Histone Deacetylase Inhibitors Activate Tristetraprolin Expression through Induction of Early Growth Response Protein 1 (EGR1) in Colorectal Cancer Cells
Abstract
:1. Introduction
2. Results
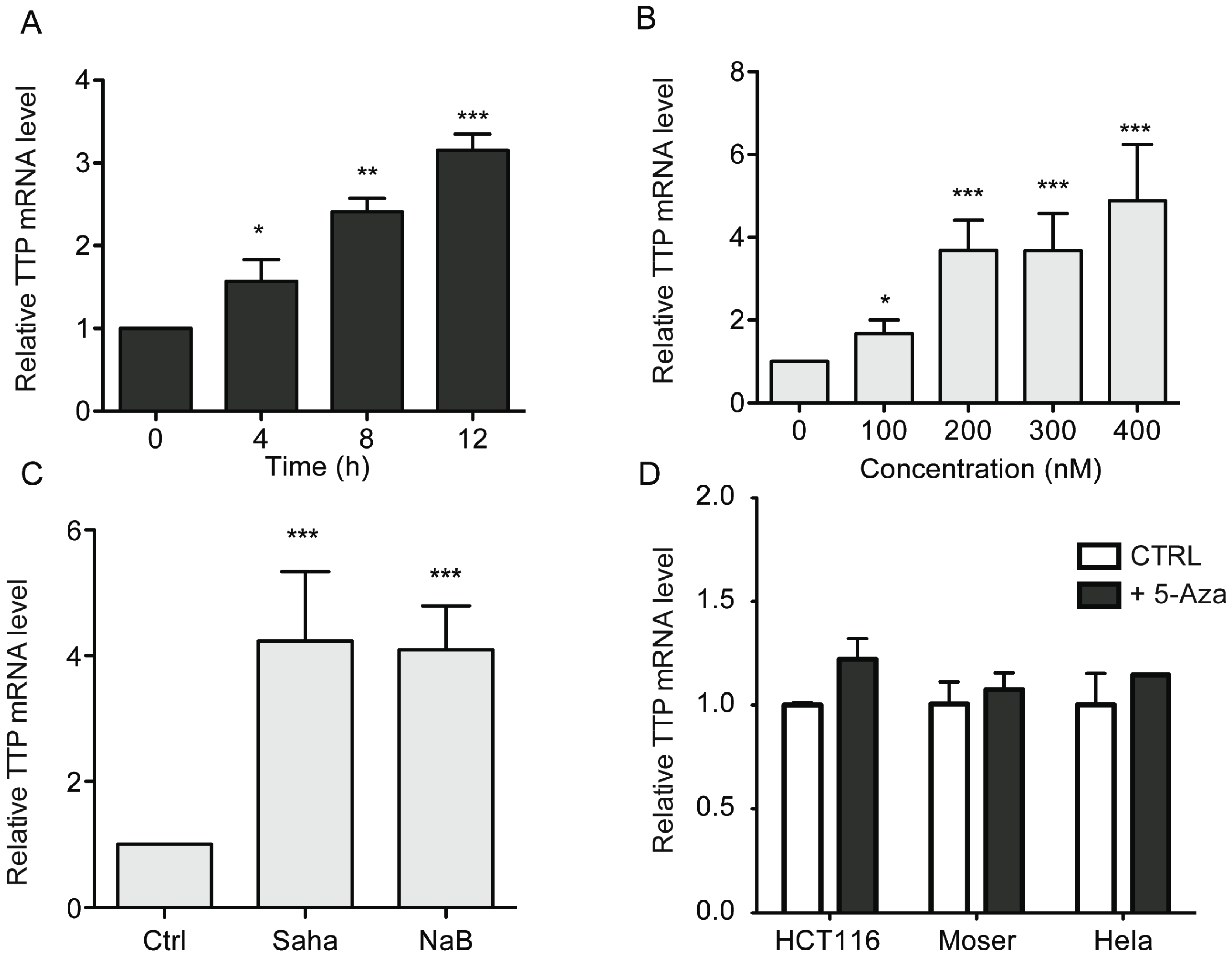
2.1. HDAC Inhibitors Promote TTP Expression in CRC Cells

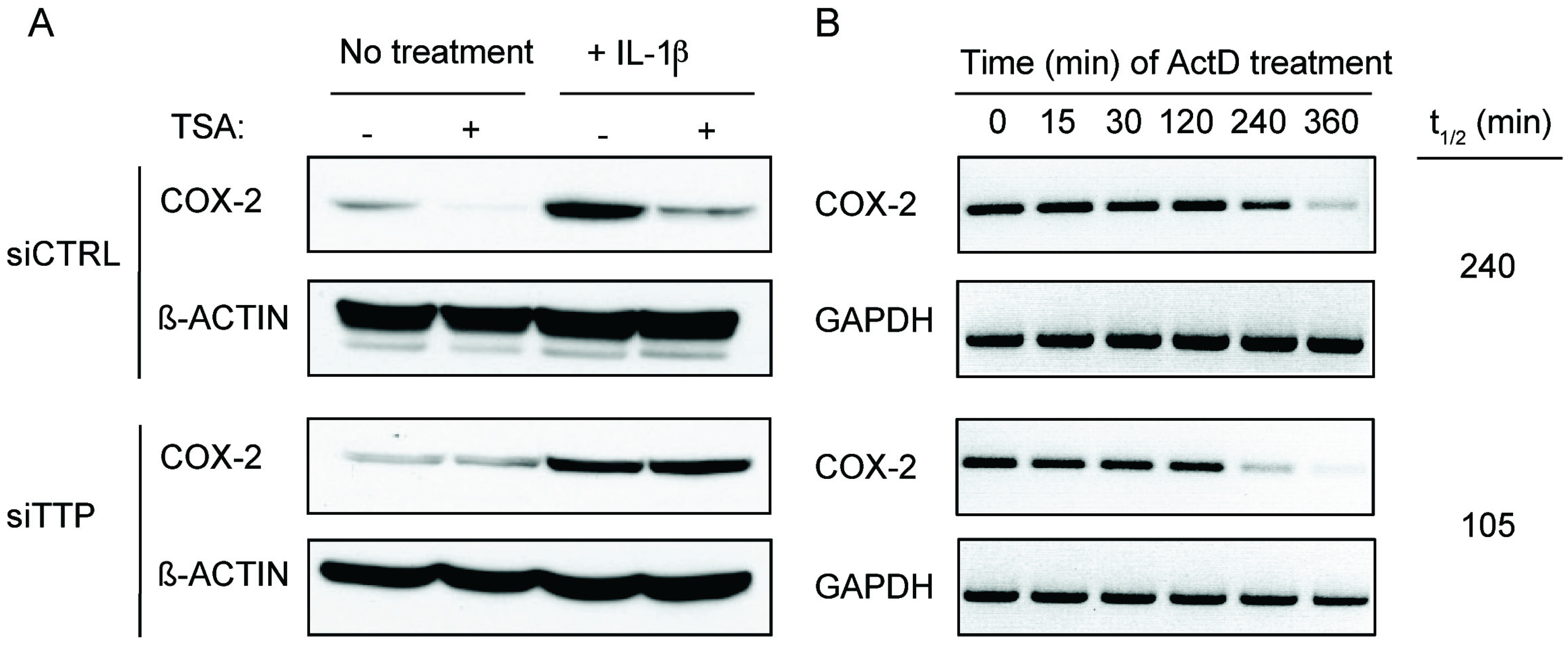
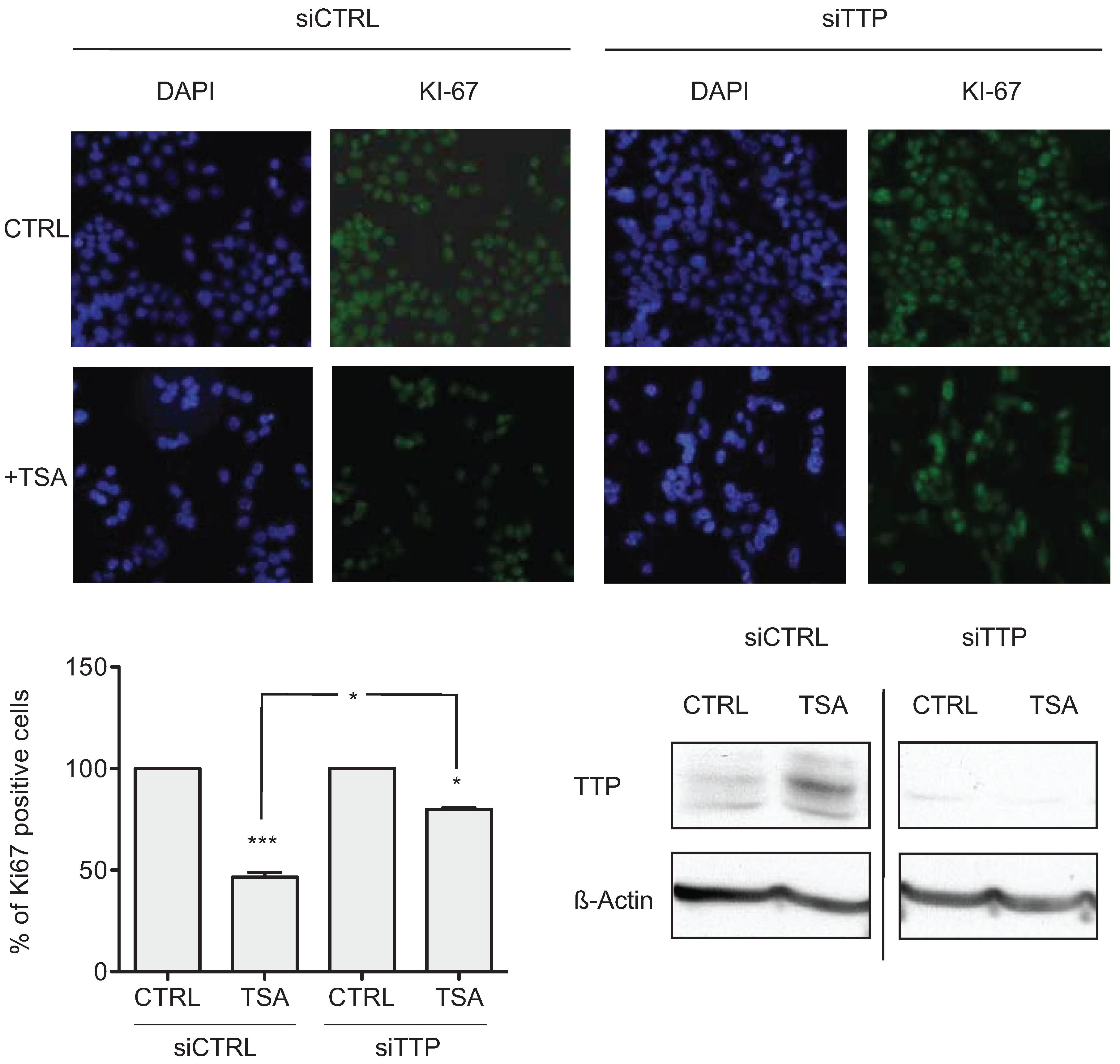
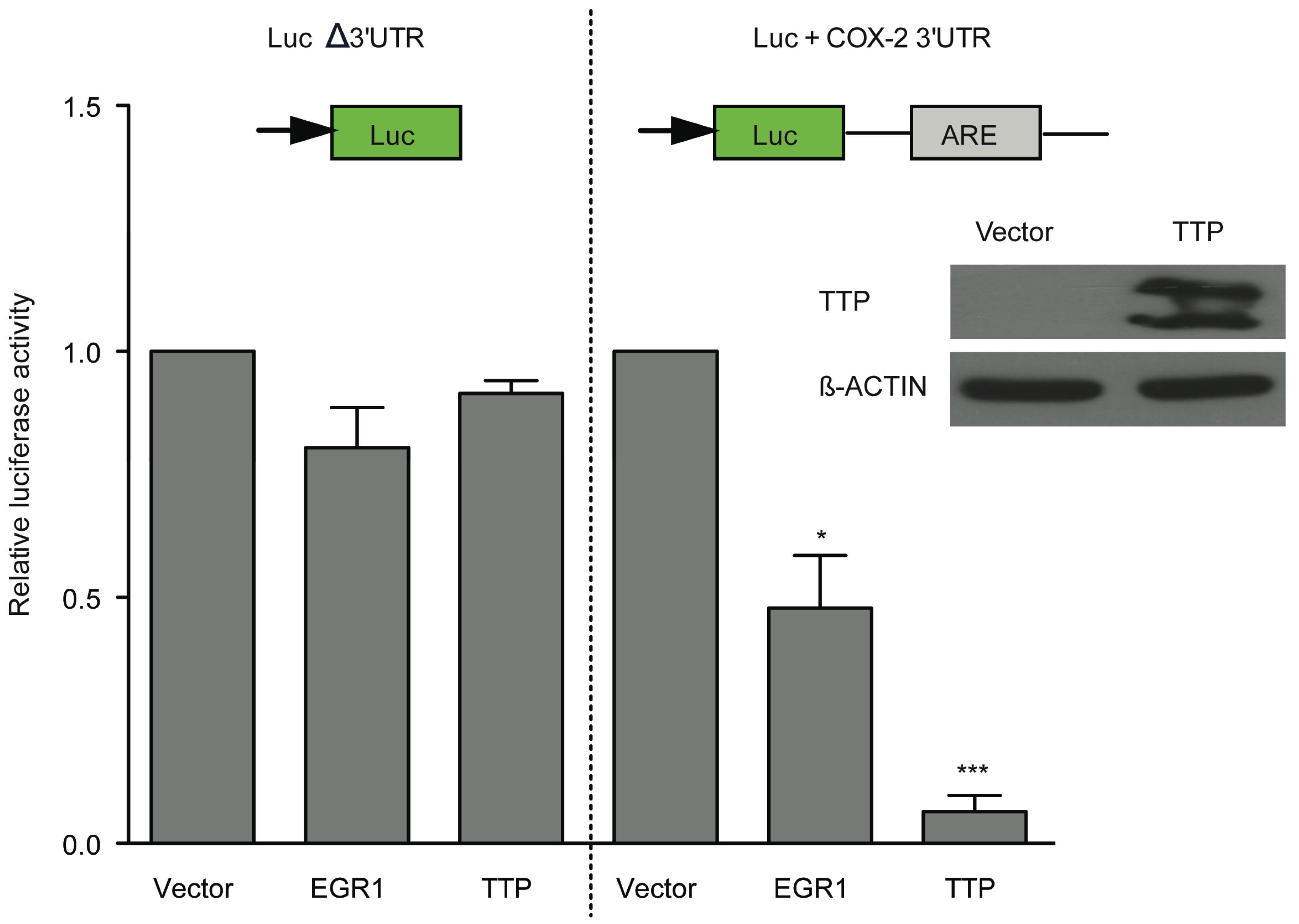
2.2. TSA-Induced TTP Expression Contributes to ARE-mRNA Decay and Growth Inhibition
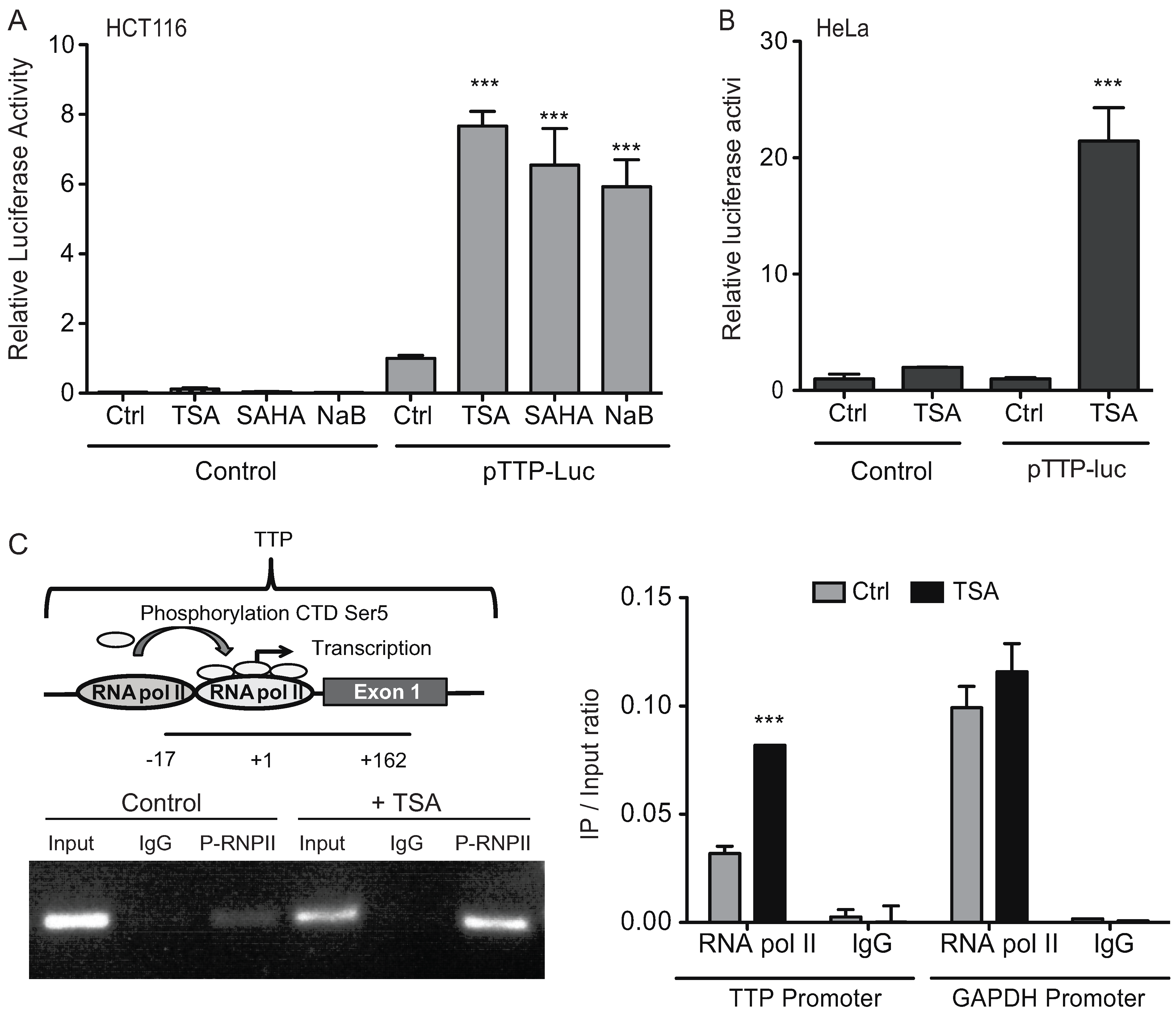
2.3. HDAC Inhibitors Promote TTP Transcription

2.4. Early Growth Response Protein 1 (EGR1) is Associated in TSA-Induced TTP Expression

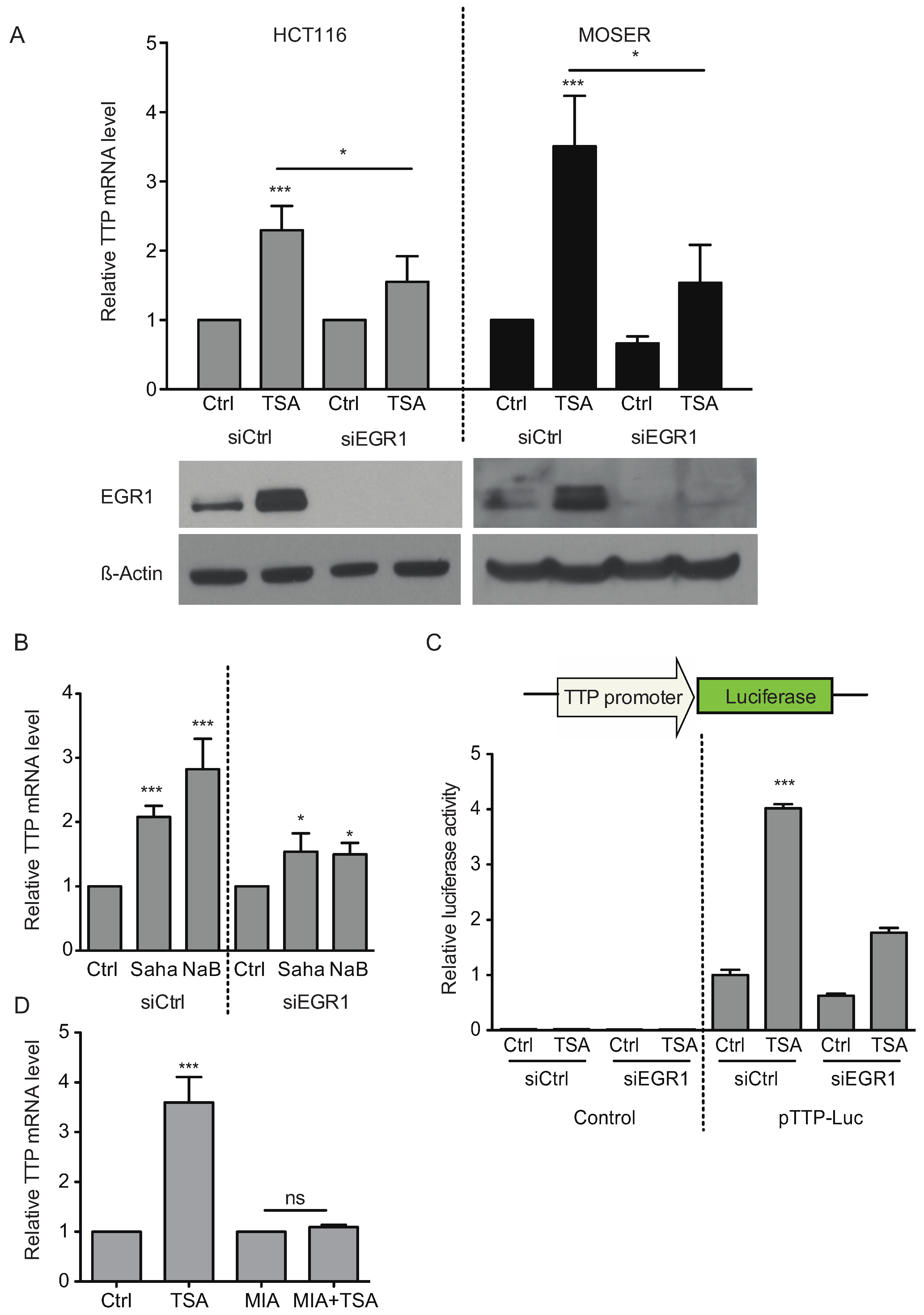
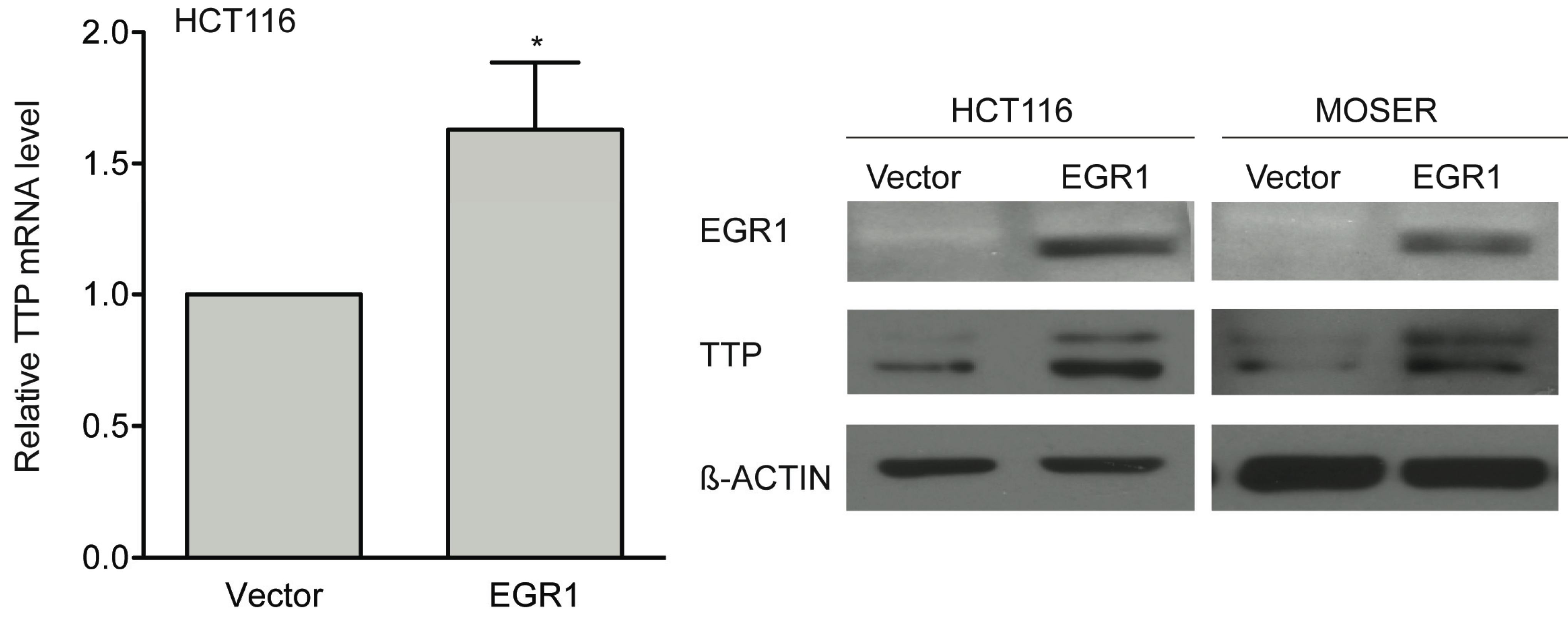
2.5. P-Bodies, TTP and EGR1 Are Decreased in Colorectal Tumors
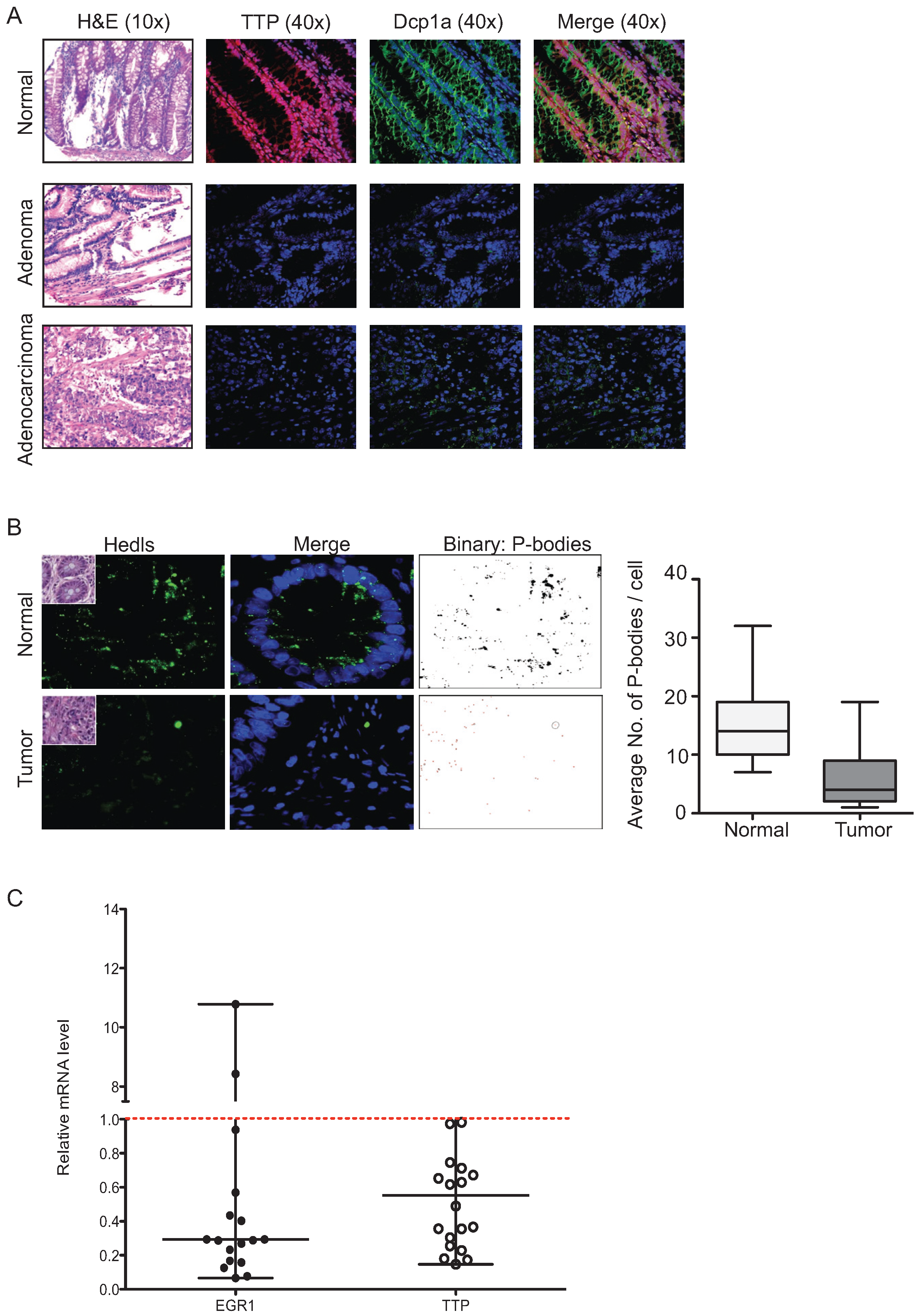
3. Discussion
4. Experimental Section
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jemal, A.; Center, M.M.; DeSantis, C.; Ward, E.M. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1893–1907. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Young, L.E.; Dixon, D.A. Posttranscriptional regulation of cyclooxygenase 2 expression in colorectal cancer. Curr. Colorectal Cancer Rep. 2010, 6, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Garneau, N.L.; Wilusz, J.; Wilusz, C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007, 8, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Kanies, C.L.; Smith, J.J.; Kis, C.; Schmidt, C.; Levy, S.; Khabar, K.S.; Morrow, J.; Deane, N.; Dixon, D.A.; Beauchamp, R.D. Oncogenic ras and transforming growth factor-beta synergistically regulate AU-rich element-containing mRNAs during epithelial to mesenchymal transition. Mol. Cancer Res. 2008, 6, 1124–1136. [Google Scholar] [CrossRef] [PubMed]
- Lopez de Silanes, I.; Fan, J.; Yang, X.; Zonderman, A.B.; Potapova, O.; Pizer, E.S.; Gorospe, M. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 2003, 22, 7146–7154. [Google Scholar] [CrossRef] [PubMed]
- Young, L.E.; Sanduja, S.; Bemis-Standoli, K.; Pena, E.A.; Price, R.L.; Dixon, D.A. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology 2009, 136, 1669–1679. [Google Scholar] [CrossRef] [PubMed]
- Sanduja, S.; Blanco, F.F.; Dixon, D.A. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip. Rev. RNA 2011, 2, 42–57. [Google Scholar] [CrossRef] [PubMed]
- Taylor, G.A.; Carballo, E.; Lee, D.M.; Lai, W.S.; Thompson, M.J.; Patel, D.D.; Schenkman, D.I.; Gilkeson, G.S.; Broxmeyer, H.E.; Haynes, B.F.; et al. A pathogenetic role for tnf alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 1996, 4, 445–454. [Google Scholar] [CrossRef]
- Carballo, E.; Lai, W.S.; Blackshear, P.J. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 1998, 281, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Phillips, K.; Kedersha, N.; Shen, L.; Blackshear, P.J.; Anderson, P. Arthritis suppressor genes tia-1 and ttp dampen the expression of tumor necrosis factor alpha, cyclooxygenase 2, and inflammatory arthritis. Proc. Natl. Acad. Sci. USA 2004, 101, 2011–2016. [Google Scholar] [CrossRef] [PubMed]
- Sawaoka, H.; Dixon, D.A.; Oates, J.A.; Boutaud, O. Tristetrapolin binds to the 3' untranslated region of cyclooxygenase-2 mRNA: A polyadenylation variant in a cancer cell line lacks the binding site. J. Biol. Chem. 2003, 278, 13928–13935. [Google Scholar] [CrossRef] [PubMed]
- Sanduja, S.; Blanco, F.F.; Young, L.E.; Kaza, V.; Dixon, D.A. The role of tristetraprolin in cancer and inflammation. Front. Biosci. 2012, 17, 174–188. [Google Scholar] [CrossRef]
- Staub, E.; Grone, J.; Mennerich, D.; Ropcke, S.; Klamann, I.; Hinzmann, B.; Castanos-Velez, E.; Mann, B.; Pilarsky, C.; Brummendorf, T.; et al. A genome-wide map of aberrantly expressed chromosomal islands in colorectal cancer. Mol. Cancer 2006, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Mariadason, J.M. HDACs and HDAC inhibitors in colon cancer. Epigenetics 2008, 3, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Egger, G.; Liang, G.; Aparicio, A.; Jones, P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004, 429, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Bassett, A.; Cooper, S.; Wu, C.; Travers, A. The folding and unfolding of eukaryotic chromatin. Curr. Opin. Genet. Dev. 2009, 19, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.J.; Byun, D.S.; Popova, N.; Murray, L.B.; L’Italien, K.; Sowa, Y.; Arango, D.; Velcich, A.; Augenlicht, L.H.; Mariadason, J.M. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J. Biol. Chem. 2006, 281, 13548–13558. [Google Scholar] [CrossRef] [PubMed]
- Brennan, S.E.; Kuwano, Y.; Alkharouf, N.; Blackshear, P.J.; Gorospe, M.; Wilson, G.M. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009, 69, 5168–5176. [Google Scholar] [CrossRef] [PubMed]
- Carrick, D.M.; Blackshear, P.J. Comparative expression of tristetraprolin (TTP) family member transcripts in normal human tissues and cancer cell lines. Arch. Biochem. Biophys. 2007, 462, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Sohn, B.H.; Park, I.Y.; Lee, J.J.; Yang, S.J.; Jang, Y.J.; Park, K.C.; Kim, D.J.; Lee, D.C.; Sohn, H.A.; Kim, T.W.; et al. Functional switching of TGF-beta1 signaling in liver cancer via epigenetic modulation of a single CpG site in TTP promoter. Gastroenterology 2010, 138, 1898–1908. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Jiang, Y.W.; Su, Y.L.; Lee, S.C.; Chang, M.S.; Chang, C.J. Transcriptional regulation of tristetraprolin by NF-κB signaling in LPS-stimulated macrophages. Mol. Biol. Rep. 2013, 40, 2867–2877. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.S.; Thompson, M.J.; Taylor, G.A.; Liu, Y.; Blackshear, P.J. Promoter analysis of ZFP-36, the mitogen-inducible gene encoding the zinc finger protein tristetraprolin. J. Biol. Chem. 1995, 270, 25266–25272. [Google Scholar] [CrossRef] [PubMed]
- Florkowska, M.; Tymoszuk, P.; Balwierz, A.; Skucha, A.; Kochan, J.; Wawro, M.; Stalinska, K.; Kasza, A. Egf activates TTP expression by activation of ELK-1 and EGR-1 transcription factors. BMC Mol. Biol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Cheng, H.; Sampaio, A.V.; Nielsen, T.O.; Underhill, T.M. Egr1 reactivation by histone deacetylase inhibitors promotes synovial sarcoma cell death through the PTEN tumor suppressor. Oncogene 2010, 29, 4352–4361. [Google Scholar] [CrossRef] [PubMed]
- Gitenay, D.; Baron, V.T. Is EGR1 a potential target for prostate cancer therapy? Future Oncol. 2009, 5, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Young, L.E.; Moore, A.E.; Sokol, L.; Meisner-Kober, N.; Dixon, D.A. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol. Cancer Res. 2012, 10, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, M.; Ozgur, S.; Stoecklin, G. On track with P-bodies. Biochem. Soc. Trans. 2010, 38, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Kedersha, N.; Ivanov, P. Stress granules, p-bodies and cancer. Biochim. Biophys. Acta Gene Regul. Mech. 2015, 1849, 861–870. [Google Scholar] [CrossRef] [PubMed]
- Blanco, F.F.; Sanduja, S.; Deane, N.G.; Blackshear, P.J.; Dixon, D.A. Transforming growth factor beta regulates P-body formation through induction of the mRNA decay factor tristetraprolin. Mol. Cell. Biol. 2014, 34, 180–195. [Google Scholar] [CrossRef] [PubMed]
- Rounbehler, R.J.; Fallahi, M.; Yang, C.; Steeves, M.A.; Li, W.; Doherty, J.R.; Schaub, F.X.; Sanduja, S.; Dixon, D.A.; Blackshear, P.J.; et al. Tristetraprolin impairs Myc-induced lymphoma and abolishes the malignant state. Cell 2012, 150, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Yim, S.; Choi, B.J.; Jang, Y.; Lee, J.J.; Sohn, B.H.; Yoo, H.S.; Yeom, Y.I.; Park, K.C. Tristetraprolin regulates the stability of HIF-1alpha mRNA during prolonged hypoxia. Biochem. Biophys. Res. Commun. 2010, 391, 963–968. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, R.; Sanduja, S.; Kaza, V.; Dixon, D.A. Genetic polymorphisms in RNA binding proteins contribute to breast cancer survival. Int. J. Cancer 2013, 132, E128–E138. [Google Scholar] [CrossRef] [PubMed]
- Sanduja, S.; Kaza, V.; Dixon, D.A. The mRNA decay factor tristetraprolin (TTP) induces senescence in human papillomavirus-transformed cervical cancer cells by targeting E6-AP ubiquitin ligase. Aging 2009, 1, 803–817. [Google Scholar] [PubMed]
- Sobolewski, C.; Cerella, C.; Dicato, M.; Ghibelli, L.; Diederich, M. The role of cyclooxygenase-2 in cell proliferation and cell death in human malignancies. Int. J. Cell Biol. 2010, 2010, 215158. [Google Scholar] [CrossRef] [PubMed]
- Mahalingam, D.; Natoni, A.; Keane, M.; Samali, A.; Szegezdi, E. Early growth response-1 is a regulator of DR5-induced apoptosis in colon cancer cells. Br. J. Cancer 2010, 102, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Sarver, A.L.; Li, L.; Subramanian, S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010, 70, 9570–9580. [Google Scholar] [CrossRef] [PubMed]
- Tice, D.A.; Soloviev, I.; Polakis, P. Activation of the wnt pathway interferes with serum response element-driven transcription of immediate early genes. J. Biol. Chem. 2002, 277, 6118–6123. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.J.; Chueh, A.C.; Togel, L.; Corner, G.A.; Ahmed, N.; Goel, S.; Byun, D.S.; Nasser, S.; Houston, M.A.; Jhawer, M.; et al. Apoptotic sensitivity of colon cancer cells to histone deacetylase inhibitors is mediated by an Sp1/Sp3-activated transcriptional program involving immediate-early gene induction. Cancer Res. 2010, 70, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K.; Chen, F.; Kim, Y.J.; Chen, Y. Transcriptional regulation of tristetraprolin by transforming growth factor-beta in human T cells. J. Biol. Chem. 2003, 278, 30373–30381. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.N.; Bai, J.X.; Zhou, Q.; Yan, B.; Qin, W.W.; Jia, L.T.; Meng, Y.L.; Jin, B.Q.; Yao, L.B.; Wang, T.; et al. TSA suppresses miR-106b-93-25 cluster expression through downregulation of Myc and inhibits proliferation and induces apoptosis in human EMC. PLoS ONE 2012, 7, e45133. [Google Scholar] [CrossRef] [PubMed]
- Gebeshuber, C.A.; Zatloukal, K.; Martinez, J. MiR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009, 10, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.K.; Law, P.T.; Lee, C.W.; Cho, C.H.; Fan, D.; Wu, K.; Yu, J.; Sung, J.J. MicroRNA in colorectal cancer: From benchtop to bedside. Carcinogenesis 2011, 32, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Bhat, A.A.; Krishnan, M.; Singh, A.B.; Dhawan, P. Trichostatin-A modulates claudin-1 mRNA stability through the modulation of Hu antigen R and tristetraprolin in colon cancer cells. Carcinogenesis 2013, 34, 2610–2621. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.P.; Fan, Y.; Ni, Z.; Mercola, D.; Adamson, E.D. Reciprocal modulation between Sp1 and EGR-1. J. Cell. Biochem. 1997, 66, 489–499. [Google Scholar] [CrossRef]
- Fu, L.; Huang, W.; Jing, Y.; Jiang, M.; Zhao, Y.; Shi, J.; Huang, S.; Xue, X.; Zhang, Q.; Tang, J.; et al. AML1-ATO triggers epigenetic activation of early growth response gene l, inducing apoptosis in t(8;21) acute myeloid leukemia. FEBS J. 2014, 281, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Deckmann, K.; Rorsch, F.; Geisslinger, G.; Grosch, S. Dimethylcelecoxib induces an inhibitory complex consisting of HDAC1/NF-kappaB(p65)RelA leading to transcriptional downregulation of mPGES-1 and EGR1. Cell. Signal. 2012, 24, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Lu, J.; Wang, X.; Han, L.; Zhang, Y.; Han, S.; Huang, B. Histone deacetylase inhibitor trichostatin A potentiates doxorubicin-induced apoptosis by up-regulating PTEN expression. Cancer 2007, 109, 1676–1688. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Kwon, H.J.; Yoon, B.I.; Kim, J.H.; Han, S.U.; Joo, H.J.; Kim, D.Y. Expression profile of histone deacetylase 1 in gastric cancer tissues. Jpn. J. Cancer Res. 2001, 92, 1300–1304. [Google Scholar] [CrossRef] [PubMed]
- Halkidou, K.; Gaughan, L.; Cook, S.; Leung, H.Y.; Neal, D.E.; Robson, C.N. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate 2004, 59, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Patra, S.K.; Patra, A.; Dahiya, R. Histone deacetylase and DNA methyltransferase in human prostate cancer. Biochem. Biophys. Res. Commun. 2001, 287, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Martin, E.; Mengwasser, J.; Schlag, P.; Janssen, K.P.; Gottlicher, M. Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell 2004, 5, 455–463. [Google Scholar] [CrossRef]
- Dixon, D.A.; Kaplan, C.D.; McIntyre, T.M.; Zimmerman, G.A.; Prescott, S.M. Post-transcriptional control of cyclooxygenase-2 gene expression. The role of the 3'-untranslated region. J. Biol. Chem. 2000, 275, 11750–11757. [Google Scholar] [CrossRef] [PubMed]
- Fenger-Gron, M.; Fillman, C.; Norrild, B.; Lykke-Andersen, J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell 2005, 20, 905–915. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobolewski, C.; Sanduja, S.; Blanco, F.F.; Hu, L.; Dixon, D.A. Histone Deacetylase Inhibitors Activate Tristetraprolin Expression through Induction of Early Growth Response Protein 1 (EGR1) in Colorectal Cancer Cells. Biomolecules 2015, 5, 2035-2055. https://doi.org/10.3390/biom5032035
Sobolewski C, Sanduja S, Blanco FF, Hu L, Dixon DA. Histone Deacetylase Inhibitors Activate Tristetraprolin Expression through Induction of Early Growth Response Protein 1 (EGR1) in Colorectal Cancer Cells. Biomolecules. 2015; 5(3):2035-2055. https://doi.org/10.3390/biom5032035
Chicago/Turabian StyleSobolewski, Cyril, Sandhya Sanduja, Fernando F. Blanco, Liangyan Hu, and Dan A. Dixon. 2015. "Histone Deacetylase Inhibitors Activate Tristetraprolin Expression through Induction of Early Growth Response Protein 1 (EGR1) in Colorectal Cancer Cells" Biomolecules 5, no. 3: 2035-2055. https://doi.org/10.3390/biom5032035





