Enzymes for N-Glycan Branching and Their Genetic and Nongenetic Regulation in Cancer
Abstract
:1. Introduction
2. Enzymes for Branch Synthesis and Their Involvement in Cancer
2.1. GnT-III (MGAT3)
2.2. GnT-V and GnT-IX (GnT-Vb) (MGAT5 and MGAT5B)
2.3. GnT-IVs (MGAT4A-C)
2.4. Fut8 (FUT8)
3. Regulation of N-glycan Branches in Cancer by Several Pathways
3.1. Genetics
3.2. Epigenetics
3.3. Nucleotide Sugar Metabolism
3.4. Subcellular Localization and the Cleavage of Branching Enzymes
4. Potential Clinical Applications as Drug Targets
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Fuc | Fucose |
| Fut | Fucosyltransferase |
| Gal | Galactose |
| GlcNAc | N-Acetylglucosamine |
| GnT | N-Acetylglucosaminyltransferase |
| miRNA | microRNA |
| Sia | Sialic acid |
References
- Moremen, K.W.; Tiemeyer, M.; Nairn, A.V. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012, 13, 448–462. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, N.; Kizuka, Y. Glycans and cancer: Role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv. Cancer Res. 2015, 126, 11–51. [Google Scholar]
- Hakomori, S. Glycosylation defining cancer malignancy: New wine in an old bottle. Proc. Natl. Acad. Sci. USA 2002, 99, 10231–10233. [Google Scholar] [CrossRef]
- Stanley, P.; Schachter, H.; Taniguchi, N. N-glycans. In Essentials of glycobiology, 2nd ed.; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor: New York, NY, USA, 2009. [Google Scholar]
- Taniguchi, N.; Korekane, H. Branched N-glycans and their implications for cell adhesion, signaling and clinical applications for cancer biomarkers and in therapeutics. BMB Rep. 2011, 44, 772–781. [Google Scholar] [CrossRef]
- Ikeda, Y.; Ihara, H.; Tsukamoto, H.; Gu, J.; Taniguchi, N. Mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase (mgat3); β1,4-N-acetylglucosaminyltransferase III (gnt-III, glcnact-III). In Handbook of Glycosyltransferases and Related Genes; Taniguchi, N., Honke, K., Fukuda, M., Narimatsu, H., Yamaguchi, Y., Angata, T., Eds.; Springer Japan: Tokyo, Japan, 2014; pp. 209–222. [Google Scholar]
- Zhao, Y.; Sato, Y.; Isaji, T.; Fukuda, T.; Matsumoto, A.; Miyoshi, E.; Gu, J.; Taniguchi, N. Branched N-glycans regulate the biological functions of integrins and cadherins. FEBS J. 2008, 275, 1939–1948. [Google Scholar] [CrossRef] [PubMed]
- Schachter, H. Biosynthetic controls that determine the branching and microheterogeneity of protein-bound oligosaccharides. Biochem. Cell Biol. 1986, 64, 163–181. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Nishikawa, A.; Tsuruoka, N.; Ohno, M.; Yamaguchi, N.; Kangawa, K.; Taniguchi, N. Purification and characterization of udp-N-acetylglucosamine: Alpha-6-d-mannoside beta 1-6n-acetylglucosaminyltransferase (N-acetylglucosaminyltransferase v) from a human lung cancer cell line. J. Biochem. 1993, 113, 614–619. [Google Scholar] [PubMed]
- Re, S.; Miyashita, N.; Yamaguchi, Y.; Sugita, Y. Structural diversity and changes in conformational equilibria of biantennary complex-type N-glycans in water revealed by replica-exchange molecular dynamics simulation. Biophys. J. 2011, 101, L44–L46. [Google Scholar] [CrossRef] [PubMed]
- Nagae, M.; Kanagawa, M.; Morita-Matsumoto, K.; Hanashima, S.; Kizuka, Y.; Taniguchi, N.; Yamaguchi, Y. Atomic visualization of a flipped-back conformation of bisected glycans bound to specific lectins. Sci. Rep. 2016, 6, 22973. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Nishiura, T.; Nishikawa, A.; Miura, R.; Taniguchi, N. Structural heterogeneity of sugar chains in immunoglobulin G. Conformation of immunoglobulin g molecule and substrate specificities of glycosyltransferases. J. Biol. Chem. 1990, 265, 6009–6018. [Google Scholar] [PubMed]
- Taniguchi, N.; Yoshimura, M.; Miyoshi, E.; Ihara, Y.; Nishikawa, A.; Fujii, S. Remodeling of cell surface glycoproteins by N-acetylglucosaminyltransferase iii gene transfection: Modulation of metastatic potentials and down regulation of hepatitis b virus replication. Glycobiology 1996, 6, 691–694. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, S. Control of glycoprotein synthesis. Udp-glcnac:Glycopeptide beta 4-N-acetylglucosaminyltransferase iii, an enzyme in hen oviduct which adds glcnac in beta 1–4 linkage to the beta-linked mannose of the trimannosyl core of N-glycosyl oligosaccharides. J. Biol. Chem. 1982, 257, 10235–10242. [Google Scholar] [PubMed]
- Gleeson, P.A.; Schachter, H. Control of glycoprotein synthesis. J. Biol. Chem. 1983, 258, 6162–6173. [Google Scholar] [PubMed]
- Nishikawa, A.; Ihara, Y.; Hatakeyama, M.; Kangawa, K.; Taniguchi, N. Purification, cdna cloning, and expression of udp-N-acetylglucosamine: Beta-d-mannoside beta-1,4n-acetylglucosaminyltransferase iii from rat kidney. J. Biol. Chem. 1992, 267, 18199–18204. [Google Scholar] [PubMed]
- Miyoshi, E.; Nishikawa, A.; Ihara, Y.; Gu, J.; Sugiyama, T.; Hayashi, N.; Fusamoto, H.; Kamada, T.; Taniguchi, N. N-acetylglucosaminyltransferase III and V messenger rna levels in lec rats during hepatocarcinogenesis. Cancer Res. 1993, 53, 3899–3902. [Google Scholar] [PubMed]
- Abbott, K.L.; Nairn, A.V.; Hall, E.M.; Horton, M.B.; McDonald, J.F.; Moremen, K.W.; Dinulescu, D.M.; Pierce, M. Focused glycomic analysis of the N-linked glycan biosynthetic pathway in ovarian cancer. Proteomics 2008, 8, 3210–3220. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, M.; Nishikawa, A.; Ihara, Y.; Nishiura, T.; Nakao, H.; Kanayama, Y.; Matuzawa, Y.; Taniguchi, N. High expression of udp-N-acetylglucosamine: Beta-d mannoside beta-1,4-N-acetylglucosaminyltransferase iii (gnt-iii) in chronic myelogenous leukemia in blast crisis. Int. J. Cancer 1995, 60, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, M.; Nishikawa, A.; Ihara, Y.; Taniguchi, S.; Taniguchi, N. Suppression of lung metastasis of b16 mouse melanoma by N-acetylglucosaminyltransferase iii gene transfection. Proc. Natl. Acad. Sci. USA 1995, 92, 8754–8758. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Aglipay, J.A.; Bernstein, J.D.; Goswami, S.; Stanley, P. The bisecting glcnac on N-glycans inhibits growth factor signaling and retards mammary tumor progression. Cancer Res. 2010, 70, 3361–3371. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, M.; Ihara, Y.; Matsuzawa, Y.; Taniguchi, N. Aberrant glycosylation of e-cadherin enhances cell-cell binding to suppress metastasis. J. Biol. Chem. 1996, 271, 13811–13815. [Google Scholar] [PubMed]
- Pinho, S.S.; Reis, C.A.; Paredes, J.; Magalhaes, A.M.; Ferreira, A.C.; Figueiredo, J.; Xiaogang, W.; Carneiro, F.; Gartner, F.; Seruca, R. The role of N-acetylglucosaminyltransferase iii and v in the post-transcriptional modifications of e-cadherin. Hum. Mol. Genet. 2009, 18, 2599–2608. [Google Scholar] [CrossRef] [PubMed]
- Iijima, J.; Zhao, Y.; Isaji, T.; Kameyama, A.; Nakaya, S.; Wang, X.; Ihara, H.; Cheng, X.; Nakagawa, T.; Miyoshi, E.; et al. Cell-cell interaction-dependent regulation of N-acetylglucosaminyltransferase iii and the bisected N-glycans in ge11 epithelial cells. Involvement of e-cadherin-mediated cell adhesion. J. Biol. Chem. 2006, 281, 13038–13046. [Google Scholar] [CrossRef] [PubMed]
- Isaji, T.; Gu, J.; Nishiuchi, R.; Zhao, Y.; Takahashi, M.; Miyoshi, E.; Honke, K.; Sekiguchi, K.; Taniguchi, N. Introduction of bisecting glcnac into integrin alpha5beta1 reduces ligand binding and down-regulates cell adhesion and cell migration. J. Biol. Chem. 2004, 279, 19747–19754. [Google Scholar] [CrossRef] [PubMed]
- Kariya, Y.; Kato, R.; Itoh, S.; Fukuda, T.; Shibukawa, Y.; Sanzen, N.; Sekiguchi, K.; Wada, Y.; Kawasaki, N.; Gu, J. N-glycosylation of laminin-332 regulates its biological functions. A novel function of the bisecting glcnac. J. Biol. Chem. 2008, 283, 33036–33045. [Google Scholar] [CrossRef] [PubMed]
- Kitada, T.; Miyoshi, E.; Noda, K.; Higashiyama, S.; Ihara, H.; Matsuura, N.; Hayashi, N.; Kawata, S.; Matsuzawa, Y.; Taniguchi, N. The addition of bisecting N-acetylglucosamine residues to e-cadherin down-regulates the tyrosine phosphorylation of beta-catenin. J. Biol. Chem. 2001, 276, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Seruca, R.; Gartner, F.; Yamaguchi, Y.; Gu, J.; Taniguchi, N.; Reis, C.A. Modulation of e-cadherin function and dysfunction by N-glycosylation. Cell Mol. Life Sci. 2011, 68, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Isaji, T.; Lu, Y.; Gu, W.; Kondo, M.; Fukuda, T.; Du, Y.; Gu, J. Roles of N-acetylglucosaminyltransferase iii in epithelial-to-mesenchymal transition induced by transforming growth factor beta1 (TGF-beta1) in epithelial cell lines. J. Biol. Chem. 2012, 287, 16563–16574. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Oliveira, P.; Cabral, J.; Carvalho, S.; Huntsman, D.; Gartner, F.; Seruca, R.; Reis, C.A.; Oliveira, C. Loss and recovery of mgat3 and GnT-III mediated e-cadherin N-glycosylation is a mechanism involved in epithelial-mesenchymal-epithelial transitions. PLoS ONE 2012, 7, e33191. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Isaji, T.; Im, S.; Fukuda, T.; Kameyama, A.; Gu, J. Expression of N-acetylglucosaminyltransferase iii suppresses alpha2,3 sialylation and its distinctive functions in cell migration are attributed to alpha2,6 sialylation levels. J. Biol. Chem. 2016, 291, 5708–5720. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Tang, J.; Rogler, C.E.; Stanley, P. Reduced hepatocyte proliferation is the basis of retarded liver tumor progression and liver regeneration in mice lacking N-acetylglucosaminyltransferase III. Cancer Res. 2003, 63, 7753–7759. [Google Scholar] [PubMed]
- Yoshimura, M.; Ihara, Y.; Ohnishi, A.; Ijuhin, N.; Nishiura, T.; Kanakura, Y.; Matsuzawa, Y.; Taniguchi, N. Bisecting N-acetylglucosamine on k562 cells suppresses natural killer cytotoxicity and promotes spleen colonization. Cancer Res. 1996, 56, 412–418. [Google Scholar] [PubMed]
- Kizuka, Y.; Kitazume, S.; Fujinawa, R.; Saito, T.; Iwata, N.; Saido, T.C.; Nakano, M.; Yamaguchi, Y.; Hashimoto, Y.; Staufenbiel, M.; et al. An aberrant sugar modification of bace1 blocks its lysosomal targeting in Alzheimer's disease. EMBO Mol. Med. 2015, 7, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Kizuka, Y.; Nakano, M.; Kitazume, S.; Saito, T.; Saido, T.C.; Taniguchi, N. Bisecting glcnac modification stabilizes bace1 protein under oxidative stress conditions. Biochem. J. 2015, 473, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Dennis, J.W.; Taniguchi, N.; Pierce, M. Mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase (mgat5). In Handbook of Glycosyltransferases and Related Genes; Taniguchi, N., Honke, K., Fukuda, M., Narimatsu, H., Yamaguchi, Y., Angata, T., Eds.; Springer Japan: Tokyo, Japan, 2014; pp. 233–246. [Google Scholar]
- Shoreibah, M.; Perng, G.S.; Adler, B.; Weinstein, J.; Basu, R.; Cupples, R.; Wen, D.; Browne, J.K.; Buckhaults, P.; Fregien, N.; et al. Isolation, characterization, and expression of a cdna encoding N-acetylglucosaminyltransferase V. J. Biol. Chem. 1993, 268, 15381–15385. [Google Scholar] [PubMed]
- Shoreibah, M.G.; Hindsgaul, O.; Pierce, M. Purification and characterization of rat kidney udp-N-acetylglucosamine: Alpha-6-d-mannoside beta-1,6-N-acetylglucosaminyltransferase. J. Biol. Chem. 1992, 267, 2920–2927. [Google Scholar] [PubMed]
- Dennis, J.W.; Nabi, I.R.; Demetriou, M. Metabolism, cell surface organization, and disease. Cell 2009, 139, 1229–1241. [Google Scholar] [CrossRef] [PubMed]
- Boscher, C.; Dennis, J.W.; Nabi, I.R. Glycosylation, galectins and cellular signaling. Curr. Opin. Cell Biol. 2011, 23, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Ihara, S.; Miyoshi, E.; Ko, J.H.; Murata, K.; Nakahara, S.; Honke, K.; Dickson, R.B.; Lin, C.Y.; Taniguchi, N. Prometastatic effect of N-acetylglucosaminyltransferase v is due to modification and stabilization of active matriptase by adding beta 1-6 glcnac branching. J. Biol. Chem. 2002, 277, 16960–16967. [Google Scholar] [CrossRef] [PubMed]
- Ihara, S.; Miyoshi, E.; Nakahara, S.; Sakiyama, H.; Ihara, H.; Akinaga, A.; Honke, K.; Dickson, R.B.; Lin, C.Y.; Taniguchi, N. Addition of beta1-6 glcnac branching to the oligosaccharide attached to asn 772 in the serine protease domain of matriptase plays a pivotal role in its stability and resistance against trypsin. Glycobiology 2004, 14, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, S.; Catarino, T.A.; Dias, A.M.; Kato, M.; Almeida, A.; Hessling, B.; Figueiredo, J.; Gartner, F.; Sanches, J.M.; Ruppert, T.; et al. Preventing e-cadherin aberrant N-glycosylation at asn-554 improves its critical function in gastric cancer. Oncogene 2016, 35, 1619–1631. [Google Scholar] [CrossRef]
- Guo, H.B.; Johnson, H.; Randolph, M.; Pierce, M. Regulation of homotypic cell-cell adhesion by branched N-glycosylation of N-cadherin extracellular ec2 and ec3 domains. J. Biol. Chem. 2009, 284, 34986–34997. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Figueiredo, J.; Cabral, J.; Carvalho, S.; Dourado, J.; Magalhaes, A.; Gartner, F.; Mendonfa, A.M.; Isaji, T.; Gu, J.; et al. E-cadherin and adherens-junctions stability in gastric carcinoma: Functional implications of glycosyltransferases involving N-glycan branching biosynthesis, N-acetylglucosaminyltransferases iii and v. Biochim. Biophys. Acta 2013, 1830, 2690–2700. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Miyoshi, E.; Sasai, K.; Nakano, N.; Eguchi, H.; Honke, K.; Taniguchi, N. A secreted type of beta 1,6-N-acetylglucosaminyltransferase v (GnT-V) induces tumor angiogenesis without mediation of glycosylation: A novel function of gnt-v distinct from the original glycosyltransferase activity. J. Biol. Chem. 2002, 277, 17002–17008. [Google Scholar] [CrossRef] [PubMed]
- Yanagi, M.; Aoyagi, Y.; Suda, T.; Mita, Y.; Asakura, H. N-acetylglucosaminyltransferase V as a possible aid for the evaluation of tumor invasiveness in patients with hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2001, 16, 1282–1289. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, K.; Marth, J.D. Glycosylation in cellular mechanisms of health and disease. Cell 2006, 126, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, P.H.; Voss, M.; Haug-Kroper, M.; Schroder, B.; Schepers, U.; Brase, S.; Haass, C.; Lichtenthaler, S.F.; Fluhrer, R. Secretome analysis identifies novel signal peptide peptidase-like 3 (SPPL3) substrates and reveals a role of SPPL3 in multiple Golgi glycosylation pathways. Mol. Cell. Proteomics 2015, 14, 1584–1598. [Google Scholar] [CrossRef] [PubMed]
- Voss, M.; Kunzel, U.; Higel, F.; Kuhn, P.H.; Colombo, A.; Fukumori, A.; Haug-Kroper, M.; Klier, B.; Grammer, G.; Seidl, A.; et al. Shedding of glycan-modifying enzymes by signal peptide peptidase-like 3 (SPPL3) regulates cellular N-glycosylation. EMBO J. 2014, 33, 2890–2905. [Google Scholar] [CrossRef] [PubMed]
- Granovsky, M.; Fata, J.; Pawling, J.; Muller, W.J.; Khokha, R.; Dennis, J.W. Suppression of tumor growth and metastasis in mgat5-deficient mice. Nat. Med. 2000, 6, 306–312. [Google Scholar] [PubMed]
- Guo, H.B.; Johnson, H.; Randolph, M.; Nagy, T.; Blalock, R.; Pierce, M. Specific posttranslational modification regulates early events in mammary carcinoma formation. Proc. Natl. Acad. Sci. USA 2010, 107, 21116–21121. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Miyoshi, E.; Kawaguchi, N.; Shaker, M.; Ito, Y.; Taniguchi, N.; Tsujimoto, M.; Matsuura, N. The implication of N-acetylglucosaminyltransferase v expression in gastric cancer. Pathobiology 2008, 75, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, Y.; Dosaka-Akita, H.; Miyoshi, E.; Shindoh, M.; Miyamoto, M.; Kinoshita, I.; Miyazaki, H.; Itoh, T.; Kondo, S.; Nishimura, M.; et al. Expression of N-acetylglucosaminyltransferase V in the development of human esophageal cancers: Immunohistochemical data from carcinomas and nearby noncancerous lesions. Oncology 2005, 69, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Nagy, T.; Pierce, M. Post-translational glycoprotein modifications regulate colon cancer stem cells and colon adenoma progression in apc(min/+) mice through altered wnt receptor signaling. J. Biol. Chem. 2014, 289, 31534–31549. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Miyoshi, E.; Sakon, M.; Takeda, T.; Noda, K.; Tsujimoto, M.; Ito, S.; Honda, H.; Takemura, F.; Wakasa, K.; et al. Elevated expression of UDP-N-acetylglucosamine: Alphamannoside beta1,6 N-acetylglucosaminyltransferase is an early event in hepatocarcinogenesis. Int. J. Cancer 2001, 91, 631–637. [Google Scholar] [CrossRef]
- Handerson, T.; Camp, R.; Harigopal, M.; Rimm, D.; Pawelek, J. Beta1,6-branched oligosaccharides are increased in lymph node metastases and predict poor outcome in breast carcinoma. Clin. Cancer Res. 2005, 11, 2969–2973. [Google Scholar] [CrossRef] [PubMed]
- Inamori, K.; Endo, T.; Ide, Y.; Fujii, S.; Gu, J.; Honke, K.; Taniguchi, N. Molecular cloning and characterization of human gnt-ix, a novel beta1,6-N-acetylglucosaminyltransferase that is specifically expressed in the brain. J. Biol. Chem. 2003, 278, 43102–43109. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, M.; Alvarez-Manilla, G.; Kamar, M.; Lee, I.; Lee, J.K.; Troupe, K.; Zhang, W.; Osawa, M.; Pierce, M. A novel beta(1,6)-N-acetylglucosaminyltransferase v (GnT-Vb)(1). FEBS Lett. 2003, 554, 515–519. [Google Scholar] [CrossRef]
- Alvarez-Manilla, G.; Troupe, K.; Fleming, M.; Martinez-Uribe, E.; Pierce, M. Comparison of the substrate specificities and catalytic properties of the sister N-acetylglucosaminyltransferases, GnT-V and GnT-Vb (IX). Glycobiology 2010, 20, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Inamori, K.; Endo, T.; Gu, J.; Matsuo, I.; Ito, Y.; Fujii, S.; Iwasaki, H.; Narimatsu, H.; Miyoshi, E.; Honke, K.; et al. N-acetylglucosaminyltransferase ix acts on the glcnac beta 1,2-man alpha 1-ser/thr moiety, forming a 2,6-branched structure in brain O-mannosyl glycan. J. Biol. Chem. 2004, 279, 2337–2340. [Google Scholar] [CrossRef] [PubMed]
- Inamori, K.-I.; Pierce, M.; Taniguchi, N. Mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme b (mgat5b). In Handbook of Glycosyltransferases and Related Genes; Taniguchi, N., Honke, K., Fukuda, M., Narimatsu, H., Yamaguchi, Y., Angata, T., Eds.; Springer Japan: Tokyo, Japan, 2014; pp. 247–255. [Google Scholar]
- Kizuka, Y.; Kitazume, S.; Okahara, K.; Villagra, A.; Sotomayor, E.M.; Taniguchi, N. Epigenetic regulation of a brain-specific glycosyltransferase N-acetylglucosaminyltransferase-ix (GnT-IX) by specific chromatin modifiers. J. Biol. Chem. 2014, 289, 11253–11261. [Google Scholar] [CrossRef] [PubMed]
- Kizuka, Y.; Kitazume, S.; Yoshida, M.; Taniguchi, N. Brain-specific expression of N-acetylglucosaminyltransferase ix (GnT-IX) is regulated by epigenetic histone modifications. J. Biol. Chem. 2011, 286, 31875–31884. [Google Scholar] [CrossRef] [PubMed]
- Kanekiyo, K.; Inamori, K.; Kitazume, S.; Sato, K.; Maeda, J.; Higuchi, M.; Kizuka, Y.; Korekane, H.; Matsuo, I.; Honke, K.; et al. Loss of branched o-mannosyl glycans in astrocytes accelerates remyelination. J. Neurosci. 2013, 33, 10037–10047. [Google Scholar] [CrossRef] [PubMed]
- Abbott, K.L.; Matthews, R.T.; Pierce, M. Receptor tyrosine phosphatase beta (RPTPbeta) activity and signaling are attenuated by glycosylation and subsequent cell surface galectin-1 binding. J. Biol. Chem. 2008, 283, 33026–33035. [Google Scholar] [CrossRef] [PubMed]
- Lommel, M.; Winterhalter, P.R.; Willer, T.; Dahlhoff, M.; Schneider, M.R.; Bartels, M.F.; Renner-Muller, I.; Ruppert, T.; Wolf, E.; Strahl, S. Protein O-mannosylation is crucial for e-cadherin-mediated cell adhesion. Proc. Natl. Acad. Sci. USA 2013, 110, 21024–21029. [Google Scholar] [CrossRef] [PubMed]
- Vester-Christensen, M.B.; Halim, A.; Joshi, H.J.; Steentoft, C.; Bennett, E.P.; Levery, S.B.; Vakhrushev, S.Y.; Clausen, H. Mining the O-mannose glycoproteome reveals cadherins as major O-mannosylated glycoproteins. Proc. Natl. Acad. Sci. USA 2013, 110, 21018–21023. [Google Scholar] [CrossRef] [PubMed]
- Endo, T. Glycobiology of alpha-dystroglycan and muscular dystrophy. J. Biochem. 2015, 157, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kanagawa, M.; Kobayashi, K.; Tajiri, M.; Manya, H.; Kuga, A.; Yamaguchi, Y.; Akasaka-Manya, K.; Furukawa, J.; Mizuno, M.; Kawakami, H.; et al. Identification of a post-translational modification with ribitol-phosphate and its defect in muscular dystrophy. Cell Rep. 2016, 14, 2209–2223. [Google Scholar] [CrossRef] [PubMed]
- Inamori, K.; Gu, J.; Ohira, M.; Kawasaki, A.; Nakamura, Y.; Nakagawa, T.; Kondo, A.; Miyoshi, E.; Nakagawara, A.; Taniguchi, N. High expression of N-acetylglucosaminyltransferase V in favorable neuroblastomas: Involvement of its effect on apoptosis. FEBS Lett. 2006, 580, 627–632. [Google Scholar] [CrossRef]
- Lange, T.; Ullrich, S.; Muller, I.; Nentwich, M.F.; Stubke, K.; Feldhaus, S.; Knies, C.; Hellwinkel, O.J.; Vessella, R.L.; Abramjuk, C.; et al. Human prostate cancer in a clinically relevant xenograft mouse model: Identification of beta(1,6)-branched oligosaccharides as a marker of tumor progression. Clin. Cancer Res. 2012, 18, 1364–1373. [Google Scholar] [CrossRef]
- Vojta, A.; Samarzija, I.; Bockor, L.; Zoldos, V. Glyco-genes change expression in cancer through aberrant methylation. Biochim. Biophys. Acta. 2016. [Google Scholar] [CrossRef]
- Ohtsubo, K.; Taniguchi, N. Mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme a,b (mgat4a,b). In Handbook of Glycosyltransferases and Related Genes; Taniguchi, N., Honke, K., Fukuda, M., Narimatsu, H., Yamaguchi, Y., Angata, T., Eds.; Springer Japan: Tokyo, Japan, 2014; pp. 223–232. [Google Scholar]
- Oguri, S.; Minowa, M.T.; Ihara, Y.; Taniguchi, N.; Ikenaga, H.; Takeuchi, M. Purification and characterization of udp-N-acetylglucosamine: Alpha1,3-d-mannoside beta1,4-N-acetylglucosaminyltransferase (N-acetylglucosaminyltransferase-iv) from bovine small intestine. J. Biol. Chem. 1997, 272, 22721–22727. [Google Scholar] [CrossRef] [PubMed]
- Minowa, M.T.; Oguri, S.; Yoshida, A.; Hara, T.; Iwamatsu, A.; Ikenaga, H.; Takeuchi, M. Cdna cloning and expression of bovine udp-N-acetylglucosamine: Alpha1, 3-d-mannoside beta1,4-N-acetylglucosaminyltransferase iv. J. Biol. Chem. 1998, 273, 11556–11562. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, S.; Antonopoulos, A.; Ohtsubo, K.; Ditto, D.; Chiba, Y.; Le, D.T.; Morris, H.R.; Haslam, S.M.; Dell, A.; Marth, J.D.; et al. Physiological and glycomic characterization of N-acetylglucosaminyltransferase-IVa and -IVb double deficient mice. Glycobiology 2010, 20, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Oguri, S.; Yoshida, A.; Minowa, M.T.; Takeuchi, M. Kinetic properties and substrate specificities of two recombinant human N-acetylglucosaminyltransferase-IV isozymes. Glycoconj. J. 2006, 23, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, K.; Takamatsu, S.; Minowa, M.T.; Yoshida, A.; Takeuchi, M.; Marth, J.D. Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell 2005, 123, 1307–1321. [Google Scholar] [CrossRef] [PubMed]
- Niimi, K.; Yamamoto, E.; Fujiwara, S.; Shinjo, K.; Kotani, T.; Umezu, T.; Kajiyama, H.; Shibata, K.; Ino, K.; Kikkawa, F. High expression of N-acetylglucosaminyltransferase iva promotes invasion of choriocarcinoma. Br. J. Cancer. 2012, 107, 1969–1977. [Google Scholar] [CrossRef] [PubMed]
- Ide, Y.; Miyoshi, E.; Nakagawa, T.; Gu, J.; Tanemura, M.; Nishida, T.; Ito, T.; Yamamoto, H.; Kozutsumi, Y.; Taniguchi, N. Aberrant expression of N-acetylglucosaminyltransferase-iva and ivb (GnT-IVa and b) in pancreatic cancer. Biochem. Biophys. Res. Commun 2006, 341, 478–482. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Wang, S.; Yu, S.; He, J.; Zheng, W.; Zhang, J. N-acetylglucosaminyltransferase IVa regulates metastatic potential of mouse hepatocarcinoma cells through glycosylation of cd147. Glycoconj. J. 2012, 29, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, K.; Chen, M.Z.; Olefsky, J.M.; Marth, J.D. Pathway to diabetes through attenuation of pancreatic beta cell glycosylation and glucose transport. Nat. Med. 2011, 17, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Ebrahim, A.H.; Alalawi, Z.; Mirandola, L.; Rakhshanda, R.; Dahlbeck, S.; Nguyen, D.; Jenkins, M.; Grizzi, F.; Cobos, E.; Figueroa, J.A.; et al. Galectins in cancer: Carcinogenesis, diagnosis and therapy. Ann. Transl. Med. 2014, 2, 88. [Google Scholar] [PubMed]
- Ihara, H.; Tsukamoto, H.; Gu, J.; Miyoshi, E.; Taniguchi, N.; Ikeda, Y. Fucosyltransferase 8. Gdp-fucose N-glycan core α6-fucosyltransferase (fut8). In Handbook of Glycosyltransferases and Related Genes; Taniguchi, N., Honke, K., Fukuda, M., Narimatsu, H., Yamaguchi, Y., Angata, T., Eds.; Springer Japan: Tokyo, Japan, 2014; pp. 581–596. [Google Scholar]
- Takahashi, M.; Kuroki, Y.; Ohtsubo, K.; Taniguchi, N. Core fucose and bisecting glcnac, the direct modifiers of the N-glycan core: Their functions and target proteins. Carbohydr. Res. 2009, 344, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Brockhausen, I.; Narasimhan, S.; Schachter, H. The biosynthesis of highly branched N-glycans: Studies on the sequential pathway and functional role of N-acetylglucosaminyltransferases I, II, III, IV, V and VI. Biochimie 1988, 70, 1521–1533. [Google Scholar] [CrossRef]
- Uozumi, N.; Yanagidani, S.; Miyoshi, E.; Ihara, Y.; Sakuma, T.; Gao, C.X.; Teshima, T.; Fujii, S.; Shiba, T.; Taniguchi, N. Purification and cDNA cloning of porcine brain GDP-L-Fuc: N-acetyl-beta-d-glucosaminide alpha1-->6fucosyltransferase. J. Biol. Chem. 1996, 271, 27810–27817. [Google Scholar] [CrossRef] [PubMed]
- Yanagidani, S.; Uozumi, N.; Ihara, Y.; Miyoshi, E.; Yamaguchi, N.; Taniguchi, N. Purification and cdna cloning of gdp-l-fuc: N-acetyl-beta-d-glucosaminide:Alpha1-6 fucosyltransferase (alpha1-6 fuct) from human gastric cancer MKN45 cells. J. Biochem. 1997, 121, 626–632. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, E.; Uozumi, N.; Noda, K.; Hayashi, N.; Hori, M.; Taniguchi, N. Expression of alpha1-6 fucosyltransferase in rat tissues and human cancer cell lines. Int. J. Cancer 1997, 72, 1117–1121. [Google Scholar] [CrossRef]
- Noda, K.; Miyoshi, E.; Uozumi, N.; Gao, C.X.; Suzuki, K.; Hayashi, N.; Hori, M.; Taniguchi, N. High expression of alpha-1-6 fucosyltransferase during rat hepatocarcinogenesis. Int. J. Cancer 1998, 75, 444–450. [Google Scholar] [CrossRef]
- Honma, R.; Kinoshita, I.; Miyoshi, E.; Tomaru, U.; Matsuno, Y.; Shimizu, Y.; Takeuchi, S.; Kobayashi, Y.; Kaga, K.; Taniguchi, N.; et al. Expression of fucosyltransferase 8 is associated with an unfavorable clinical outcome in non-small cell lung cancers. Oncology 2015, 88, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Jan, Y.H.; Juan, Y.H.; Yang, C.J.; Huang, M.S.; Yu, C.J.; Yang, P.C.; Hsiao, M.; Hsu, T.L.; Wong, C.H. Fucosyltransferase 8 as a functional regulator of nonsmall cell lung cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 630–635. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Inoue, S.; Gu, J.; Miyoshi, E.; Noda, K.; Li, W.; Mizuno-Horikawa, Y.; Nakano, M.; Asahi, M.; Takahashi, M.; et al. Dysregulation of TGF-beta1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc. Natl. Acad. Sci. USA 2005, 102, 15791–15796. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Maeno, T.; Ota, F.; Ueno, M.; Korekane, H.; Takamatsu, S.; Shirato, K.; Matsumoto, A.; Kobayashi, S.; Yoshida, K.; et al. Sensitivity of heterozygous alpha1,6-fucosyltransferase knock-out mice to cigarette smoke-induced emphysema: Implication of aberrant transforming growth factor-beta signaling and matrix metalloproteinase gene expression. J. Biol. Chem. 2012, 287, 16699–16708. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, T.; Hashimoto, H.; Okayasu, N.; Kameyama, A.; Onogi, H.; Nakagawasai, O.; Nakazawa, T.; Kurosawa, T.; Hao, Y.; Isaji, T.; et al. Alpha1,6-fucosyltransferase-deficient mice exhibit multiple behavioral abnormalities associated with a schizophrenia-like phenotype: Importance of the balance between the dopamine and serotonin systems. J. Biol. Chem. 2011, 286, 18434–18443. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Fukuda, T.; Isaji, T.; Hang, Q.; Lee, H.H.; Sakai, S.; Morise, J.; Mitoma, J.; Higashi, H.; Taniguchi, N.; et al. Loss of alpha1,6-fucosyltransferase decreases hippocampal long term potentiation: Implications for core fucosylation in the regulation of ampa receptor heteromerization and cellular signaling. J. Biol. Chem. 2015, 290, 17566–17575. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yu, R.; Ma, B.; Yang, Y.; Jiao, X.; Liu, Y.; Cao, H.; Dong, W.; Liu, L.; Ma, K.; et al. Core fucosylation of IGG b cell receptor is required for antigen recognition and antibody production. J. Immunol. 2015, 194, 2596–2606. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Shinzaki, S.; Iijima, H.; Wakamatsu, K.; Iwamoto, C.; Sobajima, T.; Kuwahara, R.; Hiyama, S.; Hayashi, Y.; Takamatsu, S.; et al. Core fucosylation on T cells, required for activation of T-cell receptor signaling and induction of colitis in mice, is increased in patients with inflammatory bowel disease. Gastroenterology 2016, 150. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gu, J.; Ihara, H.; Miyoshi, E.; Honke, K.; Taniguchi, N. Core fucosylation regulates epidermal growth factor receptor-mediated intracellular signaling. J. Biol. Chem. 2006, 281, 2572–2577. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Yen, H.Y.; Chen, C.Y.; Chen, C.H.; Cheng, P.F.; Juan, Y.H.; Chen, C.H.; Khoo, K.H.; Yu, C.J.; Yang, P.C.; et al. Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc. Natl. Acad. Sci. USA 2011, 108, 11332–11337. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fukuda, T.; Isaji, T.; Lu, J.; Gu, W.; Lee, H.H.; Ohkubo, Y.; Kamada, Y.; Taniguchi, N.; Miyoshi, E.; et al. Loss of alpha1,6-fucosyltransferase suppressed liver regeneration: Implication of core fucose in the regulation of growth factor receptor-mediated cellular signaling. Sci. Rep. 2015, 5, 8264. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fukuda, T.; Isaji, T.; Lu, J.; Im, S.; Hang, Q.; Gu, W.; Hou, S.; Ohtsubo, K.; Gu, J. Loss of alpha1,6-fucosyltransferase inhibits chemical-induced hepatocellular carcinoma and tumorigenesis by down-regulating several cell signaling pathways. FASEB J. 2015, 29, 3217–3227. [Google Scholar] [CrossRef] [PubMed]
- Potapenko, I.O.; Haakensen, V.D.; Luders, T.; Helland, A.; Bukholm, I.; Sorlie, T.; Kristensen, V.N.; Lingjaerde, O.C.; Borresen-Dale, A.L. Glycan gene expression signatures in normal and malignant breast tissue; possible role in diagnosis and progression. Mol. Oncol. 2010, 4, 98–118. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, J.; Li, Q.K.; Peskoe, S.B.; Zhang, B.; Choi, C.; Platz, E.A.; Zhang, H. Overexpression of alpha (1,6) fucosyltransferase associated with aggressive prostate cancer. Glycobiology 2014, 24, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Saldova, R.; Fan, Y.; Fitzpatrick, J.M.; Watson, R.W.; Rudd, P.M. Core fucosylation and alpha2-3 sialylation in serum N-glycome is significantly increased in prostate cancer comparing to benign prostate hyperplasia. Glycobiology 2011, 21, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Taketa, K.; Endo, Y.; Sekiya, C.; Tanikawa, K.; Koji, T.; Taga, H.; Satomura, S.; Matsuura, S.; Kawai, T.; Hirai, H. A collaborative study for the evaluation of lectin-reactive alpha-fetoproteins in early detection of hepatocellular carcinoma. Cancer Res. 1993, 53, 5419–5423. [Google Scholar] [PubMed]
- Toyoda, H.; Kumada, T.; Tada, T. Highly sensitive lens culinaris agglutinin-reactive alpha-fetoprotein: A new tool for the management of hepatocellular carcinoma. Oncology 2011, 81 Suppl 1, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Abelev, G.I. Alpha-fetoprotein in ontogenesis and its association with malignant tumors. Adv. Cancer Res. 1971, 14, 295–358. [Google Scholar] [PubMed]
- Nakagawa, T.; Miyoshi, E.; Yakushijin, T.; Hiramatsu, N.; Igura, T.; Hayashi, N.; Taniguchi, N.; Kondo, A. Glycomic analysis of alpha-fetoprotein L3 in hepatoma cell lines and hepatocellular carcinoma patients. J. Proteome Res. 2008, 7, 2222–2233. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, E.; Moriwaki, K.; Terao, N.; Tan, C.C.; Terao, M.; Nakagawa, T.; Matsumoto, H.; Shinzaki, S.; Kamada, Y. Fucosylation is a promising target for cancer diagnosis and therapy. Biomolecules 2012, 2, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, N.; Ide, Y.; Nakano, M.; Nakagawa, T.; Yamanaka, K.; Moriwaki, K.; Murata, K.; Ohigashi, H.; Yokoyama, S.; Eguchi, H.; et al. Fucosylated haptoglobin is a novel marker for pancreatic cancer: A detailed analysis of the oligosaccharide structure and a possible mechanism for fucosylation. Int. J. Cancer 2006, 118, 2803–2808. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Sugiyama, T.; Shimomura, M.; Kamada, Y.; Fujita, K.; Nonomura, N.; Miyoshi, E.; Nakano, M. Site-specific and linkage analyses of fucosylated N-glycans on haptoglobin in sera of patients with various types of cancer: Possible implication for the differential diagnosis of cancer. Glycoconj. J. 2016, 33. [Google Scholar] [CrossRef] [PubMed]
- Shuptrine, C.W.; Surana, R.; Weiner, L.M. Monoclonal antibodies for the treatment of cancer. Semin. Cancer Biol. 2012, 22, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Shields, R.L.; Lai, J.; Keck, R.; O'Connell, L.Y.; Hong, K.; Meng, Y.G.; Weikert, S.H.; Presta, L.G. Lack of fucose on human IGG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J. Biol. Chem. 2002, 277, 26733–26740. [Google Scholar] [CrossRef] [PubMed]
- Shinkawa, T.; Nakamura, K.; Yamane, N.; Shoji-Hosaka, E.; Kanda, Y.; Sakurada, M.; Uchida, K.; Anazawa, H.; Satoh, M.; Yamasaki, M.; et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IGG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 2003, 278, 3466–3473. [Google Scholar] [CrossRef] [PubMed]
- Reusch, D.; Tejada, M.L. Fc glycans of therapeutic antibodies as critical quality attributes. Glycobiology 2015, 25, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.R.; Rodrigues, M.E.; Henriques, M.; Oliveira, R.; Azeredo, J. Glycosylation: Impact, control and improvement during therapeutic protein production. Crit. Rev. Biotechnol. 2014, 34, 281–299. [Google Scholar] [CrossRef] [PubMed]
- Buckhaults, P.; Chen, L.; Fregien, N.; Pierce, M. Transcriptional regulation of N-acetylglucosaminyltransferase V by the src oncogene. J. Biol. Chem. 1997, 272, 19575–19581. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, W.; Fregien, N.; Pierce, M. The her-2/neu oncogene stimulates the transcription of N-acetylglucosaminyltransferase V and expression of its cell surface oligosaccharide products. Oncogene 1998, 17, 2087–2093. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Saito, H.; Ihara, Y.; Miyoshi, E.; Koyama, N.; Sheng, Y.; Taniguchi, N. Transcriptional regulation of the N-acetylglucosaminyltransferase V gene in human bile duct carcinoma cells (HuCC-T1) is mediated by ets-1. J. Biol. Chem. 1996, 271, 26706–26712. [Google Scholar] [PubMed]
- Ko, J.H.; Miyoshi, E.; Noda, K.; Ekuni, A.; Kang, R.; Ikeda, Y.; Taniguchi, N. Regulation of the GnT-V promoter by transcription factor Ets-1 in various cancer cell lines. J. Biol. Chem. 1999, 274, 22941–22948. [Google Scholar] [CrossRef] [PubMed]
- Lauc, G.; Essafi, A.; Huffman, J.E.; Hayward, C.; Knezevic, A.; Kattla, J.J.; Polasek, O.; Gornik, O.; Vitart, V.; Abrahams, J.L.; et al. Genomics meets glycomics-the first gwas study of human N-glycome identifies HNF1alpha as a master regulator of plasma protein fucosylation. PLoS Genet. 2010, 6, e1001256. [Google Scholar] [CrossRef] [PubMed]
- Akama, R.; Sato, Y.; Kariya, Y.; Isaji, T.; Fukuda, T.; Lu, L.; Taniguchi, N.; Ozawa, M.; Gu, J. N-acetylglucosaminyltransferase III expression is regulated by cell-cell adhesion via the e-cadherin-catenin-actin complex. Proteomics 2008, 8, 3221–3228. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Akama, R.; Isaji, T.; Lu, Y.; Hashimoto, H.; Kariya, Y.; Fukuda, T.; Du, Y.; Gu, J. Wnt/beta-catenin signaling down-regulates N-acetylglucosaminyltransferase III expression: The implications of two mutually exclusive pathways for regulation. J. Biol. Chem. 2011, 286, 4310–4318. [Google Scholar] [CrossRef] [PubMed]
- Kurimoto, A.; Kitazume, S.; Kizuka, Y.; Nakajima, K.; Oka, R.; Fujinawa, R.; Korekane, H.; Yamaguchi, Y.; Wada, Y.; Taniguchi, N. The absence of core fucose up-regulates GnT-III and WNT target genes: A possible mechanism for an adaptive response in terms of glycan function. J. Biol. Chem. 2014, 289, 11704–11714. [Google Scholar] [CrossRef] [PubMed]
- Bonasio, R.; Tu, S.; Reinberg, D. Molecular signals of epigenetic states. Science 2010, 330, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Plass, C.; Pfister, S.M.; Lindroth, A.M.; Bogatyrova, O.; Claus, R.; Lichter, P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat. Rev. Genet. 2013, 14, 765–780. [Google Scholar] [CrossRef] [PubMed]
- Bergman, Y.; Cedar, H. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013, 20, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, J.; Collignon, E.; Fuks, F. Portraits of TET-mediated DNA hydroxymethylation in cancer. Curr. Opin. Genet. Dev. 2016, 36, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Anugraham, M.; Jacob, F.; Nixdorf, S.; Everest-Dass, A.V.; Heinzelmann-Schwarz, V.; Packer, N.H. Specific glycosylation of membrane proteins in epithelial ovarian cancer cell lines: Glycan structures reflect gene expression and DNA methylation status. Mol. Cell. Proteomics 2014, 13, 2213–2232. [Google Scholar] [CrossRef] [PubMed]
- Saldova, R.; Dempsey, E.; Perez-Garay, M.; Marino, K.; Watson, J.A.; Blanco-Fernandez, A.; Struwe, W.B.; Harvey, D.J.; Madden, S.F.; Peracaula, R.; et al. 5-aza-2′-deoxycytidine induced demethylation influences N-glycosylation of secreted glycoproteins in ovarian cancer. Epigenetics 2011, 6, 1362–1372. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Lauc, G.; Vojta, A.; Zoldos, V. Epigenetic regulation of glycosylation is the quantum mechanics of biology. Biochim. Biophys. Acta. 2014, 1840, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.V.; Mattick, J.S. The rise of regulatory rna. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, J.; Kong, X.; Chen, H.; Wang, Y.; Qin, M.; Lin, Y.; Chen, H.; Xu, J.; Hong, J.; et al. Mir-198 represses tumor growth and metastasis in colorectal cancer by targeting fucosyl transferase 8. Sci. Rep. 2014, 4, 6145. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, C.; Soffientini, U.; Piacente, F.; Tonetti, M.G. Effects of micrornas on fucosyltransferase 8 (FUT8) expression in hepatocarcinoma cells. PLoS ONE 2013, 8, e76540. [Google Scholar] [CrossRef] [PubMed]
- Vaiana, C.A.; Kurcon, T.; Mahal, L.K. MicroRNA-424 predicts a role for beta-1,4 branched glycosylation in cell cycle progression. J. Biol. Chem. 2016, 291, 1529–1537. [Google Scholar] [CrossRef] [PubMed]
- Kurcon, T.; Liu, Z.; Paradkar, A.V.; Vaiana, C.A.; Koppolu, S.; Agrawal, P.; Mahal, L.K. MiRNA proxy approach reveals hidden functions of glycosylation. Proc. Natl. Acad. Sci. USA 2015, 112, 7327–7332. [Google Scholar] [CrossRef] [PubMed]
- Kasper, B.T.; Koppolu, S.; Mahal, L.K. Insights into mirna regulation of the human glycome. Biochem. Biophys. Res. Commun. 2014, 445, 774–779. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Kurcon, T.; Pilobello, K.T.; Rakus, J.F.; Koppolu, S.; Liu, Z.; Batista, B.S.; Eng, W.S.; Hsu, K.L.; Liang, Y.; et al. Mapping posttranscriptional regulation of the human glycome uncovers microrna defining the glycocode. Proc. Natl. Acad. Sci. USA 2014, 111, 4338–4343. [Google Scholar] [CrossRef] [PubMed]
- Hart, G.W.; Slawson, C.; Ramirez-Correa, G.; Lagerlof, O. Cross talk between O-glcnacylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011, 80, 825–858. [Google Scholar] [CrossRef] [PubMed]
- Sasai, K.; Ikeda, Y.; Fujii, T.; Tsuda, T.; Taniguchi, N. UDP-glcnac concentration is an important factor in the biosynthesis of beta1,6-branched oligosaccharides: Regulation based on the kinetic properties of N-acetylglucosaminyltransferase v. Glycobiology 2002, 12, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Boroughs, L.K.; DeBerardinis, R.J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 2015, 17, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.; Kitazume, S.; Angata, T.; Fujinawa, R.; Ohtsubo, K.; Miyoshi, E.; Taniguchi, N. Simultaneous determination of nucleotide sugars with ion-pair reversed-phase hplc. Glycobiology 2010, 20, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, K.; Ito, E.; Ohtsubo, K.; Shirato, K.; Takamiya, R.; Kitazume, S.; Angata, T.; Taniguchi, N. Mass isotopomer analysis of metabolically labeled nucleotide sugars and N- and O-glycans for tracing nucleotide sugar metabolisms. Mol. Cell. Proteomics 2013, 12, 2468–2480. [Google Scholar] [CrossRef] [PubMed]
- Korekane, H.; Park, J.Y.; Matsumoto, A.; Nakajima, K.; Takamatsu, S.; Ohtsubo, K.; Miyamoto, Y.; Hanashima, S.; Kanekiyo, K.; Kitazume, S.; et al. Identification of ectonucleotide pyrophosphatase/phosphodiesterase 3 (ENPP3) as a regulator of N-acetylglucosaminyltransferase GnT-IX (GnT-Vb). J. Biol. Chem. 2013, 288, 27912–27926. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, P.; Dabelsteen, S.; Madsen, F.B.; Francavilla, C.; Kopp, K.L.; Steentoft, C.; Vakhrushev, S.Y.; Olsen, J.V.; Hansen, L.; Bennett, E.P.; et al. Immature truncated O-glycophenotype of cancer directly induces oncogenic features. Proc. Natl. Acad. Sci. USA 2014, 111, E4066–E4075. [Google Scholar] [CrossRef] [PubMed]
- Gill, D.J.; Tham, K.M.; Chia, J.; Wang, S.C.; Steentoft, C.; Clausen, H.; Bard-Chapeau, E.A.; Bard, F.A. Initiation of galnac-type O-glycosylation in the endoplasmic reticulum promotes cancer cell invasiveness. Proc. Natl. Acad. Sci. USA 2013, 110, E3152–E3161. [Google Scholar] [CrossRef] [PubMed]
- Sasai, K.; Ikeda, Y.; Ihara, H.; Honke, K.; Taniguchi, N. Caveolin-1 regulates the functional localization of N-acetylglucosaminyltransferase III within the golgi apparatus. J. Biol. Chem. 2003, 278, 25295–25301. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, N.; Liu, P.; Peng, F.; Tang, H.; Chen, Q.; Xu, R.; Dai, Y.; Lin, Y.; Xie, X.; et al. Caveolin-1, a stress-related oncotarget, in drug resistance. Oncotarget 2015, 6, 37135–37150. [Google Scholar] [PubMed]
- Thaysen-Andersen, M.; Packer, N.H. Site-specific glycoproteomics confirms that protein structure dictates formation of N-glycan type, core fucosylation and branching. Glycobiology 2012, 22, 1440–1452. [Google Scholar] [CrossRef] [PubMed]
- Sethi, M.K.; Kim, H.; Park, C.K.; Baker, M.S.; Paik, Y.K.; Packer, N.H.; Hancock, W.S.; Fanayan, S.; Thaysen-Andersen, M. In-depth N-glycome profiling of paired colorectal cancer and non-tumorigenic tissues reveals cancer-, stage- and EGFR-specific protein N-glycosylation. Glycobiology 2015, 25, 1064–1078. [Google Scholar] [CrossRef] [PubMed]
- Moh, E.S.; Thaysen-Andersen, M.; Packer, N.H. Relative versus absolute quantitation in disease glycomics. Proteomics Clin. Appl. 2015, 9, 368–382. [Google Scholar] [CrossRef] [PubMed]
- Levery, S.B.; Steentoft, C.; Halim, A.; Narimatsu, Y.; Clausen, H.; Vakhrushev, S.Y. Advances in mass spectrometry driven O-glycoproteomics. Biochim. Biophys. Acta 2015, 1850, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Laughlin, S.T.; Bertozzi, C.R. Imaging the glycome. Proc. Natl. Acad. Sci. USA 2009, 106, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Y. Applications of azide-based bioorthogonal click chemistry in glycobiology. Molecules 2013, 18, 7145–7159. [Google Scholar] [CrossRef] [PubMed]
- Rabuka, D.; Hubbard, S.C.; Laughlin, S.T.; Argade, S.P.; Bertozzi, C.R. A chemical reporter strategy to probe glycoprotein fucosylation. J. Am. Chem. Soc. 2006, 128, 12078–12079. [Google Scholar] [CrossRef] [PubMed]
- Hanashima, S.; Inamori, K.; Manabe, S.; Taniguchi, N.; Ito, Y. Systematic synthesis of bisubstrate-type inhibitors of N-acetylglucosaminyltransferases. Chemistry 2006, 12, 3449–3462. [Google Scholar] [CrossRef] [PubMed]
- Hanashima, S.; Korekane, H.; Taniguchi, N.; Yamaguchi, Y. Synthesis of N-glycan units for assessment of substrate structural requirements of N-acetylglucosaminyltransferase III. Bioorg. Med. Chem. Lett. 2014, 24, 4533–4537. [Google Scholar] [CrossRef] [PubMed]
- Ihara, H.; Ikeda, Y.; Toma, S.; Wang, X.; Suzuki, T.; Gu, J.; Miyoshi, E.; Tsukihara, T.; Honke, K.; Matsumoto, A.; et al. Crystal structure of mammalian alpha1,6-fucosyltransferase, FUT8. Glycobiology 2007, 17, 455–466. [Google Scholar] [CrossRef] [PubMed]
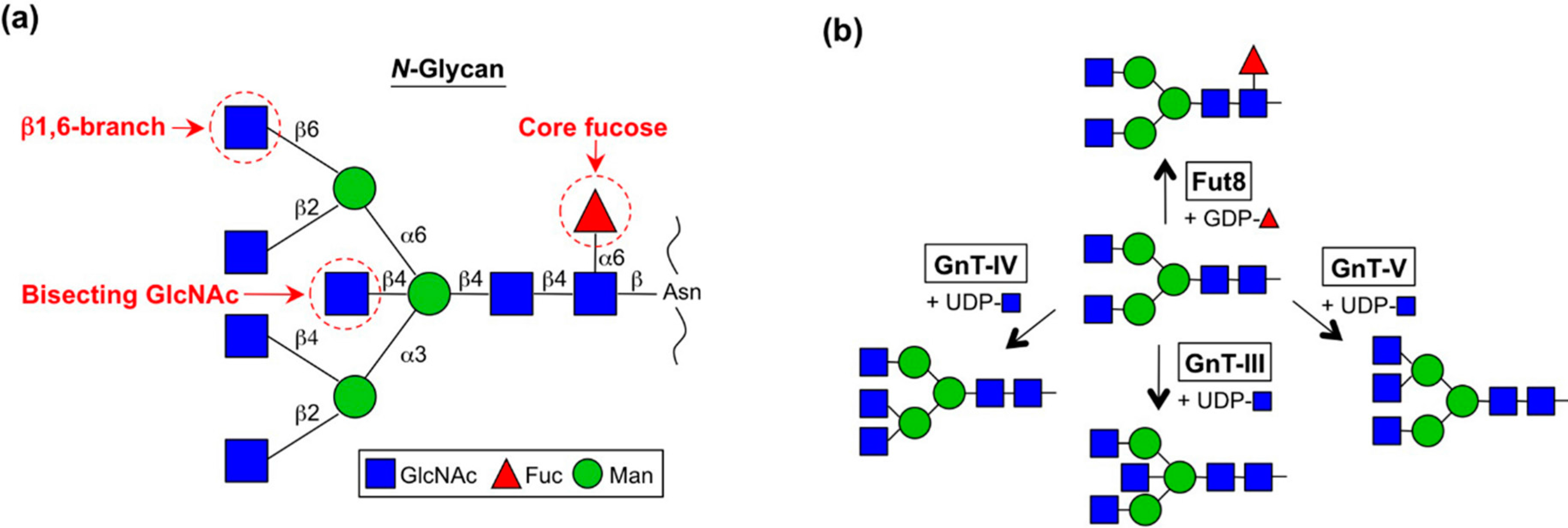
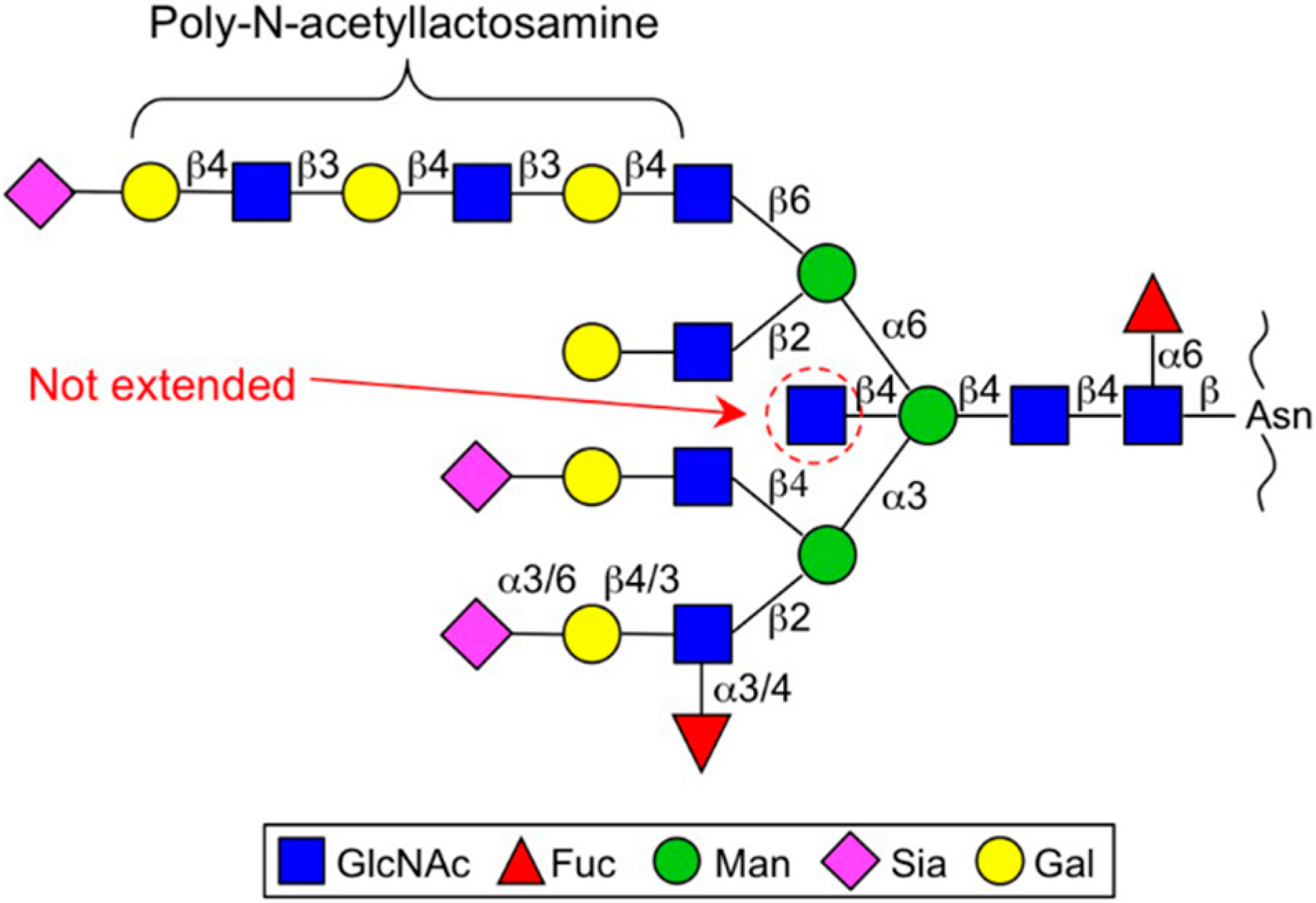

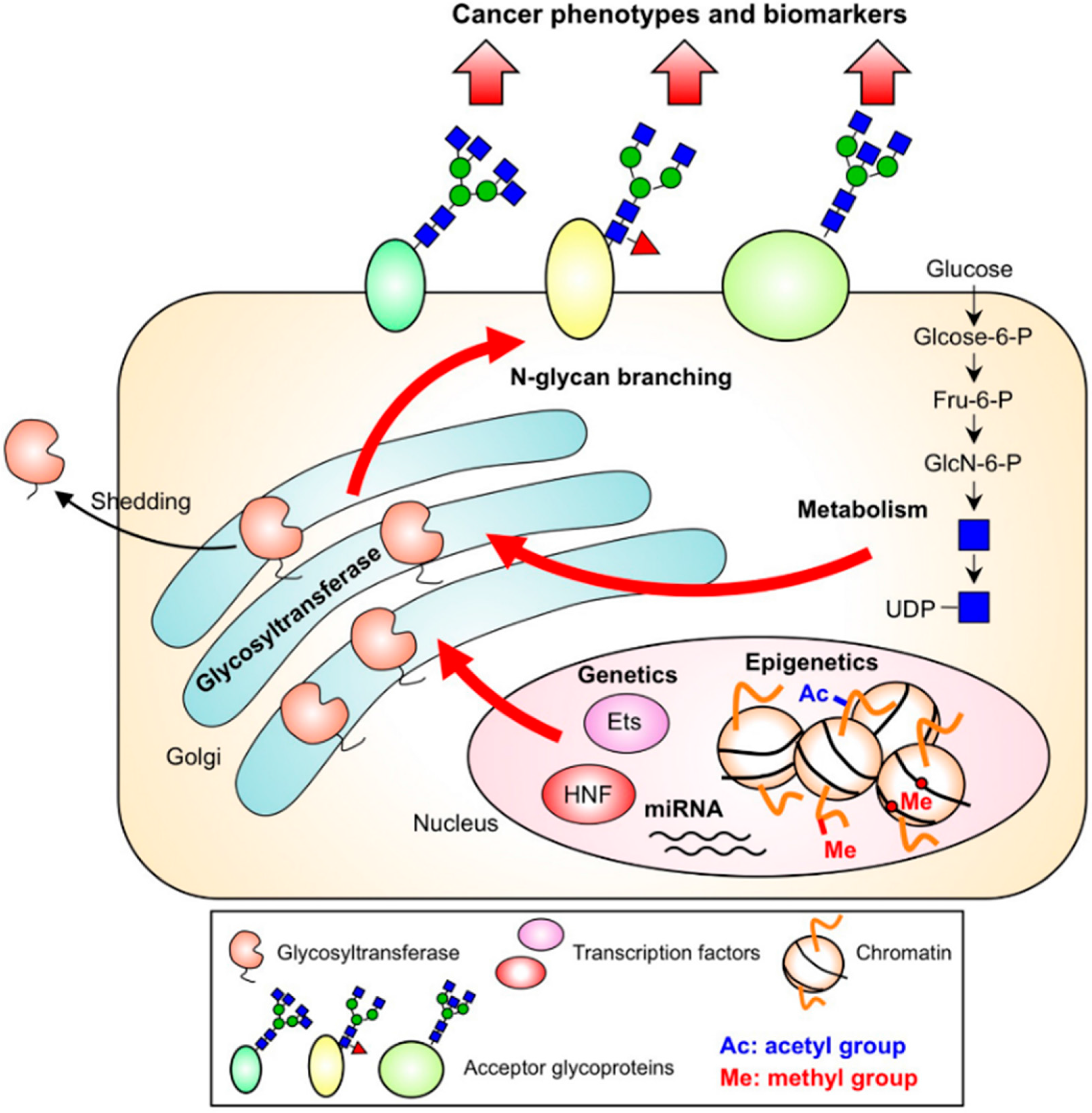
| Enzyme | Gene | Glycan | Dysregulation in cancer | Functional output in cancer |
|---|---|---|---|---|
| GnT-III | MGAT3 | Bisecting GlcNAc | Hepatoma [18], ovarian cancer [19], leukemia [20] | Suppression of metastasis [21], decrease in tumor growth [22], suppression of EMT [30,31] tumor growth [33,34] |
| GnT-IVa-c | MGAT4A-C | β1,4-branch | Choriocarcinoma [81], pancreatic cancer [82], hepatoma [83] | Invasion [81], metastasis [83] |
| GnT-V | MGAT5 | β1,6-branch on α1,6-mannose | Gastric [54], esophageal [55], colon [56], liver [57] cancers | Angiogenesis [47], tumor growth [52,53] |
| GnT-Vb (IX) | MGAT5b | β1,6-branch on O-mannose | Neuroblastoma [72], prostate cancer [73] | Decreased adhesion, increased migration [67] |
| Fut8 | FUT8 | Core fucose | Hepatoma [92], lung [93,94], breast [105], prostate [106,107] cancers | Tumor growth [93,94,102,104], tumor biomarker [108], antibody therapy (ADCC) [116,117] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kizuka, Y.; Taniguchi, N. Enzymes for N-Glycan Branching and Their Genetic and Nongenetic Regulation in Cancer. Biomolecules 2016, 6, 25. https://doi.org/10.3390/biom6020025
Kizuka Y, Taniguchi N. Enzymes for N-Glycan Branching and Their Genetic and Nongenetic Regulation in Cancer. Biomolecules. 2016; 6(2):25. https://doi.org/10.3390/biom6020025
Chicago/Turabian StyleKizuka, Yasuhiko, and Naoyuki Taniguchi. 2016. "Enzymes for N-Glycan Branching and Their Genetic and Nongenetic Regulation in Cancer" Biomolecules 6, no. 2: 25. https://doi.org/10.3390/biom6020025





