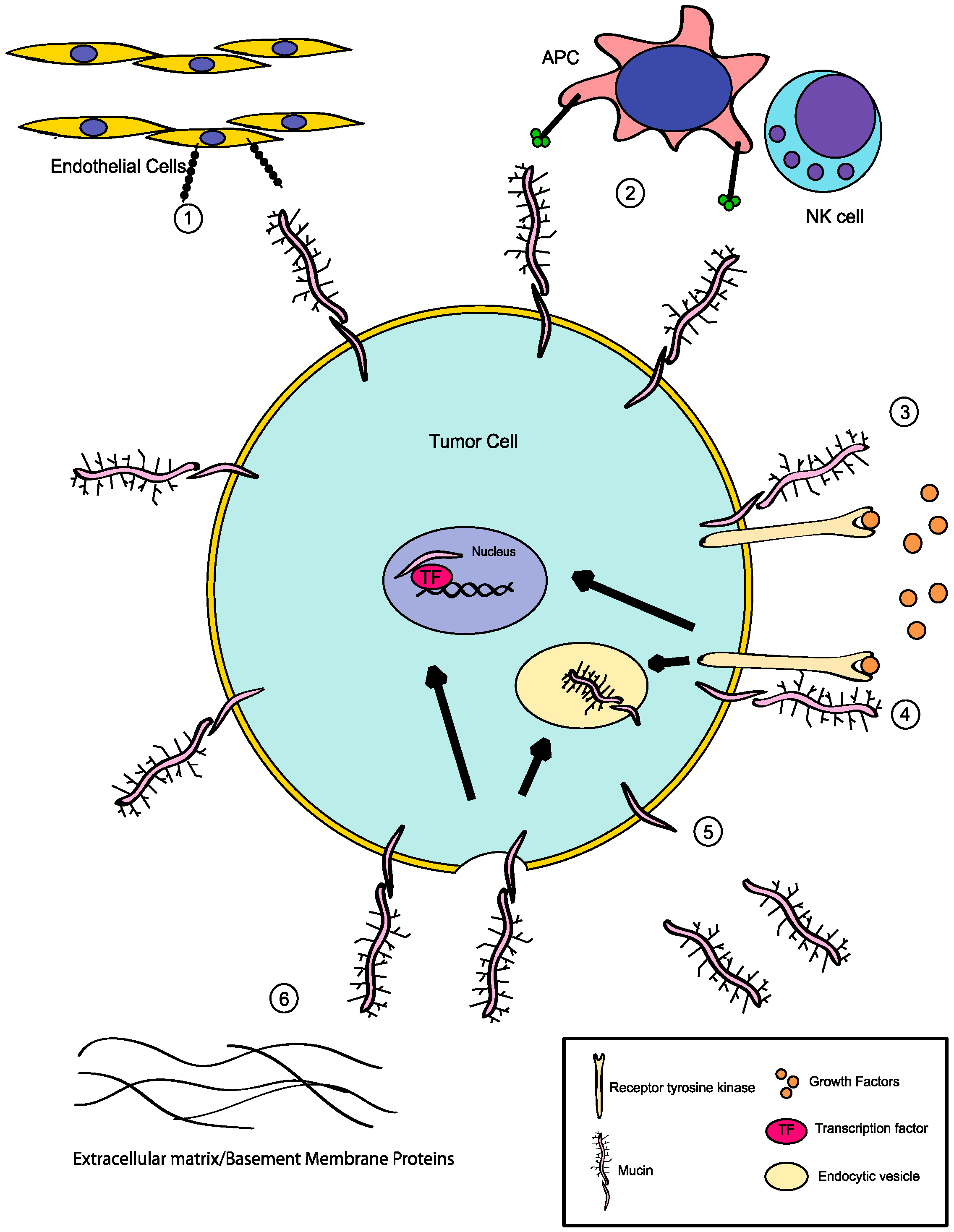
:1. Mucins: Structure and Function
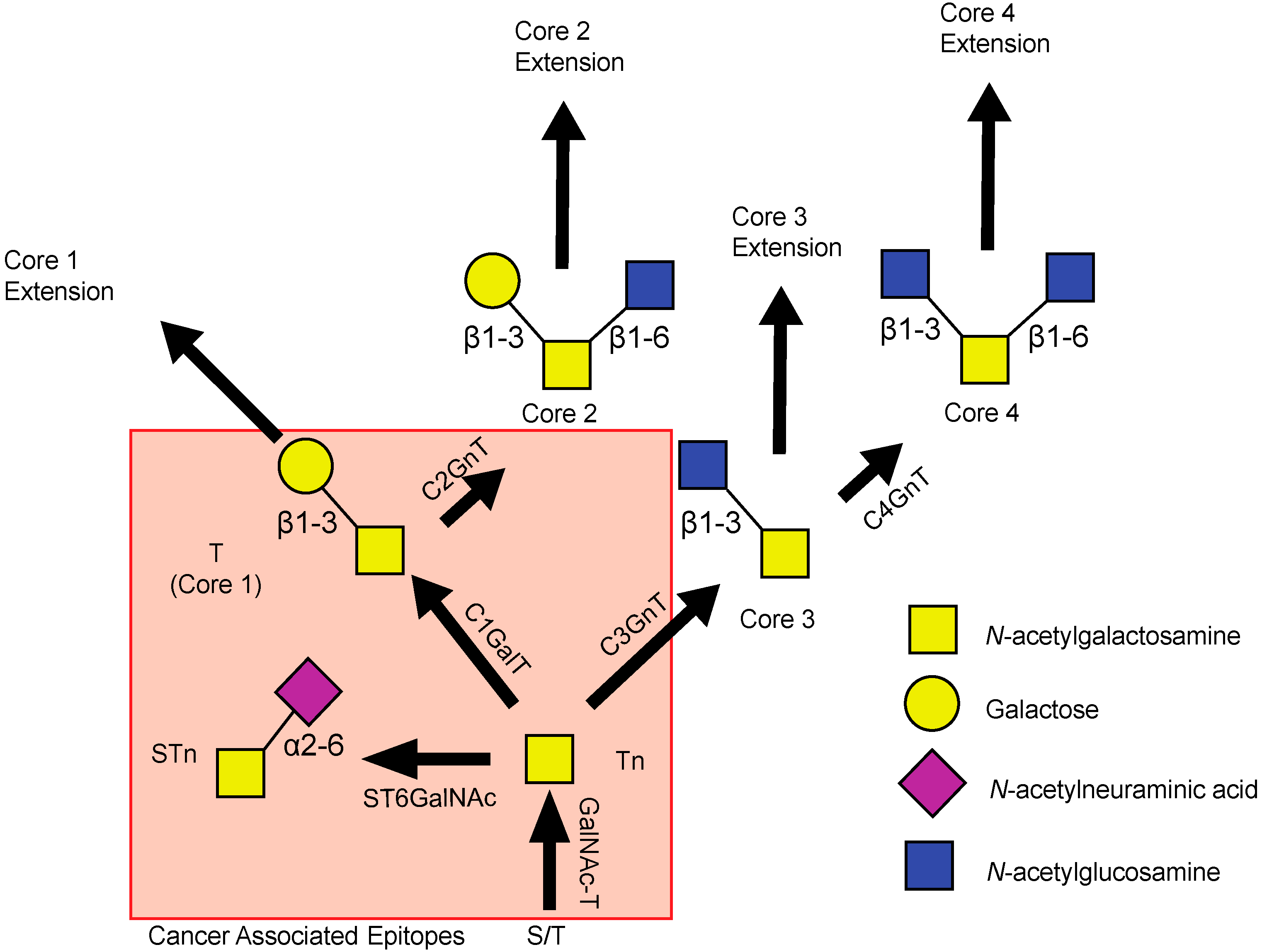
2. Deregulation of Mucin Expression and O-Type Glycosylation in Cancer
3. Signaling Through the Cytoplasmic Tail
3.1. MUC1
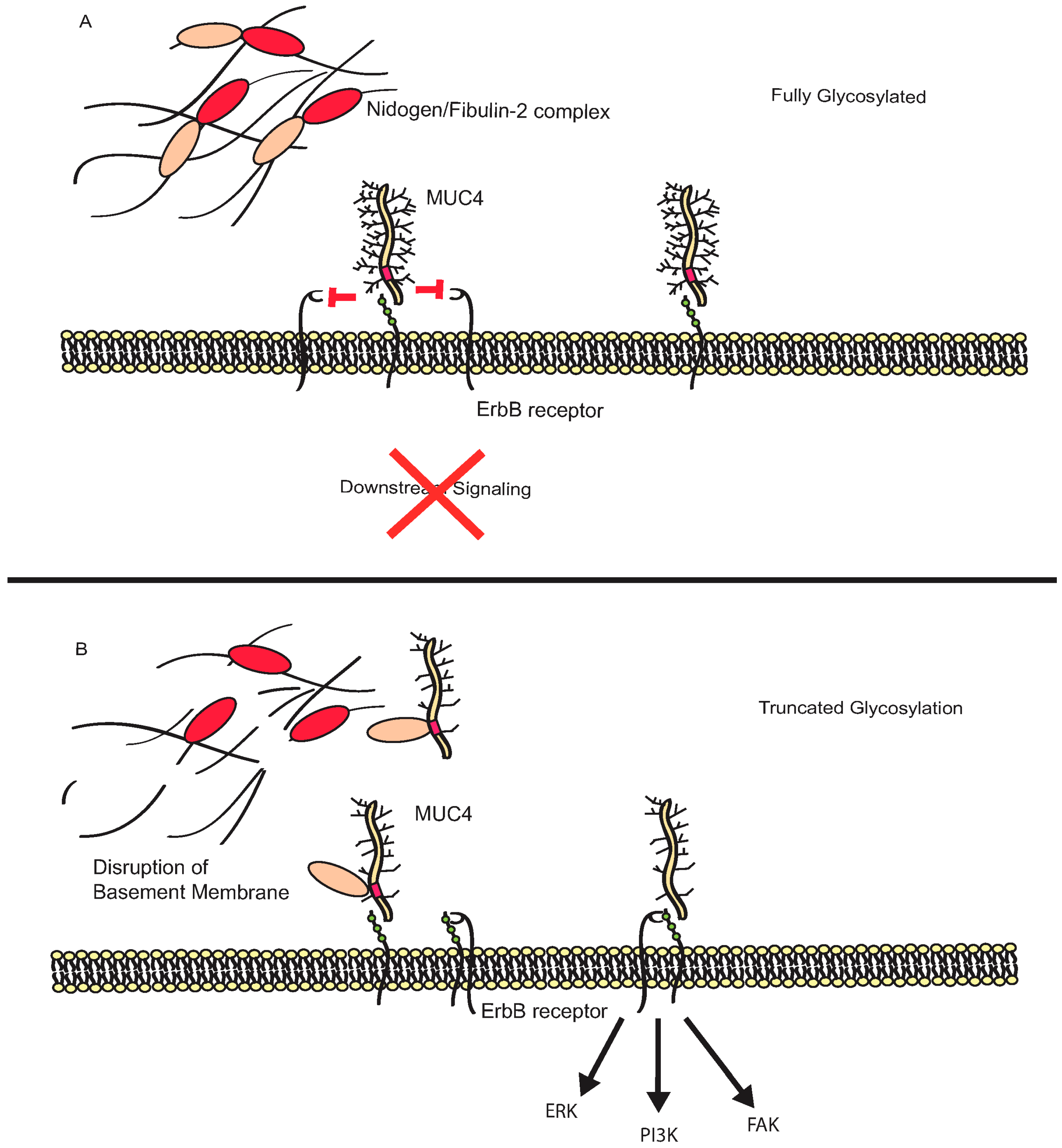
3.2. MUC4
3.3. MUC16
4. Additional Roles for Aberrant Glycosylation in Tumor Progression
5. Summary
Conflicts of Interest
References
- Hollingsworth, M.A.; Swanson, B.J. Mucins in cancer: Protection and control of the cell surface. Nat. Rev. Cancer 2004, 4, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Hattrup, C.L.; Gendler, S.J. Structure and Function of the Cell Surface (Tethered) Mucins. Annu. Rev. Physiol. 2008, 70, 431–457. [Google Scholar] [CrossRef] [PubMed]
- Thornton, D.J.; Rousseau, K.; McGuckin, M.A. Structure and Function of the Polymeric Mucins in Airways Mucus. Annu. Rev. Physiol. 2008, 70, 459–486. [Google Scholar] [CrossRef] [PubMed]
- Argueso, P.; Gipson, I.K. Epithelial Mucins of the Ocular Surface: Structure, Biosynthesis and Function. Exp. Eye Res. 2001, 73, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Seregni, E.; Botti, C.; Massaron, S.; Lombardo, C.; Capobianco, A.; Bogni, A.; Bombardieri, E. Structure, function and gene expression of epithelial mucins. Tumori 1997, 83, 625–632. [Google Scholar] [PubMed]
- Andrianifahanana, M.; Moniaux, N.; Batra, S.K. Regulation of mucin expression: Mechanistic aspects and implications for cancer and inflammatory diseases. Biochim. Biophys. Acta 2006, 1765, 189–222. [Google Scholar] [CrossRef] [PubMed]
- Voynow, J.A.; Gendler, S.J.; Rose, M.C. Regulation of mucin genes in chronic inflammatory airway diseases. Am. J. Respir. Cell Mol. Biol. 2006, 34, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Hollingsworth, M.A. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006, 16, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Timpte, C.S.; Eckhardt, A.E.; Abernethy, J.L.; Hill, R.L. Porcine Submaxillary Gland Apomucin Contains Tandemly Repeated, Identical Sequences of 81 Residues. J. Biol. Chem. 1988, 263, 1081–1088. [Google Scholar] [PubMed]
- Gupta, R.; Jentoft, N. Subunit structure of porcine submaxillary mucin. Biochemistry 1989, 28, 6114–6121. [Google Scholar] [CrossRef] [PubMed]
- Carlstedt, I.; Sheehan, J.K.; Corfield, A.P.; Gallagher, J.T. Mucous glycoproteins: A gel of a problem. Essays Biochem. 1985, 20, 40–76. [Google Scholar] [PubMed]
- McDermott, K.M.; Crocker, P.R.; Harris, A.; Burdick, M.D.; Hinoda, Y.; Hayashi, T.; Imai, K.; Hollingsworth, M.A. Overexpression of MUC1 reconfigures the binding properties of tumor cells. Int. J. Cancer 2001, 94, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Bennett, E.P.; Mandel, U.; Clausen, H.; Gerken, T.A.; Fritz, T.A.; Tabak, L.A. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology 2012, 22, 736–756. [Google Scholar] [CrossRef] [PubMed]
- Carraway, K.L.; Hull, S.R. O-glycosylation pathway for mucin-type glycoproteins. BioEssays 1989, 10, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Hanisch, F.G. O-Glycosylation of the Mucin Type. Biol. Chem. 2001, 382, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.H.; Kolarich, D.; Packer, N.H. Mucin-type O-glycosylation--putting the pieces together. FEBS J. 2010, 277, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Clausen, H.; Bennett, E.P. A family of UDP-GalNAc: polypeptide N-acetylgalactosaminyl-transferases control the initiation of mucin-type O-linked glycosylation. Glycobiology 1996, 6, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.W.; Radhakrishnan, P. Mucin O-Glycan Branching Enzymes: structure, Function, and Gene Regulation. Adv. Exp. Med. Biol. 2011, 705, 465–492. [Google Scholar] [PubMed]
- Ju, T.; Brewer, K.; D’Souza, A.; Cummings, R.D.; Canfield, W.M. Cloning and Expression of Human Core 1 β1,3-Galactosyltransferase. J. Biol. Chem. 2002, 277, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Bierhuizen, M.F.; Fukuda, M. Expression cloning of a cDNA encoding UDP-GlcNAc:Galβ1-3-GalNAc-R (GlcNAc to GalNAc) β1-6GlcNAc transferase by gene transfer into CHO cells expressing polyoma large tumor antigen. Proc. Natl. Acad. Sci. USA 1992, 89, 9326–9330. [Google Scholar] [CrossRef] [PubMed]
- Schwientek, T.; Nomoto, M.; Levery, S.B.; Merkx, G.; van Kessel, A.G.; Bennett, E.P.; Hollingsworth, M.A.; Clausen, H. Control of O-Glycan Branch Formation. Molecular Cloning of Human cDNA Encoding a Novel β1,6-N-Acetylglucosaminyltransferase Forming Core 2 and Core 4. J. Biol. Chem. 1999, 274, 4504–4512. [Google Scholar] [CrossRef] [PubMed]
- Tarp, M.A.; Clausen, H. Mucin-type O-glycosylation and its potential use in drug and vaccine development. Biochim. Biophys. Acta 2008, 1780, 546–563. [Google Scholar] [CrossRef] [PubMed]
- Iwai, T.; Inaba, N.; Naundorf, A.; Zhang, Y.; Gotoh, M.; Iwasaki, H.; Kudo, T.; Togayachi, A.; Ishizuka, Y.; Nakanishi, H.; et al. Molecular Cloning and Characterization of a Novel UDP-GlcNAc:GalNAc-peptide β1,3-N-acetylglucosaminyltransferase (β3Gn-T6), an Enzyme Synthesizing the Core 3 Structure of O-Glycans. J. Biol. Chem. 2002, 277, 12802–12809. [Google Scholar] [CrossRef] [PubMed]
- Krishn, S.R.; Kaur, S.; Smith, L.M.; Johansson, S.L.; Jain, M.; Patel, A.; Gautam, S.K.; Hollingsworth, M.A.; Mandel, U.; Clausen, H.; et al. Mucins and associated glycan signatures in colon adenoma-carcinoma sequence: Prospective pathological implication(s) for early diagnosis of colon cancer. Cancer Lett. 2016, 374, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Gendler, S.J. MUC1, The Renaissance Molecule. J. Mammary Gland Biol. Neoplasia 2001, 6, 339–353. [Google Scholar] [CrossRef] [PubMed]
- Awaya, H.; Takeshima, Y.; Yamasaki, M.; Inai, K. Expression of MUC1, MUC2, MUC5AC, and MUC6 in Atypical Adenomatous Hyperplasia, Bronchioloalveolar Carcinoma, Adenocarcinoma With Mixed Subtypes, and Mucinous Bronchioloalveolar Carcinoma of the Lung. Am. J. Clin. Pathol. 2004, 121, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.P.; Chauhan, S.C.; Bafna, S.; Johansson, S.L.; Smith, L.M.; Moniaux, N.; Lin, M.F.; Batra, S.K. Aberrant expression of transmembrane mucins, MUC1 and MUC4, in human prostate carcinomas. Prostate 2006, 66, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Andrianifahanana, M.; Moniaux, N.; Schmied, B.M.; Ringel, J.; Friess, H.; Hollingsworth, M.A.; Buchler, M.W.; Aubert, J.P.; Batra, S.K. Mucin (MUC) Gene Expression in Human Pancreatic Adenocarcinoma and Chronic Pancreatitis: A potential role of MUC4 as a tumor marker of diagnostic significance. Clin. Cancer Res. 2001, 7, 4033–4040. [Google Scholar] [PubMed]
- Schwartz, P.E.; Chambers, S.K.; Chambers, J.T.; Gutmann, J.; Katopodis, N.; Foemmel, R. Circulating Tumor Markers in the Monitoring of Gynecologic Malignancies. Cancer 1987, 60, 353–361. [Google Scholar] [CrossRef]
- Yin, B.W.; Dnistrian, A.; Lloyd, K.O. Ovarian cancer antigen CA125 is encoded by the MUC16 mucin gene. Int. J. Cancer 2002, 98, 737–740. [Google Scholar] [CrossRef] [PubMed]
- Higashi, M.; Yokoyama, S.; Yamamoto, T.; Goto, Y.; Kitazono, I.; Hiraki, T.; Taguchi, H.; Hashimoto, S.; Fukukura, Y.; Koriyama, C.; et al. Mucin Expression in Endoscopic Ultrasound-Guided Fine-Needle Aspiration Specimens Is a Useful Prognostic Factor in Pancreatic Ductal Adenocarcinoma. Pancreas 2015, 44, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Haridas, D.; Chakraborty, S.; Ponnusamy, M.P.; Lakshmanan, I.; Rachagani, S.; Cruz, E.; Kumar, S.; Das, S.; Lele, S.M.; Anderson, J.M.; et al. Pathobiological Implications of MUC16 Expression in Pancreatic Cancer. PLoS ONE 2011, 6, e26839. [Google Scholar] [CrossRef] [PubMed]
- Remmers, N.; Anderson, J.M.; Linde, E.M.; DiMaio, D.J.; Lazenby, A.J.; Wandall, H.H.; Mandel, U.; Clausen, H.; Yu, F.; Hollingsworth, M.A. Aberrant Expression of Mucin Core Proteins and O-Linked Glycans Associated with Progression of Pancreatic Cancer. Clin. Cancer Res. 2013, 19, 1981–1993. [Google Scholar] [CrossRef] [PubMed]
- Hinoda, Y.; Ikematsu, Y.; Horinochi, M.; Sato, S.; Yamamoto, K.; Nakano, T.; Fukui, M.; Suehiro, Y.; Hamanaka, Y.; Nishikawa, Y.; et al. Increased expression of MUC1 in advanced pancreatic cancer. J. Gastroenterol. 2003, 38, 1162–1166. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wang, X.; Lu, S.M.; Chen, C.; Wang, J.; Zheng, Y.Y.; Ren, B.H.; Xu, L. Clinicopathological and prognostic significance of MUC4 expression in cancers: Evidence from meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 10274–10283. [Google Scholar] [PubMed]
- Ghosh, S.K.; Pantazopoulos, P.; Medarova, Z.; Moore, A. Expression of Underglycosylated MUC1 Antigen in Cancerous and Adjacent Normal Breast Tissues. Clin. Breast Cancer 2013, 13, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Rakha, E.A.; Boyce, R.W.; Abd El-Rehim, D.; Kurien, T.; Green, A.R.; Paish, E.C.; Robertson, J.F.; Ellis, I.O. Expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Mod. Pathol. 2005, 18, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, P.; Chakraborty, S.; Ponnusamy, M.P.; Lakshmanan, I.; Jain, M.; Batra, S.K. Mucins in the pathogenesis of breast cancer: Implications in diagnosis, prognosis and therapy. Biochim. Biophys. Acta 2011, 1815, 224–240. [Google Scholar] [CrossRef] [PubMed]
- Terada, T. An immunohistochemical study of primary signet-ring cell carcinoma of the stomach and colorectum: II. Expression of MUC1, MUC2, MUC5AC, and MUC6 in normal mucosa and in 42 cases. Int. J. Clin. Exp. Pathol. 2013, 6, 613–621. [Google Scholar] [PubMed]
- Kwon, K.Y.; Ro, J.Y.; Singhal, N.; Killen, D.E.; Sienko, A.; Allen, T.C.; Zander, D.S.; Barrios, R.; Haque, A.; Cagle, P.T. MUC4 Expression in Non-Small Cell Lung Carcinomas: Relationship to Tumor Histology and Patient Survival. Arch. Pathol. Lab. Med. 2007, 131, 593–598. [Google Scholar] [PubMed]
- Yin, B.W.; Lloyd, K.O. Molecular Cloning of the CA125 Ovarian Cancer Antigen: Identification as a New Mucin, MUC16. J. Biol. Chem. 2001, 276, 27371–27375. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, S.C.; Singh, A.P.; Ruiz, F.; Johansson, S.L.; Jain, M.; Smith, L.M.; Moniaux, N.; Batra, S.K. Aberrant expression of MUC4 in ovarian carcinoma: diagnostic significance alone and in combination with MUC1 and MUC16 (CA125). Mod. Pathol. 2006, 19, 1386–1394. [Google Scholar] [CrossRef] [PubMed]
- Osunkoya, A.O.; Adsay, N.V.; Cohen, C.; Epstein, J.I.; Smith, S.L. MUC2 expression in primary mucinous and nonmucinous adenocarcinoma of the prostate: An analysis of 50 cases on radical prostatectomy. Mod. Pathol. 2008, 21, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Hakomori, S. Aberrant Glycosylation in Cancer Cell Membranes as Focused on Glycolipids: Overview and Perspectives. Cancer Res. 1985, 45, 2405–2414. [Google Scholar] [PubMed]
- Springer, G.F. T and Tn, general carcinoma autoantigens. Science 1984, 224, 1198–1206. [Google Scholar] [CrossRef] [PubMed]
- Ju, T.; Lanneau, G.S.; Gautam, T.; Wang, Y.; Xia, B.; Stowell, S.R.; Willard, M.T.; Wang, W.; Xia, J.Y.; Zuna, R.E.; et al. Human Tumor Antigens Tn and Sialyl Tn Arise from Mutations in Cosmc. Cancer Res. 2008, 68, 1636–1646. [Google Scholar] [CrossRef] [PubMed]
- Ju, T.; Cummings, R.D. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 β3-galactosyltransferase. Proc. Natl. Acad. Sci. USA 2002, 99, 16613–16618. [Google Scholar] [CrossRef] [PubMed]
- Ju, T.; Aryal, R.P.; Stowell, C.J.; Cummings, R.D. Regulation of protein O-glycosylation by the endoplasmic reticulum-localized molecular chaperone Cosmc. J. Cell Biol. 2008, 182, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Ju, T.; Cummings, R.D. The Transmembrane Domain of the Molecular Chaperone Cosmc Directs Its Localization to the Endoplasmic Reticulum. J. Biol. Chem. 2011, 286, 11529–11542. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Du, Z.; Sun, X.; Shi, C.; Zhang, H.; Hu, T. Aberrant Cosmc genes result in Tn antigen expression in human colorectal carcinoma cell line HT-29. Int. J. Clin. Exp. Pathol. 2015, 8, 2590–2602. [Google Scholar] [PubMed]
- Hofmann, B.T.; Schluter, L.; Lange, P.; Mercanoglu, B.; Ewald, F.; Folster, A.; Picksak, A.S.; Harder, S.; El Gammal, A.T.; Grupp, K.; et al. Cosmc knockdown mediated aberrant O-glycosylation promotes oncogenic properties in pancreatic cancer. Mol. Cancer 2015, 14, 109. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, P.; Dabelsteen, S.; Madsen, F.B.; Francavilla, C.; Kopp, K.L.; Steentoft, C.; Vakhrushev, S.Y.; Olsen, J.V.; Hansen, L.; Bennett, E.P.; et al. Immature truncated O-glycophenotype of cancer directly induces oncogenic features. Proc. Natl. Acad. Sci. USA 2014, 111, E4066–4075. [Google Scholar] [CrossRef] [PubMed]
- Mi, R.; Song, L.; Wang, Y.; Ding, X.; Zeng, J.; Lehoux, S.; Aryal, R.P.; Wang, J.; Crew, V.K.; van Die, I.; et al. Epigenetic Silencing of the Chaperone Cosmc in Human Leukocytes Expressing Tn Antigen. J. Biol. Chem. 2012, 287, 41523–41533. [Google Scholar] [CrossRef] [PubMed]
- Schneider, F.; Kemmner, W.; Haensch, W.; Franke, G.; Gretschel, S.; Karsten, U.; Schlag, P.M. Overexpression of Sialyltransferase CMP-Sialic Acid:Galβ1,3GalNAc-R α6-Sialyltransferase Is Related to Poor Patient sSurvival in Human Colorectal Carcinomas. Cancer Res. 2001, 61, 4605–4611. [Google Scholar] [PubMed]
- Georgopoulou, N.; Breen, K.C. Overexpression of α2,3 sialyltransferase in neuroblastoma cells results in an upset in the glycosylation process. Glycoconj. J. 1999, 16, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Brockhausen, I.; Yang, J.M.; Burchell, J.; Whitehouse, C.; Taylor-Papadimitriou, J. Mechanisms Underlying Aberrant Glycosylation of MUC1 Mucin in Breast Cancer Cells. Eur. J. Biochem. 1995, 233, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, J.; Inoue, H.; Koide, S. Expression of α-1,3-Fucosyltransferase Type IV and VII Genes Is Related to Poor Prognosis in Lung Cancer. Cancer Res. 1996, 56, 325–329. [Google Scholar] [PubMed]
- Barthel, S.R.; Wiese, G.K.; Cho, J.; Opperman, M.J.; Hays, D.L.; Siddiqui, J.; Pienta, K.J.; Furie, B.; Dimitroff, C.J. Alpha 1,3 fucosyltransferases are master regulators of prostate cancer cell trafficking. Proc. Natl. Acad. Sci. USA 2009, 106, 19491–19496. [Google Scholar] [CrossRef] [PubMed]
- Hauselmann, I.; Borsig, L. Altered tumor-cell glycosylation promotes metastasis. Front. Oncol. 2014, 4, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwai, T.; Kudo, T.; Kawamoto, R.; Kubota, T.; Togayachi, A.; Hiruma, T.; Okada, T.; Kawamoto, T.; Morozumi, K.; Narimatsu, H. Core 3 synthase is down-regulated in colon carcinoma and profoundly suppresses the metastatic potential of carcinoma cells. Proc. Natl. Acad. Sci. USA 2005, 102, 4572–4577. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.C.; Chen, H.Y.; Huang, H.C.; Huang, J.; Liang, J.T.; Shen, T.L.; Lin, N.Y.; Ho, C.C.; Cho, I.M.; Hsu, S.M. C2GnT-M is downregulated in colorectal cancer and its re-expression causes growth inhibition of colon cancer cells. Oncogene 2006, 25, 3267–3276. [Google Scholar] [CrossRef] [PubMed]
- Gill, D.J.; Chia, J.; Senewiratne, J.; Bard, F. Regulation of O-glycosylation through Golgi-to-ER relocation of initiation enzymes. J. Cell Biol. 2010, 189, 843–858. [Google Scholar] [CrossRef] [PubMed]
- Gill, D.J.; Clausen, H.; Bard, F. Location, location, location: New insights into O-GalNAc protein glycosylation. Trends Cell Biol. 2011, 21, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Alving, K.; Peter-Katalinic, J.; Zachara, N.; Gooley, A.A.; Hanisch, F.G. High Density O-Glycosylation on Tandem Repeat Peptide from Secretory MUC1 of T47D Breast Cancer Cells. J. Biol. Chem. 1999, 274, 18165–18172. [Google Scholar] [CrossRef] [PubMed]
- Springer, G.F. Tn epitope (N-acetyl-d-galactosamine α-O-serine/threonine) density in primary breast carcinoma: A functional predictor of aggressiveness. Mol. Immunol. 1989, 26, 1–5. [Google Scholar] [CrossRef]
- Gerken, T.A.; Owens, C.L.; Pasumarthy, M. Site-specific Core 1 O-Glycosylation Pattern of the Porcine Submaxillary Gland Mucin Tandem Repeat. Evidence for The Modulation of Glycan Length by Peptide Sequence. J. Biol. Chem. 1998, 273, 26580–26588. [Google Scholar] [CrossRef] [PubMed]
- Yeatman, T.J. A renaissance for SRC. Nat. Rev. Cancer 2004, 4, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Hasemann, C.A.; Capra, J.D. High-level production of a functional immunoglobulin heterodimer in a baculovirus expression system. Proc. Natl. Acad. Sci. USA 1990, 87, 3942–3946. [Google Scholar] [CrossRef] [PubMed]
- Frenzel, A.; Hust, M.; Schirrmann, T. Expression of recombinant antibodies. Front. Immunol. 2013, 4, 217. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, P.; Grandgenett, P.M.; Mohr, A.M.; Bunt, S.K.; Yu, F.; Chowdhury, S.; Hollingsworth, M.A. Expression of core 3 synthase in human pancreatic cancer cells suppresses tumor growth and metastasis. Int. J. Cancer 2013, 133, 2824–2833. [Google Scholar] [CrossRef] [PubMed]
- Macao, B.; Johansson, D.G.; Hansson, G.C.; Hard, T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat. Struct. Mol. Biol. 2006, 13, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Levitin, F.; Stern, O.; Weiss, M.; Gil-Henn, C.; Ziv, R.; Prokocimer, Z.; Smorodinsky, N.I.; Rubinstein, D.B.; Wreschner, D.H. The MUC1 SEA Module Is a Self-cleaving Domain. J. Biol. Chem. 2005, 280, 33374–33386. [Google Scholar] [CrossRef] [PubMed]
- Palmai-Pallag, T.; Khodabukus, N.; Kinarsky, L.; Leir, S.H.; Sherman, S.; Hollingsworth, M.A.; Harris, A. The role of the SEA (sea urchin sperm protein, enterokinase and agrin) module in cleavage of membrane-tethered mucins. FEBS J. 2005, 272, 2901–2911. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Behrens, M.E.; Eggers, J.P.; Cerny, R.L.; Bailey, J.M.; Shanmugam, K.; Gendler, S.J.; Bennett, E.P.; Hollingsworth, M.A. Phosphorylation of MUC1 by Met Modulates Interaction with p53 and MMP1 Expression. J. Biol. Chem. 2008, 283, 26985–26995. [Google Scholar] [CrossRef] [PubMed]
- Behrens, M.E.; Grandgenett, P.M.; Bailey, J.M.; Singh, P.K.; Yi, C.H.; Yu, F.; Hollingsworth, M.A. The reactive tumor microenvironment: MUC1 signaling directly reprograms transcription of CTGF. Oncogene 2010, 29, 5667–5677. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, J.A.; Thompson, M.C.; Gardner, M.M.; Gendler, S.J. Transgenic MUC1 Interacts with Epidermal Growth Factor Receptor and Correlates with Mitogen-activated Protein Kinase Activation in the Mouse Mammary Gland. J. Biol. Chem. 2001, 276, 13057–13064. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Wen, Y.; Swanson, B.J.; Shanmugam, K.; Kazlauskas, A.; Cerny, R.L.; Gendler, S.J.; Hollingsworth, M.A. Platelet-Derived Growth Factor Receptor β-Mediated Phosphorylation of MUC1 Enhances Invasiveness in Pancreatic Adenocarcinoma Cells. Cancer Res. 2007, 67, 5201–5210. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Bharti, A.; Chen, D.; Gong, J.; Kufe, D. Interaction of Glycogen Synthase Kinase 3β with the DF3/MUC1 Carcinoma-Associated Antigen and β-Catenin. Mol. Cell. Biol. 1998, 18, 7216–7224. [Google Scholar] [CrossRef] [PubMed]
- Mertins, P.; Yang, F.; Liu, T.; Mani, D.R.; Petyuk, V.A.; Gillette, M.A.; Clauser, K.R.; Qiao, J.W.; Gritsenko, M.A.; Moore, R.J.; et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol. Cell. Proteom. 2014, 13, 1690–1704. [Google Scholar] [CrossRef] [PubMed]
- Rikova, K.; Guo, A.; Zeng, Q.; Possemato, A.; Yu, J.; Haack, H.; Nardone, J.; Lee, K.; Reeves, C.; Li, Y.; et al. Global Survey of Phosphotyrosine Signaling Identifies Oncogenic Kinases in Lung Cancer. Cell 2007, 131, 1190–1203. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.L.; Deng, X.; Huang, F.; Tucker, M.; Crosby, K.; Rimkunas, V.; Wang, Y.; Deng, G.; Zhu, L.; Tan, Z.; et al. Survey of Tyrosine Kinase Signaling Reveals ROS Kinase Fusions in Human Cholangiocarcinoma. PLoS ONE 2011, 6, e15640. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Kharbanda, S.; Kufe, D. Association of the DF3/MUC1 Breast Cancer Antigen with Grb2 and the Sos/Ras Exchange Protein. Cancer Res. 1995, 55, 4000–4003. [Google Scholar] [PubMed]
- Meerzaman, D.; Shapiro, P.S.; Kim, K.C. Involvement of the MAP kinase ERK2 in MUC1 mucin signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 2001, 281, L86–91. [Google Scholar] [PubMed]
- Trehoux, S.; Duchene, B.; Jonckheere, N.; Van Seuningen, I. The MUC1 oncomucin regulates pancreatic cancer cell biological properties and chemoresistance. Implication of p42–44 MAPK, Akt, Bcl-2 and MMP13 pathways. Biochem. Biophys. Res. Commun. 2015, 456, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Hanson, R.L.; Brown, R.B.; Steele, M.M.; Grandgenett, P.M.; Grunkemeyer, J.A.; Hollingsworth, M.A. Identification of FRA1 as a novel player in pancreatic cancer in cooperation with a MUC1: ERK signaling axis. Oncotarget 2016. [Google Scholar] [CrossRef] [PubMed]
- Bitler, B.G.; Goverdhan, A.; Schroeder, J.A. MUC1 regulates nuclear localization and function of the epidermal growth factor receptor. J. Cell Sci. 2010, 123, 1716–1723. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Xu, H.; Kufe, D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell 2005, 7, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Caffrey, T.C.; Steele, M.M.; Mohr, A.; Singh, P.K.; Radhakrishnan, P.; Kelly, D.L.; Wen, Y.; Hollingsworth, M.A. MUC1 regulates cyclin D1 gene expression through p120 catenin and β-catenin. Oncogenesis 2014, 3, e107. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kufe, D. The Human DF3/MUC1 Carcinoma-Associated Antigen Signals Nuclear Localization of the Catenin p120ctn. Biochem. Biophys. Res. Commun. 2001, 281, 440–443. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yi, C.; Wen, Y.; Radhakrishnan, P.; Tremayne, J.R.; Dao, T.; Johnson, K.R.; Hollingsworth, M.A. Interactions between MUC1 and p120 catenin regulate dynamic features of cell adhesion, motility, and metastasis. Cancer Res. 2014, 74, 1609–1620. [Google Scholar] [CrossRef] [PubMed]
- Kinlough, C.L.; Poland, P.A.; Bruns, J.B.; Harkleroad, K.L.; Hughey, R.P. MUC1 Membrane Trafficking Is Modulated by Multiple Interactions. J. Biol. Chem. 2004, 279, 53071–53077. [Google Scholar] [CrossRef] [PubMed]
- Kinlough, C.L.; McMahan, R.J.; Poland, P.A.; Bruns, J.B.; Harkleroad, K.L.; Stremple, R.J.; Kashlan, O.B.; Weixel, K.M.; Weisz, O.A.; Hughey, R.P. Recycling of MUC1 Is Dependent on Its Palmitoylation. J. Biol. Chem. 2006, 281, 12112–12122. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Lu, W.; Kai, H.; Kim, K.C. Phosphoinositide 3-kinase is activated by MUC1 but not responsible for MUC1-induced suppression of Toll-like receptor 5 signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007, 293, L686–692. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Li, Y.; Kufe, D. Protein Kinase C δ Regulates Function of the DF3/MUC1 Carcinoma Antigen in β-Catenin Signaling. J. Biol. Chem. 2002, 277, 17616–17622. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kuwahara, H.; Ren, J.; Wen, G.; Kufe, D. The c-Src Tyrosine Kinase Regulates Signaling of the Human DF3/MUC1 Carcinoma-Associated Antigen with GSK3β and β-Catenin. J. Biol. Chem. 2001, 276, 6061–6064. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Majhi, P.D.; Al-Mugotir, M.H.; Rachagani, S.; Sorgen, P.; Batra, S.K. Membrane proximal ectodomain cleavage of MUC16 occurs in the acidifying Golgi/post-Golgi compartments. Sci. Rep. 2015, 5, 9759. [Google Scholar] [CrossRef] [PubMed]
- Akita, K.; Tanaka, M.; Tanida, S.; Mori, Y.; Toda, M.; Nakada, H. CA125/MUC16 interacts with Src family kinases, and over-expression of its C-terminal fragment in human epithelial cancer cells reduces cell-cell adhesion. Eur. J. Cell Biol. 2013, 92, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Herzog, B.H.; Fu, J.; Sheng, M.; Bergstrom, K.; McDaniel, J.M.; Kondo, Y.; McGee, S.; Cai, X.; Li, P.; et al. Loss of Core 1-derived O-Glycans Decreases Breast Cancer Development in Mice. J. Biol. Chem. 2015, 290, 20159–20166. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.H.; Huang, M.J.; Chen, C.H.; Shyu, M.K.; Huang, J.; Hung, J.S.; Huang, C.S.; Huang, M.C. Up-regulation of C1GALT1 promotes breast cancer cell growth through MUC1-C signaling pathway. Oncotarget 2015, 6, 6123–6135. [Google Scholar] [CrossRef] [PubMed]
- Solatycka, A.; Owczarek, T.; Piller, F.; Piller, V.; Pula, B.; Wojciech, L.; Podhorska-Okolow, M.; Dziegiel, P.; Ugorski, M. MUC1 in human and murine mammary carcinoma cells decreases the expression of core 2 β1,6-N-acetylglucosaminyltransferase and β-galactoside α2,3-sialyltransferase. Glycobiology 2012, 22, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Cascio, S.; Farkas, A.M.; Hughey, R.P.; Finn, O.J. Altered glycosylation of MUC1 influences its association with CIN85: the role of this novel complex in cancer cell invasion and migration. Oncotarget 2013, 4, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
- Kowanetz, K.; Szymkiewicz, I.; Haglund, K.; Kowanetz, M.; Husnjak, K.; Taylor, J.D.; Soubeyran, P.; Engstrom, U.; Ladbury, J.E.; Dikic, I. Identification of a Novel Proline-Arginine Motif Involved in CIN85-dependent Clustering of Cbl and Down-regulation of Epidermal Growth Factor Receptors. J. Biol. Chem. 2003, 278, 39735–39746. [Google Scholar] [CrossRef] [PubMed]
- Kowanetz, K.; Husnjak, K.; Holler, D.; Kowanetz, M.; Soubeyran, P.; Hirsch, D.; Schmidt, M.H.; Pavelic, K.; De Camilli, P.; Randazzo, P.A.; et al. CIN85 Associates with Multiple Effectors Controlling Intracellular Trafficking of Epidermal Growth Factor Receptors. Mol. Biol. Cell 2004, 15, 3155–3166. [Google Scholar] [CrossRef] [PubMed]
- Kufe, D.W. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene 2013, 32, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Kharbanda, A.; Rajabi, H.; Jin, C.; Tchaicha, J.; Kikuchi, E.; Wong, K.K.; Kufe, D. Targeting the oncogenic MUC1-C protein inhibits mutant EGFR-mediated signaling and survival in non-small cell lung cancer cells. Clin. Cancer Res. 2014, 20, 5423–5434. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, G.; Yuan, H.; Wang, J.; Guo, Y.; Chen, T.; Zhai, R.; Shao, D.; Ni, W.; Tai, G. Mucin1 shifts Smad3 signaling from the tumor-suppressive pSmad3C/p21WAF1 pathway to the oncogenic pSmad3L/c-Myc pathway by activating JNK in human hepatocellular carcinoma cells. Oncotarget 2015, 6, 4253–4265. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, G.; Li, Q.; Wang, F.; Xie, F.; Zhai, R.; Guo, Y.; Chen, T.; Zhang, N.; Ni, W.; et al. Mucin1 promotes the migration and invasion of hepatocellular carcinoma cells via JNK-mediated phosphorylation of Smad2 at the C-terminal and linker regions. Oncotarget 2015, 6, 19264–19278. [Google Scholar] [CrossRef] [PubMed]
- Merlin, J.; Stechly, L.; de Beauce, S.; Monte, D.; Leteurtre, E.; van Seuningen, I.; Huet, G.; Pigny, P. Galectin-3 regulates MUC1 and EGFR cellular distribution and EGFR downstream pathways in pancreatic cancer cells. Oncogene 2011, 30, 2514–2525. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, J.T.; de Matos, A.J.; Santos, A.L.; Pinto, R.; Gomes, J.; Hespanhol, V.; Chammas, R.; Manninen, A.; Bernardes, E.S.; Albuquerque Reis, C.; et al. Sialylation regulates galectin-3/ligand interplay during mammary tumour progression - a case of targeted uncloaking. Int. J. Dev. Biol. 2011, 55, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.C.; Yegorova, S.; Pitteloud, J.P.; Chavaroche, A.E.; Andre, S.; Arda, A.; Minond, D.; Jimenez-Barbero, J.; Gabius, H.J.; Cudic, M. Thermodynamic Switch in Binding of Adhesion/Growth Regulatory Human Galectin-3 to Tumor-Associated TF Antigen (CD176) and MUC1 Glycopeptides. Biochemistry 2015, 54, 4462–4474. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.G.; Andrews, N.; Zhao, Q.; McKean, D.; Williams, J.F.; Connor, L.J.; Gerasimenko, O.V.; Hilkens, J.; Hirabayashi, J.; Kasai, K.; et al. Galectin-3 Interaction with Thomsen-Friedenreich Disaccharide on Cancer-associated MUC1 Causes Increased Cancer Cell Endothelial Adhesion. J. Biol. Chem. 2007, 282, 773–781. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Guo, X.; Nash, G.B.; Stone, P.C.; Hilkens, J.; Rhodes, J.M.; Yu, L.G. Circulating Galectin-3 Promotes Metastasis by Modifying MUC1 Localization on Cancer Cell Surface. Cancer Res. 2009, 69, 6799–6806. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Barclay, M.; Hilkens, J.; Guo, X.; Barrow, H.; Rhodes, J.M.; Yu, L.G. Interaction between circulating galectin-3 and cancer-associated MUC1 enhances tumour cell homotypic aggregation and prevents anoikis. Mol. Cancer 2010, 9, 154. [Google Scholar] [CrossRef] [PubMed]
- Altschuler, Y.; Kinlough, C.L.; Poland, P.A.; Bruns, J.B.; Apodaca, G.; Weisz, O.A.; Hughey, R.P. Clathrin-mediated Endocytosis of MUC1 Is Modulated by Its Glycosylation State. Mol. Biol. Cell 2000, 11, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Kufe, D.W. Functional targeting of the MUC1 oncogene in human cancers. Cancer Biol. Ther. 2009, 8, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Duraisamy, S.; Ramasamy, S.; Kharbanda, S.; Kufe, D. Distinct evolution of the human carcinoma-associated transmembrane mucins, MUC1, MUC4 and MUC16. Gene 2006, 373, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Escande, F.; Lemaitre, L.; Moniaux, N.; Batra, S.K.; Aubert, J.P.; Buisine, M.P. Genomic organization of MUC4 mucin gene. Towards the characterization of splice variants. Eur. J. Biochem. 2002, 269, 3637–3644. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, P.; Singh, A.P.; Batra, S.K. Structure, evolution, and biology of the MUC4 mucin. FASEB J. 2008, 22, 966–981. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, M.P.; Singh, A.P.; Jain, M.; Chakraborty, S.; Moniaux, N.; Batra, S.K. MUC4 activates HER2 signalling and enhances the motility of human ovarian cancer cells. Br. J. Cancer 2008, 99, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, P.; Lakshmanan, I.; Ponnusamy, M.P.; Chakraborty, S.; Jain, M.; Pai, P.; Smith, L.M.; Lele, S.M.; Batra, S.K. MUC4 Overexpression Augments Cell Migration and Metastasis through EGFR Family Proteins in Triple Negative Breast Cancer Cells. PLoS ONE 2013, 8, e54455. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, I.; Seshacharyulu, P.; Haridas, D.; Rachagani, S.; Gupta, S.; Joshi, S.; Guda, C.; Yan, Y.; Jain, M.; Ganti, A.K.; et al. Novel HER3/MUC4 oncogenic signaling aggravates the tumorigenic phenotypes of pancreatic cancer cells. Oncotarget 2015, 6, 21085–21099. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, M.P.; Seshacharyulu, P.; Vaz, A.; Dey, P.; Batra, S.K. MUC4 stabilizes HER2 expression and maintains the cancer stem cell population in ovarian cancer cells. J. Ovarian Res. 2011, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, P.; Singh, A.P.; Chakraborty, S.; Chauhan, S.C.; Bafna, S.; Meza, J.L.; Singh, P.K.; Hollingsworth, M.A.; Mehta, P.P.; Batra, S.K. MUC4 Mucin Interacts with and Stabilizes the HER2 Oncoprotein in Human Pancreatic Cancer Cells. Cancer Res. 2008, 68, 2065–2070. [Google Scholar] [CrossRef] [PubMed]
- Jonckheere, N.; Perrais, M.; Mariette, C.; Batra, S.K.; Aubert, J.P.; Pigny, P.; Van Seuningen, I. A role for human MUC4 mucin gene, the ErbB2 ligand, as a target of TGF-β in pancreatic carcinogenesis. Oncogene 2004, 23, 5729–5738. [Google Scholar] [CrossRef] [PubMed]
- Jepson, S.; Komatsu, M.; Haq, B.; Arango, M.E.; Huang, D.; Carraway, C.A.; Carraway, K.L. MUC4/sialomucin complex, the intramembrane ErbB2 ligand, induces specific phosphorylation of ErbB2 and enhances expression of p27(kip), but does not activate mitogen-activated kinase or protein kinase B/Akt pathways. Oncogene 2002, 21, 7524–7532. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Jepson, S.; Arango, M.E.; Carothers Carraway, C.A.; Carraway, K.L. MUC4/sialomucin complex, an intramembrane modulator of ErbB2/HER2/Neu, potentiates primary tumor growth and suppresses apoptosis in a xenotransplanted tumor. Oncogene 2001, 20, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.; Friedlander, E.; Tanner, M.; Kapanen, A.I.; Carraway, K.L.; Isola, J.; Jovin, T.M. Decreased Accessibility and Lack of Activation of ErbB2 in JIMT-1, a Herceptin-Resistant, MUC4-Expressing Breast Cancer Cell Line. Cancer Res. 2005, 65, 473–482. [Google Scholar] [PubMed]
- Workman, H.C.; Sweeney, C.; Carraway, K.L., 3rd. The Membrane Mucin Muc4 Inhibits Apoptosis Induced by Multiple Insults via ErbB2-Dependent and ErbB2-Independent Mechanisms. Cancer Res. 2009, 69, 2845–2852. [Google Scholar] [CrossRef] [PubMed]
- Zhi, X.; Tao, J.; Xie, K.; Zhu, Y.; Li, Z.; Tang, J.; Wang, W.; Xu, H.; Zhang, J.; Xu, Z. MUC4-induced nuclear translocation of β-catenin: A novel mechanism for growth, metastasis and angiogenesis in pancreatic cancer. Cancer Lett. 2014, 346, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Zhu, Y.; Xie, K.; Zhang, X.; Zhi, X.; Wang, W.; Li, Z.; Zhang, Q.; Wang, L.; Wang, J.; et al. The role of the AMOP domain in MUC4/Y-promoted tumour angiogenesis and metastasis in pancreatic cancer. J. Exp. Clin. Cancer Res. 2016, 35, 91. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, P.; Singh, A.P.; Moniaux, N.; Senapati, S.; Chakraborty, S.; Meza, J.L.; Batra, S.K. MUC4 Mucin Potentiates Pancreatic Tumor Cell Proliferation, Survival, and Invasive Properties and Interferes with Its Interaction to Extracellular Matrix Proteins. Mol. Cancer Res. 2007, 5, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Senapati, S.; Gnanapragassam, V.S.; Moniaux, N.; Momi, N.; Batra, S.K. Role of MUC4-NIDO domain in the MUC4-mediated metastasis of pancreatic cancer cells. Oncogene 2012, 31, 3346–3356. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, T.J.; Beard, J.B.; Underwood, L.J.; Shigemasa, K. The CA 125 Gene: A Newly Discovered Extension of the Glycosylated N-terminal Domain Doubles the Size of This Extracellular Superstructure. Tumour Biol. 2002, 23, 154–169. [Google Scholar] [PubMed]
- Haridas, D.; Ponnusamy, M.P.; Chugh, S.; Lakshmanan, I.; Seshacharyulu, P.; Batra, S.K. MUC16: molecular analysis and its functional implications in benign and malignant conditions. FASEB J. 2014, 28, 4183–4199. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, T.J.; Beard, J.B.; Underwood, L.J.; Dennis, R.A.; Santin, A.D.; York, L. The CA 125 Gene: An Extracellular Superstructure Dominated by Repeat Sequences. Tumour Biol. 2001, 22, 348–366. [Google Scholar] [PubMed]
- Rao, T.D.; Tian, H.; Ma, X.; Yan, X.; Thapi, S.; Schultz, N.; Rosales, N.; Monette, S.; Wang, A.; Hyman, D.M.; et al. Expression of the Carboxy-Terminal Portion of MUC16/CA125 Induces Transformation and Tumor Invasion. PLoS ONE 2015, 10, e0126633. [Google Scholar] [CrossRef] [PubMed]
- Bruney, L.; Conley, K.C.; Moss, N.M.; Liu, Y.; Stack, M.S. Membrane-type I matrix metalloproteinase-dependent ectodomain shedding of mucin16/CA-125 on ovarian cancer cells modulates adhesion and invasion of peritoneal mesothelium. Biol. Chem. 2014, 395, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, I.; Ponnusamy, M.P.; Das, S.; Chakraborty, S.; Haridas, D.; Mukhopadhyay, P.; Lele, S.M.; Batra, S.K. MUC16 induced rapid G2/M transition via interactions with JAK2 for increased proliferation and anti-apoptosis in breast cancer cells. Oncogene 2012, 31, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Rachagani, S.; Torres-Gonzalez, M.P.; Lakshmanan, I.; Majhi, P.D.; Smith, L.M.; Wagner, K.U.; Batra, S.K. Carboxyl-terminal domain of MUC16 imparts tumorigenic and metastatic functions through nuclear translocation of JAK2 to pancreatic cancer cells. Oncotarget 2015, 6, 5772–5787. [Google Scholar] [CrossRef] [PubMed]
- Theriault, C.; Pinard, M.; Comamala, M.; Migneault, M.; Beaudin, J.; Matte, I.; Boivin, M.; Piche, A.; Rancourt, C. MUC16 (CA125) regulates epithelial ovarian cancer cell growth, tumorigenesis and metastasis. Gynecol. Oncol. 2011, 121, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Comamala, M.; Pinard, M.; Theriault, C.; Matte, I.; Albert, A.; Boivin, M.; Beaudin, J.; Piche, A.; Rancourt, C. Downregulation of cell surface CA125/MUC16 induces epithelial-to-mesenchymal transition and restores EGFR signalling in NIH:OVCAR3 ovarian carcinoma cells. Br. J. Cancer 2011, 104, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Giannakouros, P.; Comamala, M.; Matte, I.; Rancourt, C.; Piche, A. MUC16 mucin (CA125) regulates the formation of multicellular aggregates by altering β-catenin signaling. Am. J. Cancer Res. 2015, 5, 219–230. [Google Scholar] [PubMed]
- Burford, B.; Gentry-Maharaj, A.; Graham, R.; Allen, D.; Pedersen, J.W.; Nudelman, A.S.; Blixt, O.; Fourkala, E.O.; Bueti, D.; Dawnay, A.; et al. Autoantibodies to MUC1 glycopeptides cannot be used as a screening assay for early detection of breast, ovarian, lung or pancreatic cancer. Br. J. Cancer 2013, 108, 2045–2055. [Google Scholar] [CrossRef] [PubMed]
- Wandall, H.H.; Blixt, O.; Tarp, M.A.; Pedersen, J.W.; Bennett, E.P.; Mandel, U.; Ragupathi, G.; Livingston, P.O.; Hollingsworth, M.A.; Taylor-Papadimitriou, J.; et al. Cancer Biomarkers Defined by Autoantibody Signatures to Aberrant O-Glycopeptide Epitopes. Cancer Res. 2010, 70, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Pathangey, L.B.; Lakshminarayanan, V.; Suman, V.J.; Pockaj, B.A.; Mukherjee, P.; Gendler, S.J. Aberrant Glycosylation of Anchor-Optimized MUC1 Peptides Can Enhance Antigen Binding Affinity and Reverse Tolerance to Cytotoxic T Lymphocytes. Biomolecules 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Posey, A.D., Jr.; Schwab, R.D.; Boesteanu, A.C.; Steentoft, C.; Mandel, U.; Engels, B.; Stone, J.D.; Madsen, T.D.; Schreiber, K.; Haines, K.M.; et al. Engineered CAR T Cells Targeting the Cancer-Associated Tn-Glycoform of the Membrane Mucin MUC1 Control Adenocarcinoma. Immunity 2016, 44, 1444–1454. [Google Scholar] [CrossRef] [PubMed]
- Rachagani, S.; Torres, M.P.; Moniaux, N.; Batra, S.K. Current status of mucins in the diagnosis and therapy of cancer. Biofactors 2009, 35, 509–527. [Google Scholar] [CrossRef] [PubMed]
- Felder, M.; Kapur, A.; Gonzalez-Bosquet, J.; Horibata, S.; Heintz, J.; Albrecht, R.; Fass, L.; Kaur, J.; Hu, K.; Shojaei, H.; et al. MUC16 (CA125): Tumor biomarker to cancer therapy, a work in progress. Mol. Cancer 2014, 13, 129. [Google Scholar] [CrossRef] [PubMed]
- Kawa, S.; Kato, M.; Oguchi, H.; Hsue, G.L.; Kobayashi, T.; Koiwai, T.; Tokoo, M.; Furuta, S.; Ichikawa, T.; Kanai, M. Clinical Evaluation of Pancreatic Cancer-Associated Mucin Expressing CA19–9, CA50, SPAN-1, sialyl SSEA-1, and Dupan-2. Scand. J. Gastroenterol. 1992, 27, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Cooper, E.H.; Forbes, M.A.; Taylor, M. An evaluation of DUPAN-2 in pancreatic cancer and gastrointestinal disease. Br. J. Cancer 1990, 62, 1004–1005. [Google Scholar] [CrossRef] [PubMed]
- Beatson, R.; Maurstad, G.; Picco, G.; Arulappu, A.; Coleman, J.; Wandell, H.H.; Clausen, H.; Mandel, U.; Taylor-Papadimitriou, J.; Sletmoen, M.; et al. The Breast Cancer-Associated Glycoforms of MUC1, MUC1-Tn and sialyl-Tn, Are Expressed in COSMC Wild-Type Cells and Bind the C-type Lectin MGL. PLoS ONE 2015, 10, e0125994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Napoletano, C.; Rughetti, A.; Agervig Tarp, M.P.; Coleman, J.; Bennett, E.P.; Picco, G.; Sale, P.; Denda-Nagai, K.; Irimura, T.; Mandel, U.; et al. Tumor-Associated Tn-MUC1 Glycoform Is Internalized through the Macrophage Galactose-Type C-Type Lectin and Delivered to the HLA Class I and II Compartments in Dendritic Cells. Cancer Res. 2007, 67, 8358–8367. [Google Scholar] [CrossRef] [PubMed]
- Saeland, E.; van Vliet, S.J.; Backstrom, M.; van den Berg, V.C.; Geijtenbeek, T.B.; Meijer, G.A.; van Kooyk, Y. The C-type lectin MGL expressed by dendritic cells detects glycan changes on MUC1 in colon carcinoma. Cancer Immunol. Immunother. 2007, 56, 1225–1236. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, S.J.; Gringhuis, S.I.; Geijtenbeek, T.B.; van Kooyk, Y. Regulation of effector T cells by antigen-presenting cells via interaction of the C-type lectin MGL with CD45. Nat. Immunol. 2006, 7, 1200–1208. [Google Scholar] [CrossRef] [PubMed]
- Gubbels, J.A.; Felder, M.; Horibata, S.; Belisle, J.A.; Kapur, A.; Holden, H.; Petrie, S.; Migneault, M.; Rancourt, C.; Connor, J.P.; et al. MUC16 provides immune protection by inhibiting synapse formation between NK and ovarian tumor cells. Mol. Cancer 2010, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Menon, B.B.; Kaiser-Marko, C.; Spurr-Michaud, S.; Tisdale, A.S.; Gipson, I.K. Suppression of Toll-like receptor-mediated innate immune responses at the ocular surface by the membrane-associated mucins MUC1 and MUC16. Mucosal Immunol. 2015, 8, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Madsen, C.B.; Lavrsen, K.; Steentoft, C.; Vester-Christensen, M.B.; Clausen, H.; Wandall, H.H.; Pedersen, A.E. Glycan Elongation Beyond the Mucin Associated Tn Antigen Protects Tumor Cells from Immune-Mediated Killing. PLoS ONE 2013, 8, e72413. [Google Scholar] [CrossRef]
- Geng, Y.; Yeh, K.; Takatani, T.; King, M.R. Three to Tango: MUC1 as a Ligand for Both E-Selectin and ICAM-1 in the Breast Cancer Metastatic Cascade. Front. Oncol. 2012, 2, 76. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Dallas, M.R.; Balzer, E.M.; Konstantopoulos, K. Mucin 16 is a functional selectin ligand on pancreatic cancer cells. FASEB J. 2012, 26, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Coupland, L.A.; Parish, C.R. Platelets, Selectins, and the Control of Tumor Metastasis. Semin. Oncol. 2014, 41, 422–434. [Google Scholar] [CrossRef] [PubMed]

| Mucin | Cancer | Reference |
|---|---|---|
| MUC1, MUC4, MUC5AC, MUC6, MUC16 | Pancreatic ductal adenocarcinoma | Remmers et al. [33] |
| Hinoda et al. [34] | ||
| Huang et al. [35] | ||
| Haridas et al. [32] | ||
| Higashi et al. [31] | ||
| MUC1, MUC2, MUC3, MUC4, MUC5AC, MUC5B, MUC6 | Breast cancer | Ghosh et al. [36] |
| Rakha et al. [37] | ||
| Mukhopadhyay et al. [38] | ||
| MUC1, MUC2, MUC4, MUC5AC, MUC5B, MUC6, MUC17 | Colon cancer | Terada et al. [39] |
| Krishn et al. [24] | ||
| MUC1, MUC2, MUC4, MUC5AC, MUC6 | Lung cancer | Awaya et al. [26] |
| Kwon et al. [40] | ||
| MUC1, MUC4, MUC16 | Ovarian cancer | Yin et al. [41] |
| Chauhan et al. [42] | ||
| MUC1, MUC2, MUC4 | Prostate cancer | Singh et al. [27] |
| Osunkoya et al. [43] |
| Mucin | Phosphorylation Site | Function (if Known) | Reference |
|---|---|---|---|
| MUC1 | N/A | Rikova et al. [80] | |
| MUC1 | N/A | Gu et al. [81] | |
| MUC1 | Interaction with AP1G, AP2M1, PIK3R1, p53 and increased cell motility | Singh et al. [77] | |
| Kinlough et al. [91,92] | |||
| Kato et al. [93] | |||
| MUC1 | N/A | Mertins et al. [79] | |
| MUC1 | N/A | Gu et al. [81] | |
| MUC1 | N/A | Mertins et al. [79] | |
| MUC1 | N/A | Rikova et al. [80] | |
| Gu et al. [81] | |||
| MUC1 | N/A | Gu et al. [81] | |
| MUC1 | N/A | Rikova et al. [80] | |
| Gu et al. [81] | |||
| MUC1 | Growth altered | Ren et al. [94] | |
| MUC1 | Inhibits interaction with β-catenin | Li et al. [95] | |
| MUC1 | Promotes association with β-catenin and inhibits association with GSK3β | Singh et al. [77] | |
| Li et al. [95] | |||
| MUC1 | N/A | Mertins et al. [79] | |
| MUC1 | Interaction with GRB2 | Kinlough et al. [91] | |
| MUC4 contains no confirmed sites of phosphorylation in the cytoplasmic tail | |||
| MUC16 | May regulate turnover | Das et al. [96] | |
| Akita et al. [97] | |||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanson, R.L.; Hollingsworth, M.A. Functional Consequences of Differential O-glycosylation of MUC1, MUC4, and MUC16 (Downstream Effects on Signaling). Biomolecules 2016, 6, 34. https://doi.org/10.3390/biom6030034
Hanson RL, Hollingsworth MA. Functional Consequences of Differential O-glycosylation of MUC1, MUC4, and MUC16 (Downstream Effects on Signaling). Biomolecules. 2016; 6(3):34. https://doi.org/10.3390/biom6030034
Chicago/Turabian StyleHanson, Ryan L., and Michael A. Hollingsworth. 2016. "Functional Consequences of Differential O-glycosylation of MUC1, MUC4, and MUC16 (Downstream Effects on Signaling)" Biomolecules 6, no. 3: 34. https://doi.org/10.3390/biom6030034