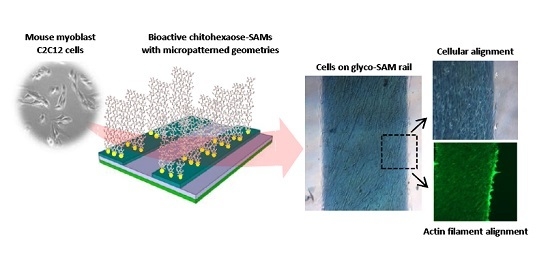
Chitooligomer-Immobilized Biointerfaces with Micropatterned Geometries for Unidirectional Alignment of Myoblast Cells
Abstract
:1. Introduction
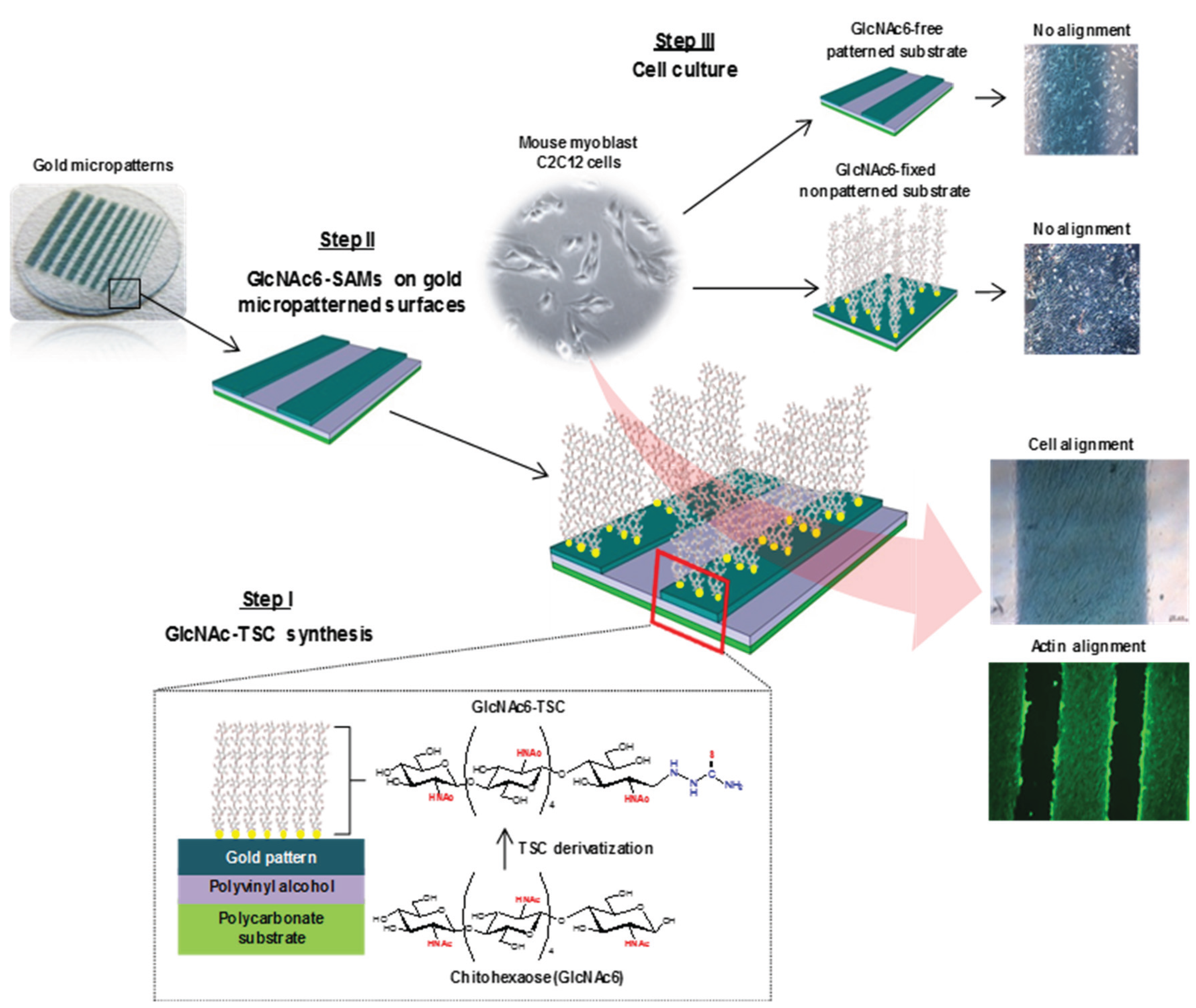
2. Results and Discussion
2.1. Self-Assembly Immobilization of Chitooligomers on Micropatterned Gold Surfaces
2.2. Surface Sugar Densities of Micropatterned Glyco-SAMs
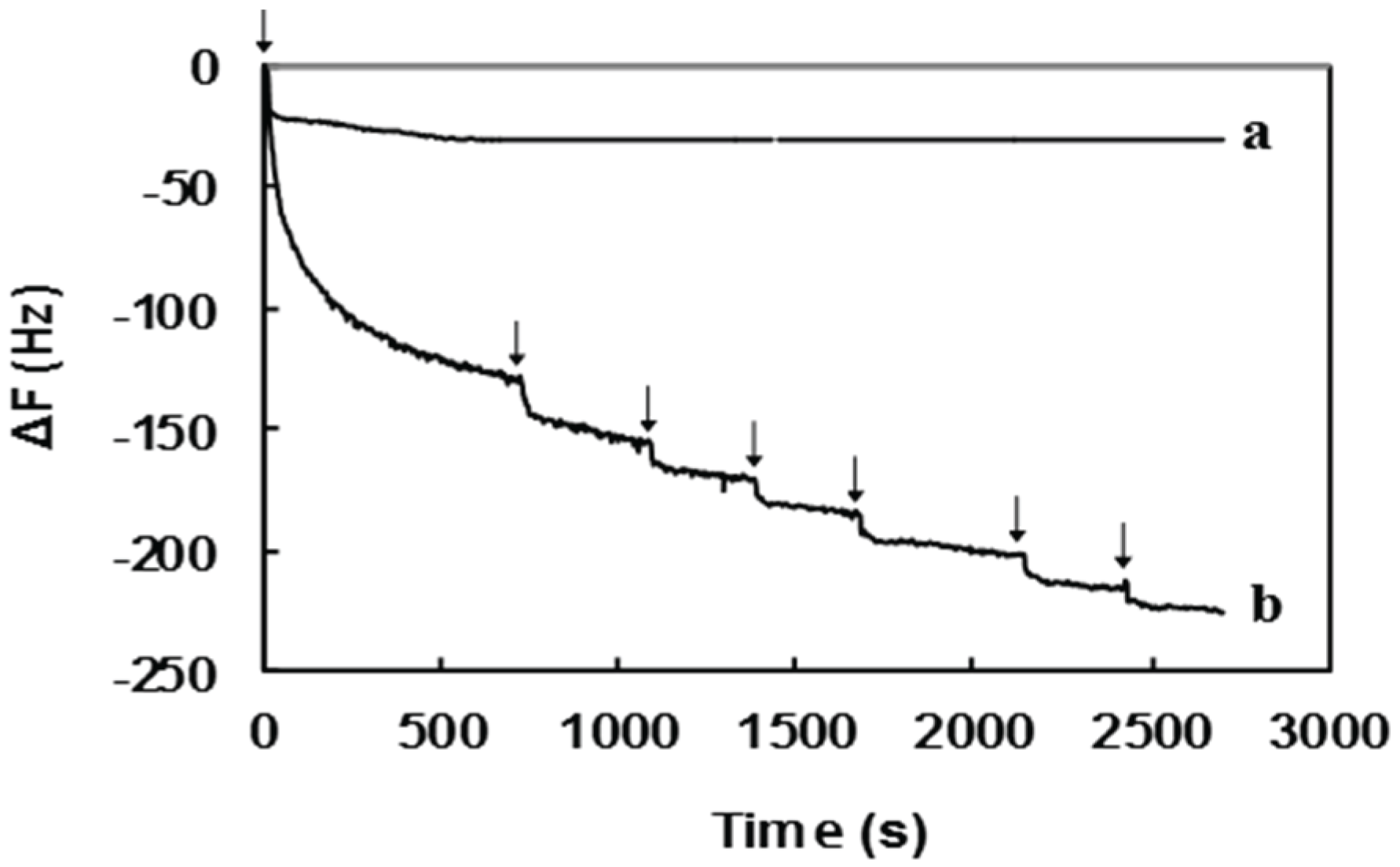
| Sample | QCM Data | GlcNAc6 Density (Chains nm−2) | |
|---|---|---|---|
| ∆F (Hz) | Mass (ng) | ||
| Pure GlcNAc6 | −30.7 | 0.92 | 0.09 |
| GlcNAc6-TSC | −225.5 | 6.77 | 0.68 |
2.3. Cell Morphology and Biofunctional Behavior on Micropatterned Biointerfaces
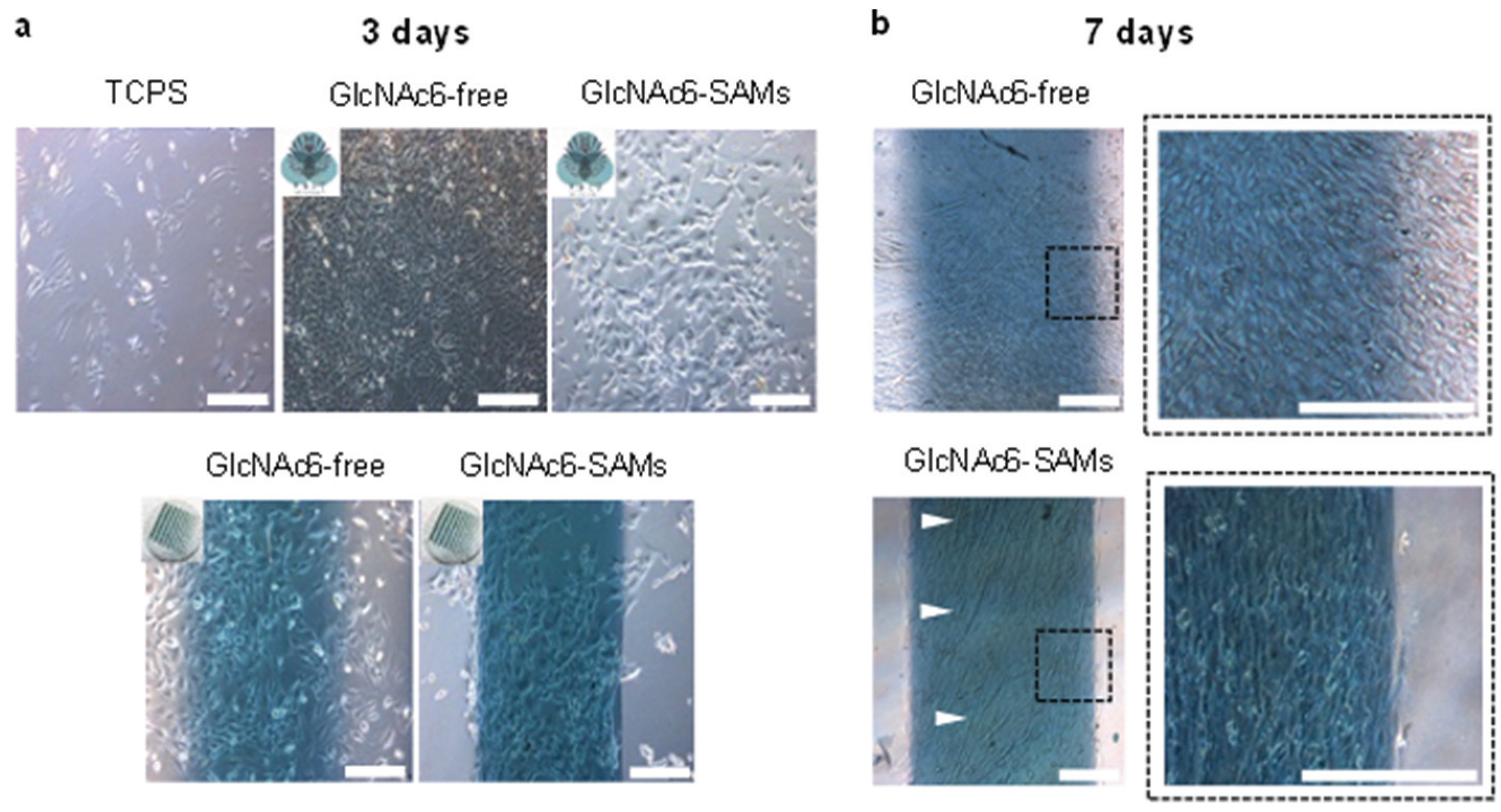
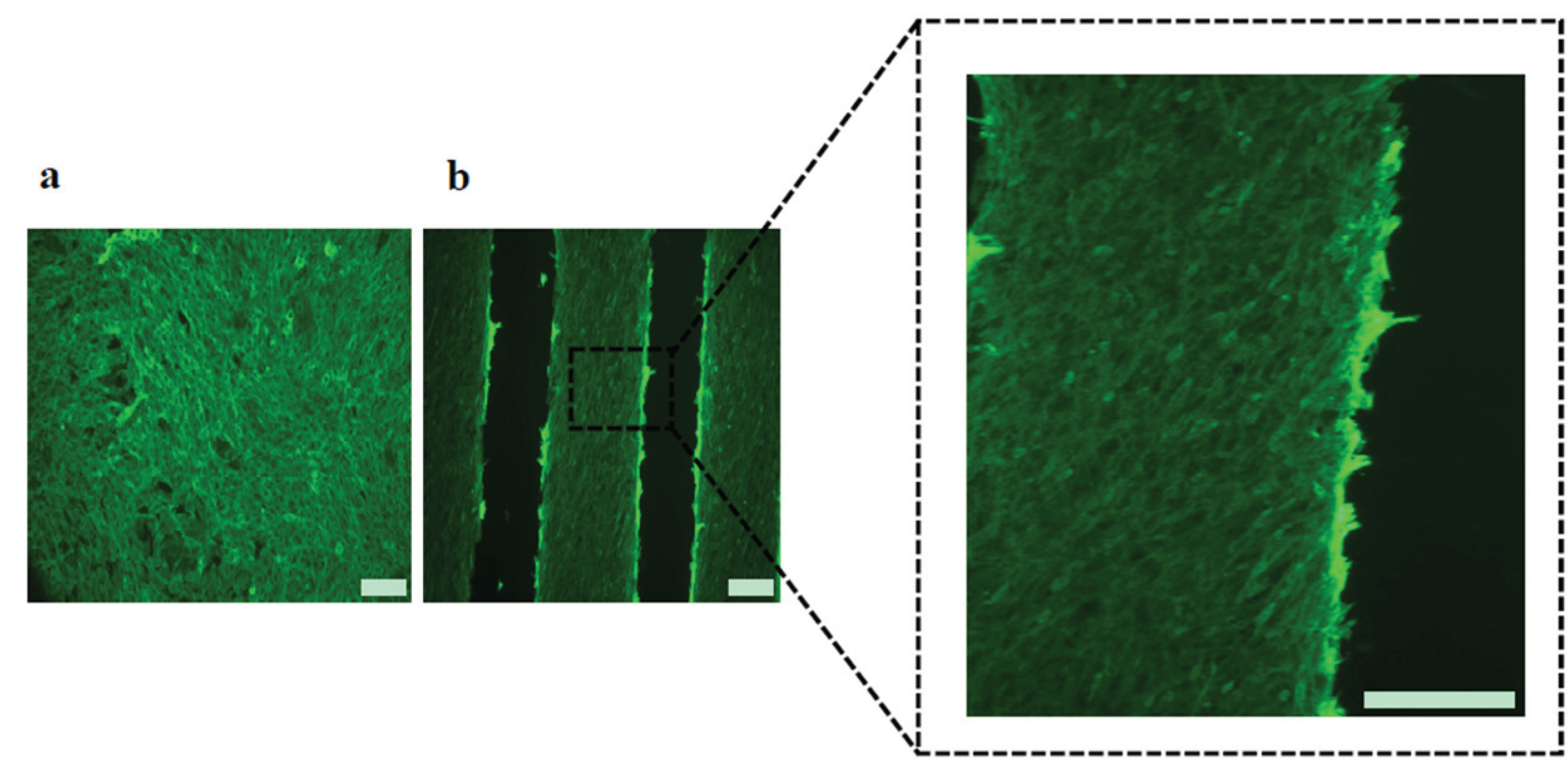
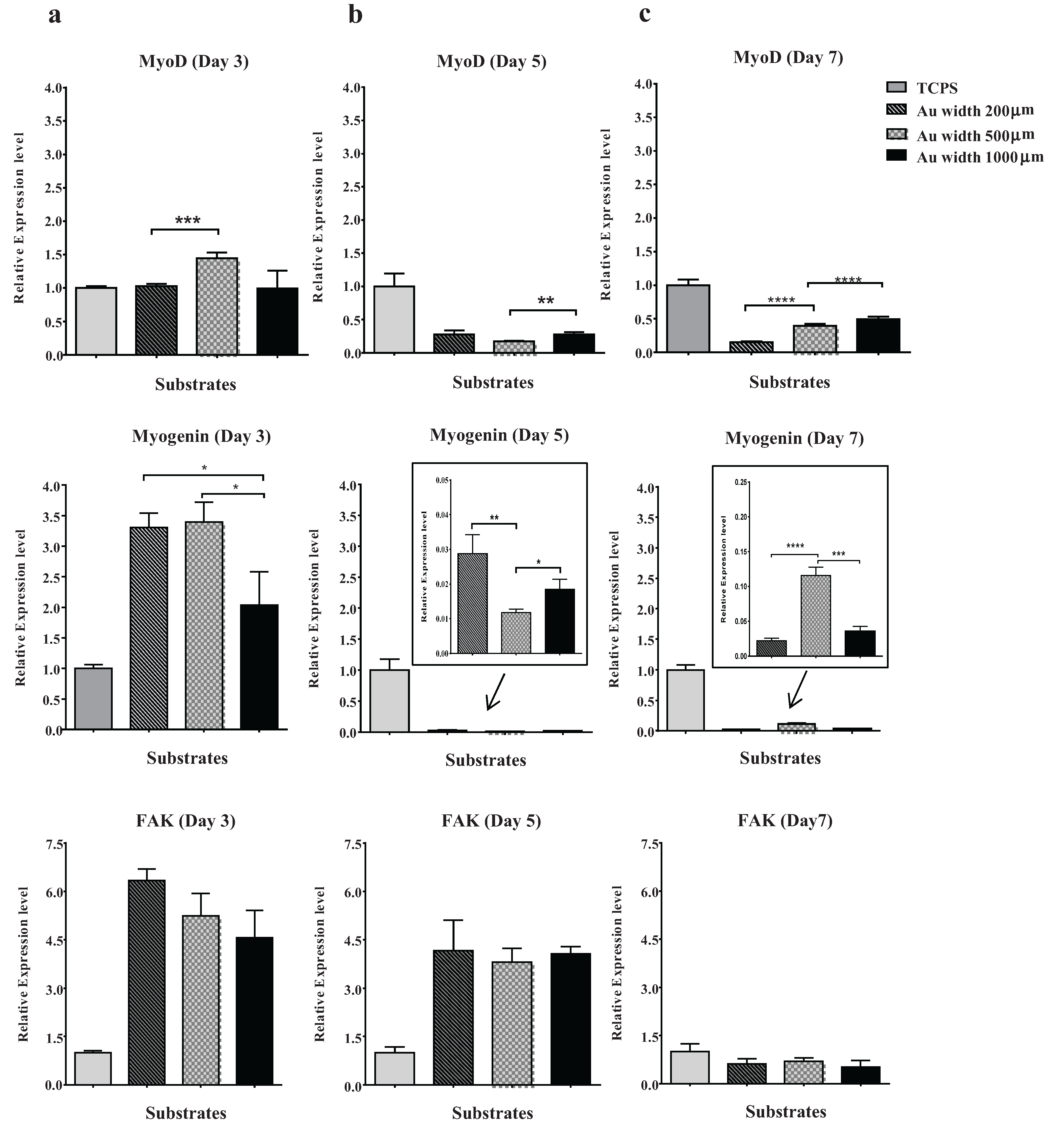
2.4. Effect of Micropattern Width on Gene Expression in C2C12 Cells
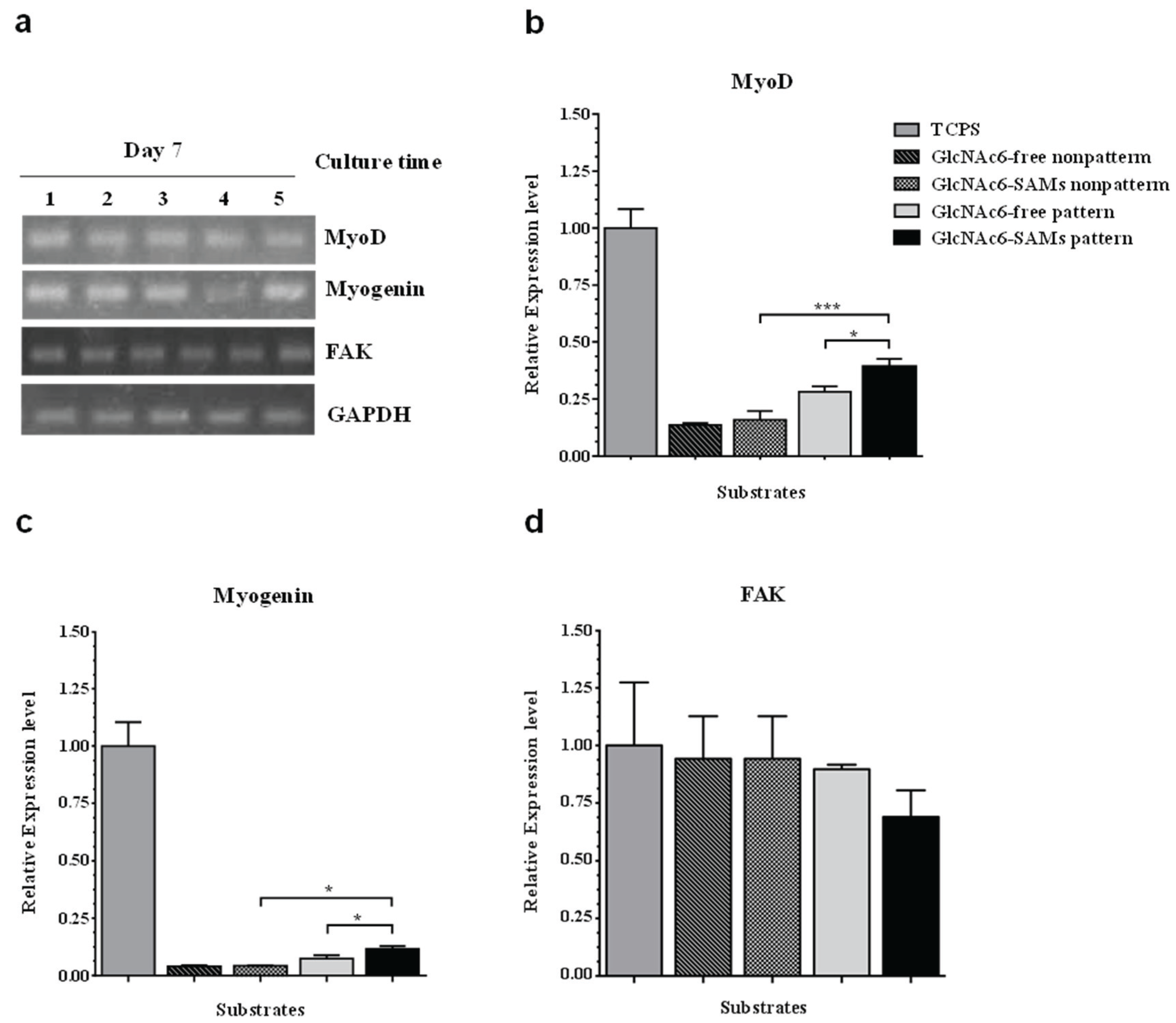
3. Experimental Section
3.1. Materials
3.2. Preparation of Micropatterned Glyco-SAMs on Gold Surfaces
3.3. Characterization of Micropatterned Glyco-SAMs
3.3.1. NMR Analysis
3.3.2. Quartz Crystal Microbalance Analysis (QCM)
3.3.3. X-Ray Photoelectron Spectroscopy (XPS)
3.4. Cell Culture Assays and Microscopic Observations
3.5. Biological Characterization
3.5.1. F-Actin Staining
3.5.2. RNA Extraction and Quantitative Real-Time PCR
3.5.3. Statistical Analysis
4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bertozzi, C.R. Chemical Glycobiology. Science. 2001, 291, 2357–2364. [Google Scholar] [CrossRef] [PubMed]
- Iijima, J.; Zhao, Y.; Isaji, T.; Kameyama, A.; Nakaya, S.; Wang, X.; Ihara, H.; Cheng, X.; Nakagawa, T.; Miyoshi, E.; et al. Cell-cell interaction-dependent regulation of N-acetylglucosaminyltransferase III and the bisected N-glycans in GE11 epithelial cells. Involvement of E-cadherin-mediated cell adhesion. J. Biol. Chem. 2006, 281, 13038–13046. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.K.; Kumar, S.; Tupperwar, N.C.; Vaidya, T.; George, A.; Rath, S.; Bal, V.; Ravindran, B. Chitohexaose activates macrophages by alternate pathway through TLR4 and blocks endotoxemia. PLoS Pathog. 2012, 8, e1002717. [Google Scholar] [CrossRef] [PubMed]
- Zachara, N.E.; Hart, G.W. Cell signaling, the essential role of O-GlcNAc. Biochim. Biophys. Acta 2006, 1761, 599–617. [Google Scholar] [CrossRef] [PubMed]
- Lowe, J.B. Glycosylation in the control of selectin counter-receptor structure and function. Immunol. Rev. 2002, 186, 19–36. [Google Scholar] [CrossRef] [PubMed]
- Lundquist, J.J.; Toone, E.J. The cluster glycoside effect. Chem. Rev. 2002, 102, 555–578. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhang, X.; Zhao, Y.; Liang, S.; Xu, A.; Gao, X.; Deng, F.; Fang, J.; Wei, S. Peptide-decorated polyvinyl alcohol/hyaluronan nanofibers for human induced pluripotent stem cell culture. Carbohydr. Polym. 2014, 101, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Hortensius, R.A.; Harley, B.A.C. The use of bioinspired alterations in the glycosaminoglycan content of collagen-GAG scaffolds to regulate cell activity. Biomaterials 2013, 34, 7645–7652. [Google Scholar] [CrossRef] [PubMed]
- Kitaoka, T.; Yoshiyama, C.; Uemura, F. Hybrid immobilization of galactosyl lactose and cellobiose on a gold substrate to modulate biological responses. Carbohydr. Polym. 2013, 92, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Mizofuchi, H.; Kobayashi, Y.; Tsuzuki, G.; Yamamoto, M.; Wada, S.; Kamemura, K. Terminal differentiation program of skeletal myogenesis is negatively regulated by O-GlcNAc glycosylation. Biochim. Biophys. Acta 2012, 1820, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Gillies, A.R.; Lieber, R.L. Structure and function of the skeletal muscle extracellular matrix. Muscle Nerve 2011, 44, 318–331. [Google Scholar] [CrossRef] [PubMed]
- Hume, S.L.; Hoyt, S.M.; Walker, J.S.; Sridhar, B.V.; Ashley, J.F.; Bowman, C.N.; Bryant, S.J. Alignment of multi-layered muscle cells within three-dimensional hydrogel macrochannels. Acta Biomater. 2012, 8, 2193–2202. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Fujita, H.; Nagamori, E. Alignment of skeletal muscle myoblasts and myotubes using linear micropatterned surfaces ground with abrasives. Biotechnol. Bioeng. 2009, 103, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, G.; Zhang, X.; Wang, L.; Du, Y.; Lu, T.J.; Xu, F. Engineering cell alignment in vitro. Biotechnol. Adv. 2014, 32, 347–365. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Song, J.; Yang, P.; Zou, R.; Fan, X.; Zhao, Z. Establishment of a three-dimensional culture and mechanical loading system for skeletal myoblasts. Cell Biol. Int. 2009, 33, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.F.; Patel, S.; Thakar, R.G.; Wu, J.; Hsiao, B.S.; Chu, B.; Lee, R.J.; Li, S. Myotube assembly on nanofibrous and micropatterned polymers. Nano Lett. 2006, 6, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zeng, H.; Nam, J.; Agarwal, S. Fabrication of skeletal muscle constructs by topographic activation of cell alignment. Biotechnol. Bioeng. 2009, 102, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Hinds, S.; Bian, W.; Dennis, R.G.; Bursac, N. The role of extracellular matrix composition in structure and function of bioengineered skeletal muscle. Biomaterials 2011, 32, 3575–3583. [Google Scholar] [CrossRef] [PubMed]
- Ker, E.D.F.; Nain, A.S.; Weiss, L.E.; Wang, J.; Suhan, J.; Amon, C.H.; Campbell, P.G. Bioprinting of growth factors onto aligned sub-micron fibrous scaffolds for simultaneous control of cell differentiation and alignment. Biomaterials 2011, 32, 8097–8107. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.W.; Chien, H.W.; Tsai, W.B. Fabrication of poly(N-isopropylacrylamide) films containing submicrometer grooves for constructing aligned cell sheets. Langmuir 2013, 29, 14351–14355. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Duffy, R.; Lee, A.; Feinberg, A.W. Optimizing the structure and contractility of engineered skeletal muscle thin films. Acta Biomater. 2013, 9, 7885–7894. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Nakayama, M.; Itoga, K.; Yamato, M.; Okano, T. Micropatterned thermoresponsive polymer brush surfaces for fabricating cell sheets with well-controlled orientational structures. Biomacromolecules 2011, 12, 1414–1418. [Google Scholar] [CrossRef] [PubMed]
- Yoshiike, Y.; Yokota, S.; Tanaka, N.; Kitaoka, T.; Wariishi, H. Preparation and cell culture behavior of self-assembled monolayers composed of chitohexaose and chitosan hexamer. Carbohydr. Polym. 2010, 82, 21–27. [Google Scholar] [CrossRef]
- Powell, C.A.; Smiley, B.L.; Mills, J. Mechanical stimulation improves tissue-engineered human skeletal muscle. Am. J. Physiol. Cell Physiol. 2002, 283, 1557–1565. [Google Scholar] [CrossRef] [PubMed]
- Yokota, S.; Kitaoka, T.; Wariishi, H. Biofunctionality of self-assembled nanolayers composed of cellulosic polymers. Carbohydr. Polym. 2008, 74, 666–672. [Google Scholar] [CrossRef]
- Tanaka, N.; Yoshiike, Y.; Yoshiyama, C.; Kitaoka, T. Self-assembly immobilization of hyaluronan thiosemicarbazone on a gold surface for cell culture applications. Carbohydr. Polym. 2010, 82, 100–105. [Google Scholar] [CrossRef]
- Yoshiike, Y.; Kitaoka, T. Tailoring hybrid glyco-nanolayers composed of chitohexaose and cellohexaose for cell culture applications. J. Mater. Chem. 2011, 21, 11150–11158. [Google Scholar] [CrossRef]
- Fyrner, T.; Ederth, T.; Aili, D.; Liedberg, B.; Konradsson, P. Synthesis of oligo(lactose)-based thiols and their self-assembly onto gold surfaces. Colloids Surf. B Biointerfaces 2013, 105, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, K.; Du, D.; Sun, Y.; He, L. Recognition of glycoprotein peroxidase via Con A-carrying self-assembly layer on gold. Biomacromolecules 2007, 8, 2142–2148. [Google Scholar] [CrossRef] [PubMed]
- Walgren, J.L.E.; Vincent, T.S.; Schey, K.L.; Buse, M.G. High glucose and insulin promote O-GlcNAc modification of proteins, including alpha-tubulin. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E424–E434. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Furukawa, S.; Morita, K.; Ishihara, T.; Shiotani, M.; Matsushita, Y.; Matsuda, M.; Shimomura, I. Glucosamine induces lipid accumulation and adipogenic change in C2C12 myoblasts. Biochem. Biophys. Res. Commun. 2005, 328, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Sakakibara, Y.; Kamemura, K. Requirement of decreased O-GlcNAc glycosylation of Mef2D for its recruitment to the myogenin promoter. Biochem. Biophys. Res. Commun. 2013, 433, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Vaz, R.; Martins, G.G.; Thorsteinsdóttir, S.; Rodrigues, G. Fibronectin promotes migration, alignment and fusion in an in vitro myoblast cell model. Cell Tissue Res. 2012, 348, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Grigola, M.S.; Dyck, C.L.; Babacan, D.S.; Joaquin, D.N.; Hsia, K.J. Myoblast alignment on 2D wavy patterns: Dependence on feature characteristics and cell–cell interaction. Biotechnol. Bioeng. 2014, 111, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Weber, G.F.; Menko, A.S. Actin filament organization regulates the induction of lens cell differentiation and survival. Dev. Biol. 2006, 295, 714–729. [Google Scholar] [CrossRef] [PubMed]
- Ting, H.J.; Chang, C. Actin associated proteins function as androgen receptor coregulators: An implication of androgen receptor’s roles in skeletal muscle. J. Steroid Biochem. Mol. Biol. 2008, 111, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Galli, C.; Piemontese, M.; Lumetti, S.; Ravanetti, F.; Macaluso, G.M.; Passeri, G. Actin cytoskeleton controls activation of Wnt/β-catenin signaling in mesenchymal cells on implant surfaces with different topographies. Acta Biomater. 2012, 8, 2963–2968. [Google Scholar] [CrossRef] [PubMed]
- Yaffe, D.; Feldman, M. The formation of hybrid multinucleated muscle fibers from myoblasts of different genetic origin. Dev. Biol. 1965, 11, 300–317. [Google Scholar] [CrossRef]
- Kim, J.A.; Shon, Y.H.; Lim, J.O.; Yoo, J.J.; Shin, H.I.; Park, E.K. MYOD mediates skeletal myogenic differentiation of human amniotic fluid stem cells and regeneration of muscle injury. Stem Cell Res. Ther. 2013. [Google Scholar] [CrossRef] [PubMed]
- Wood, W.M.; Etemad, S.; Yamamoto, M.; Goldhamer, D.J. MyoD-expressing progenitors are essential for skeletal myogenesis and satellite cell development. Dev. Biol. 2013, 384, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Dudek, S.M.; Chiang, E.T.; Camp, S.M.; Guo, Y.; Zhao, J.; Brown, M.E.; Singleton, P.; Wang, L.; Desai, A.; Arce, F.T.; et al. Focal adhesion kinase signaling regulates the expression of caveolin 3 and 1 integrin, genes essential for normal myoblast fusion. Mol. Biol. Cell 2010, 21, 4042–4056. [Google Scholar] [CrossRef] [PubMed]
- Keselowsky, B.G.; Collard, D.M.; Garcia, A.J. Integrin binding specificity regulates biomaterial surface chemistry effects on cell differentiation. Proc. Natl. Acad. Sci. USA 2005, 102, 5953–5957. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.F.; Lee, R.J.; Li, S. Engineering of aligned skeletal muscle by micropatterning. Am. J. Transl. Res. 2010, 2, 43–55. [Google Scholar] [PubMed]
- Kobayashi, S.; Makino, A.; Matsumoto, H.; Kunii, S.; Ohmae, M.; Kiyosada, T.; Makiguchi, K.; Matsumoto, A.; Horie, M.; Shoda, S.I. Enzymatic polymerization to novel polysaccharides having a glucose-N-acetylglucosamine repeating unit, a cellulose-chitin hybrid polysaccharide. Biomacromolecules 2006, 7, 1644–1656. [Google Scholar] [CrossRef] [PubMed]
- Ihara, H.; Hanashima, S.; Tsukamoto, H.; Yamaguchi, Y.; Taniguchi, N.; Ikeda, Y. Difucosylation of chitooligosaccharides by eukaryote and prokaryote α1,6-fucosyltransferases. Biochim. Biophys. Acta 2013, 1830, 4482–4490. [Google Scholar] [CrossRef] [PubMed]
- Gross, T.; Lippitz, A.; Unger, W.E.S. Chemical and elemental depth profiling of very thin organic layers by constant kinetic energy XPS: A new Synchrotron XPS analysis strategy. Anal. Chem. 2012, 84, 5984–5991. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poosala, P.; Kitaoka, T. Chitooligomer-Immobilized Biointerfaces with Micropatterned Geometries for Unidirectional Alignment of Myoblast Cells. Biomolecules 2016, 6, 12. https://doi.org/10.3390/biom6010012
Poosala P, Kitaoka T. Chitooligomer-Immobilized Biointerfaces with Micropatterned Geometries for Unidirectional Alignment of Myoblast Cells. Biomolecules. 2016; 6(1):12. https://doi.org/10.3390/biom6010012
Chicago/Turabian StylePoosala, Pornthida, and Takuya Kitaoka. 2016. "Chitooligomer-Immobilized Biointerfaces with Micropatterned Geometries for Unidirectional Alignment of Myoblast Cells" Biomolecules 6, no. 1: 12. https://doi.org/10.3390/biom6010012