The 3' to 5' Exoribonuclease DIS3: From Structure and Mechanisms to Biological Functions and Role in Human Disease
Abstract
:1. Introduction
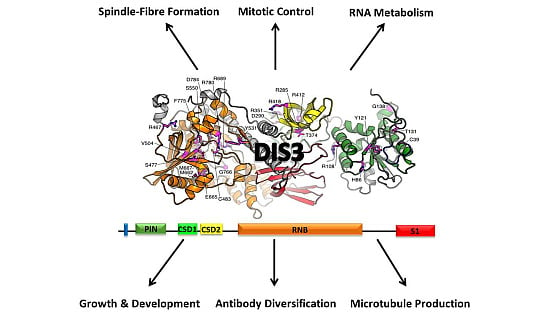
2. Conservation, Structure, Mechanistic Functions and Sub-Cellular Localisation of DIS3
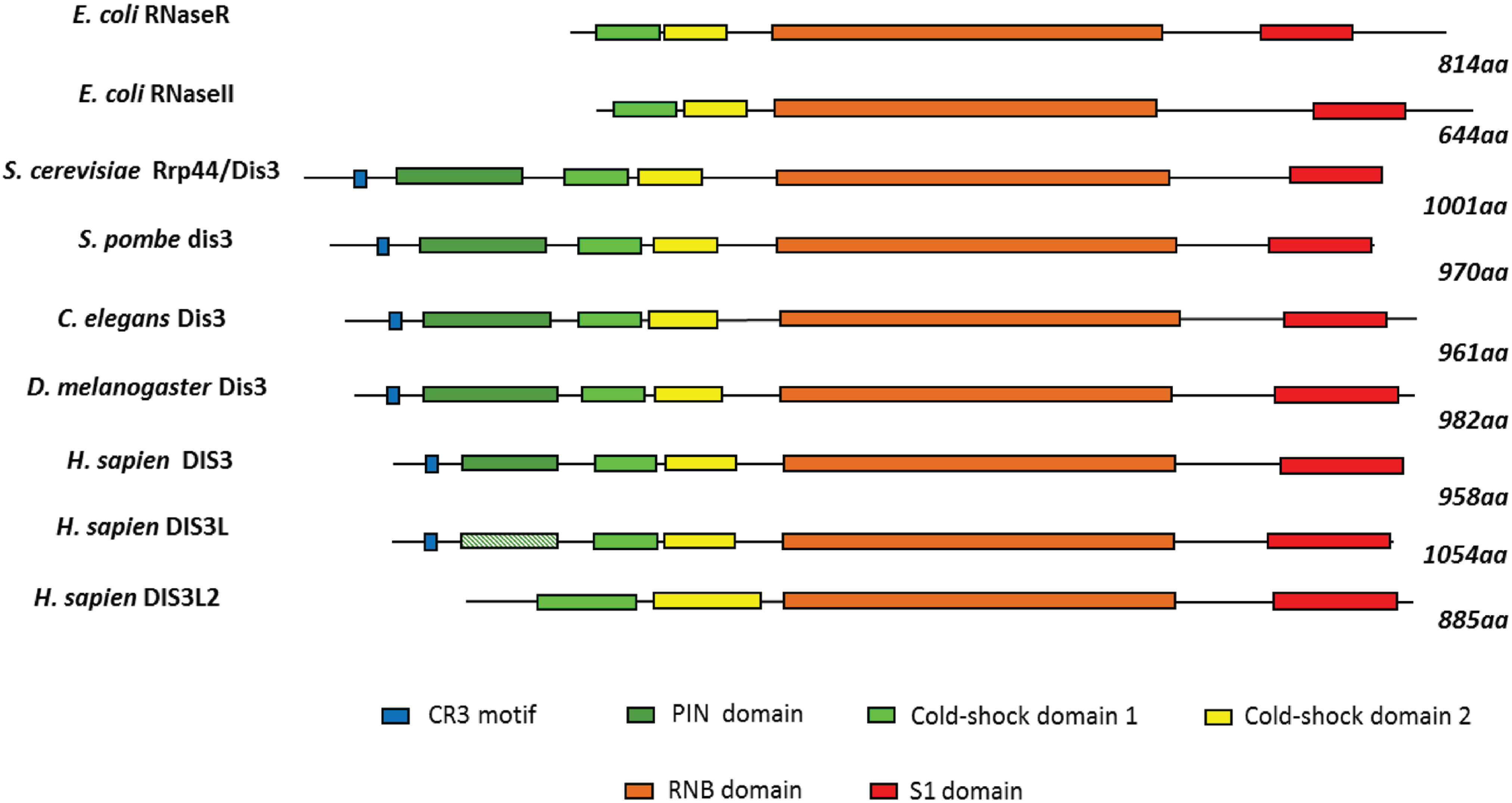
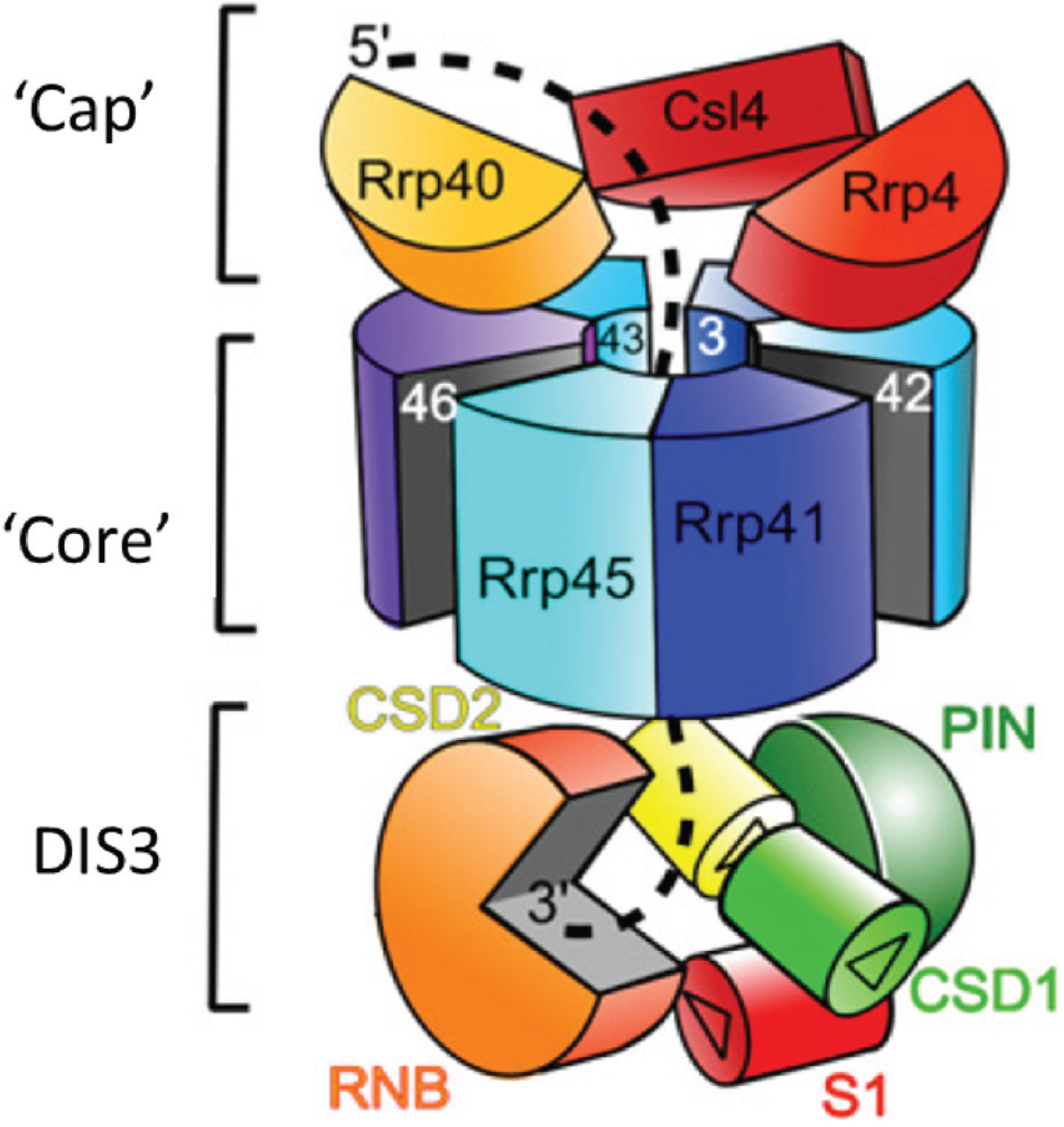
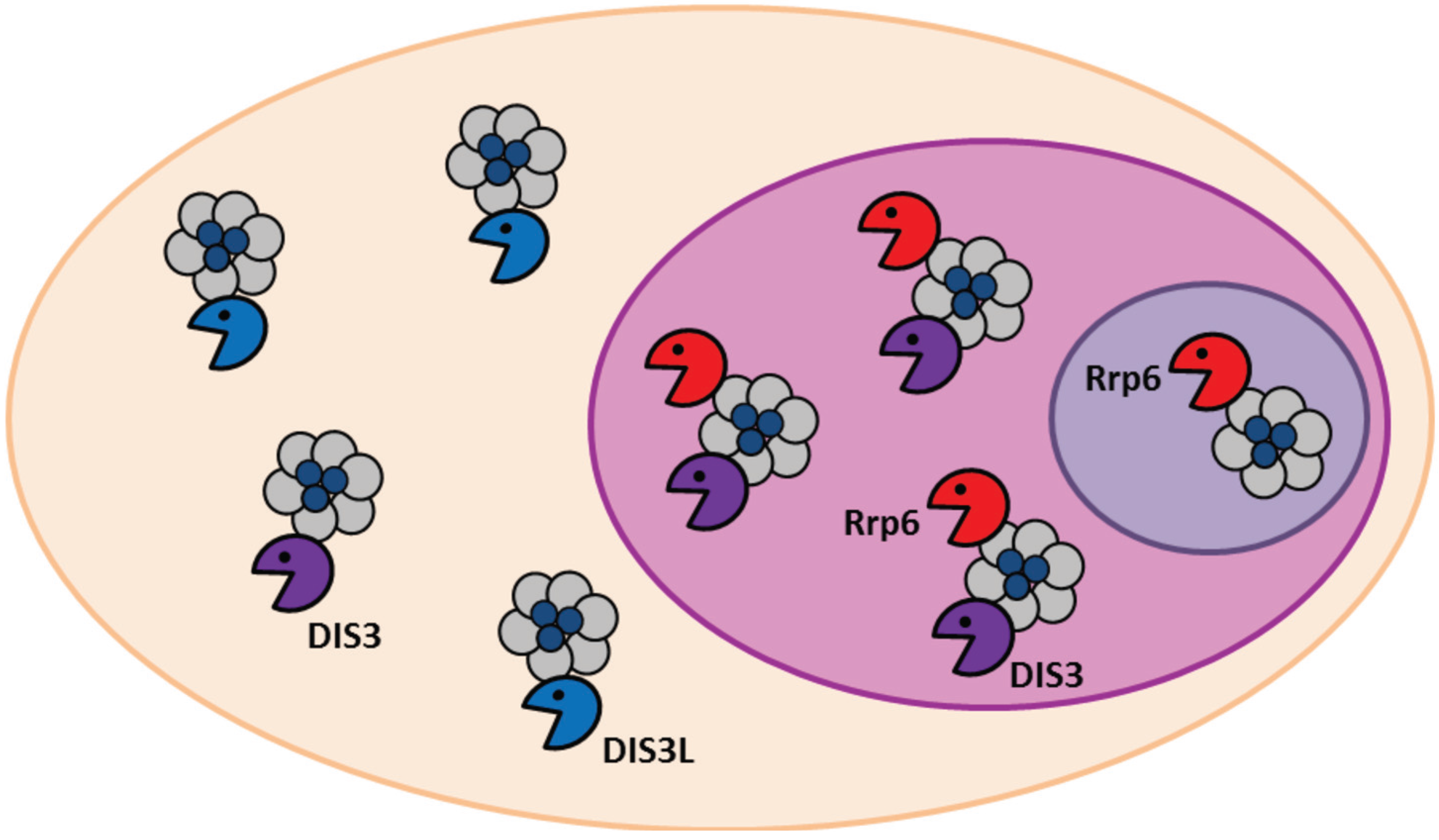
3. Molecular Functions of DIS3
3.1. Role of DIS3 in mRNA Decay
3.1.1. Role of DIS3 in RNA Quality-Control Pathways

3.1.2. Role of DIS3 in Regulated mRNA-Decay Pathways
3.2. Role of DIS3 in Small-RNA Processing and Decay
4. Biological Functions of DIS3
4.1. Role of DIS3 in Cell-Cycle Regulation
| Phenotype/Process Affected | Organism/Cell Line | Knock-Down or Mutant (AA Position) | Corresponding Human DIS3 AA Position | Conserved in Humans? | Domain | DIS3/Exosome Subunit Referred to as | Refs. |
|---|---|---|---|---|---|---|---|
| Non-separation of sister chromatids | S. pombe | P509L | P509—Conserved (based on predicted S. pombe sequence) | Yes | RNB | dis3 | [12,13] |
| Mitotic control and interaction with Ran GTPase | S. cerevisiae | G562D, E565K, V566G | G562 E565 V566 | Yes, all three | RNB | Dis3sc | [14,91] |
| Aneuploidy, spindle assembly, metaphase to anaphase transition and kinetochore function | S. pombe | P509L | P509—Conserved (based on predicted S. pombe sequence) | Yes | RNB | dis3 | [16] |
| Cell-cycle regulation and microtubule production | S. cerevisiae D. melanogaster | G562D, E565K, V566G Knock-down | G562 E565 V566 | YesN/A | RNBN/A | Dis3 | [14,66,93] |
| Larval lethality, no wings | D. melanogaster | Knock-down | N/A | N/A | N/A | Dis3 | [80] |
| Centromeric transcript turnover and heterochromatin silencing | S. pombe, S. cerevisiae | P509L | P509 | Yes | RNB | Dis3/Rrp44/Rrp4 | [16,94,95] |
| Antibody diversification | M. musculus CH12F3 lymphoma cells, Human Ramos B lymphoma cells, HEK-293 cells | Knock-down | N/A | N/A | N/A | Dis3/Rrp44/Rrp40, exosome subunit | [17] |

4.2. Role of DIS3 in Generating Antibody Diversity
5. DIS3 and Disease
5.1. DIS3 and Cancer
5.2. DIS3 and Multiple Myeloma
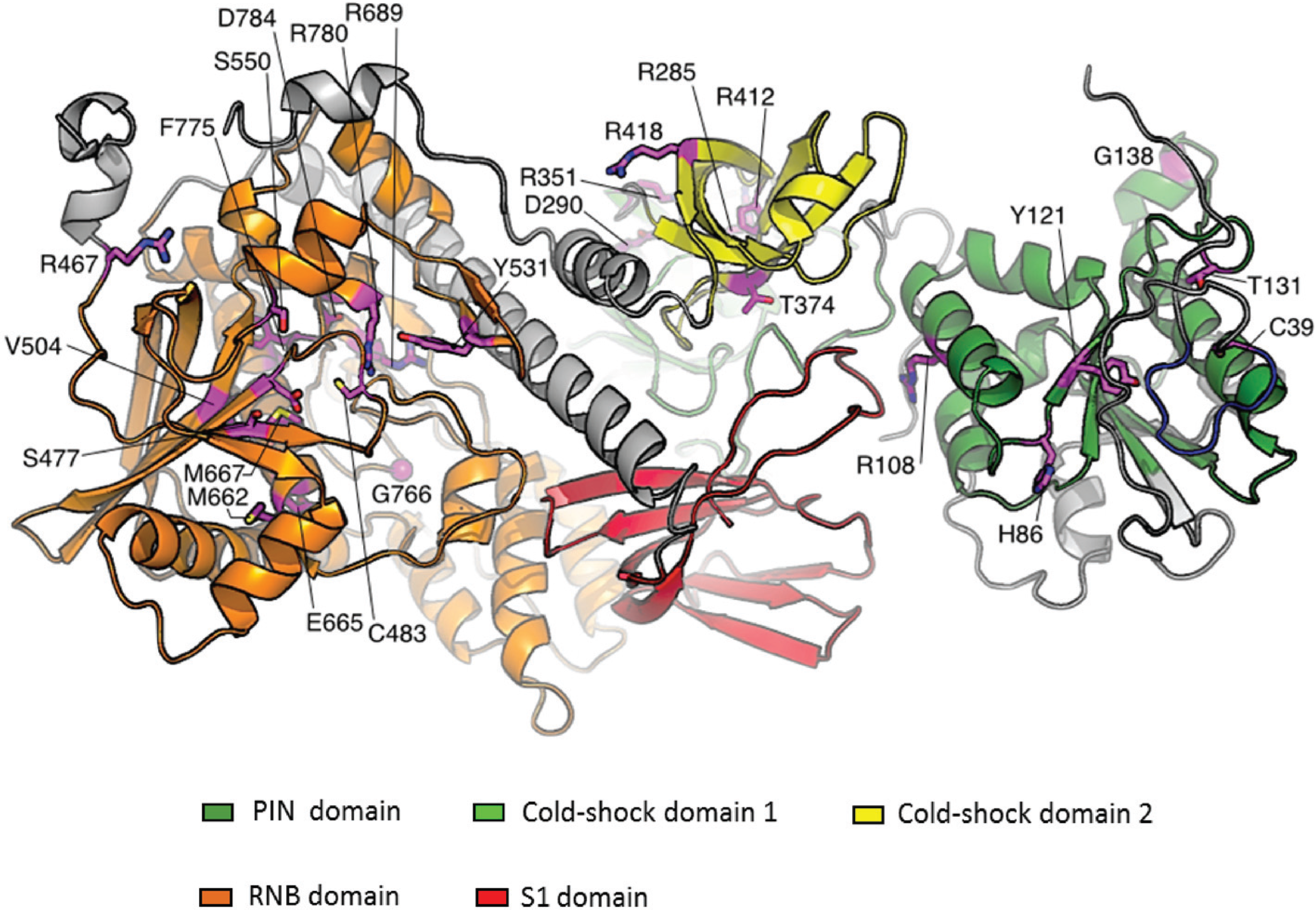
6. Conclusions and Future Prospects
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cairrão, F.; Arraiano, C.; Newbury, S. Drosophila gene tazman, an orthologue of the yeast exosome component Rrp44p/Dis3, is differentially expressed during development. Dev. Dyn. 2005, 232, 733–737. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.I.; Zabolotskaya, M.V.; Newbury, S.F. The 5'→3' exoribonuclease XRN1/Pacman and its functions in cellular processes and development. Wiley Interdiscip. Rev. RNA 2012, 3, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Waldron, J.A.; Jones, C.I.; Towler, B.P.; Pashler, A.L.; Grima, D.P.; Hebbes, S.; Crossman, S.H.; Zabolotskaya, M.V.; Newbury, S.F. Xrn1/Pacman affects apoptosis and regulates expression of hid and reaper. Biol. Open 2015. [CrossRef] [PubMed]
- Jones, C.I.; Grima, D.P.; Waldron, J.A.; Jones, S.; Parker, H.N.; Newbury, S.F. The 5'-3' exoribonuclease Pacman (Xrn1) regulates expression of the heat shock protein Hsp67Bc and the microRNA miR-277-3p in Drosophila wing imaginal discs. RNA Biol. 2013, 10, 1345–1355. [Google Scholar] [CrossRef] [PubMed]
- Wasmuth, E.V.; Lima, C.D. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol. Cell 2012, 48, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Bousquet-Antonelli, C.; Presutti, C.; Tollervey, D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 2000, 102, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Milligan, L.; Torchet, C.; Allmang, C.; Shipman, T.; Tollervey, D. A nuclear surveillance pathway for mRNAs with defective polyadenylation. Mol. Cell. Biol. 2005, 25, 9996–10004. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, D.; Gao, M.; O’Connor, J.P.; Raijmakers, R.; Pruijn, G.; Lutz, C.S.; Wilusz, J. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002, 21, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Gherzi, R.; Ong, S.E.; Chan, E.L.; Raijmakers, R.; Pruijn, G.J.; Stoecklin, G.; Moroni, C.; Mann, M.; Karin, M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 2001, 107, 451–464. [Google Scholar] [CrossRef]
- Allmang, C.; Kufel, J.; Chanfreau, G.; Mitchell, P.; Petfalski, E.; Tollervey, D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999, 18, 5399–5410. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.; Petfalski, E.; Shevchenko, A.; Mann, M.; Tollervey, D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3'→5' exoribonucleases. Cell 1997, 91, 457–466. [Google Scholar] [CrossRef]
- Ohkura, H.; Adachi, Y.; Kinoshita, N.; Niwa, O.; Toda, T.; Yanagida, M. Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J. 1988, 7, 1465–1473. [Google Scholar] [PubMed]
- Kinoshita, N.; Goebl, M.; Yanagida, M. The fission yeast Dis3+ gene encodes a 110-kDa essential protein implicated in mitotic control. Mol. Cell. Biol. 1991, 11, 5839–5847. [Google Scholar] [PubMed]
- Noguchi, E.; Hayashi, N.; Azuma, Y.; Seki, T.; Nakamura, M.; Nakashima, N.; Yanagida, M.; He, X.; Mueller, U.; Sazer, S.; et al. Dis3, implicated in mitotic control, binds directly to Ran and enhances the GEF activity of RCC1. EMBO J. 1996, 15, 5595–5605. [Google Scholar] [PubMed]
- Smith, S.B.; Kiss, D.L.; Turk, E.; Tartakoff, A.M.; Andrulis, E.D. Pronounced and extensive microtubule defects in a Saccharomyces cerevisiae DIS3 mutant. Yeast 2011, 28, 755–769. [Google Scholar] [CrossRef] [PubMed]
- Murakami, H.; Goto, D.B.; Toda, T.; Chen, E.S.; Grewal, S.I.; Martienssen, R.A.; Yanagida, M. Ribonuclease activity of Dis3 is required for mitotic progression and provides a possible link between heterochromatin and kinetochore function. PLoS ONE 2007, 2, e317. [Google Scholar] [CrossRef] [PubMed]
- Basu, U.; Meng, F.L.; Keim, C.; Grinstein, V.; Pefanis, E.; Eccleston, J.; Zhang, T.; Myers, D.; Wasserman, C.R.; Wesemann, D.R.; et al. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell 2011, 144, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Kuroki, T.; Ozaki, K.; Kohsaki, H.; Yamori, T.; Tsuruo, T.; Nakamori, S.; Imaoka, S.; Endo, M.; Nakamura, Y. Isolation of murine and human homologues of the fission-yeast Dis3+ gene encoding a mitotic-control protein and its overexpression in cancer cells with progressive phenotype. Cancer Res. 1997, 57, 921–925. [Google Scholar] [PubMed]
- Liang, L.; Qu, L.; Ding, Y. Protein and mRNA characterization in human colorectal carcinoma cell lines with different metastatic potentials. Cancer Investig. 2007, 25, 427–434. [Google Scholar] [CrossRef]
- De Groen, F.L.; Krijgsman, O.; Tijssen, M.; Vriend, L.E.; Ylstra, B.; Hooijberg, E.; Meijer, G.A.; Steenbergen, R.D.; Carvalho, B. Gene-dosage dependent overexpression at the 13q amplicon identifies DIS3 as candidate oncogene in colorectal cancer progression. Genes Chromosomes Cancer 2014, 53, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Rozenblum, E.; Vahteristo, P.; Sandberg, T.; Bergthorsson, J.T.; Syrjakoski, K.; Weaver, D.; Haraldsson, K.; Johannsdottir, H.K.; Vehmanen, P.; Nigam, S.; et al. A genomic map of a 6-Mb region at 13q21-q22 implicated in cancer development: Identification and characterization of candidate genes. Hum. Genet. 2002, 110, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Ng, D.; Toure, O.; Wei, M.H.; Arthur, D.C.; Abbasi, F.; Fontaine, L.; Marti, G.E.; Fraumeni, J.F.; Goldin, L.R.; Caporaso, N.; et al. Identification of a novel chromosome region, 13q21.33-q22.2, for susceptibility genes in familial chronic lymphocytic leukemia. Blood 2007, 109, 916–925. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.E.; Poliseno, L.; Wang, J.; Clark, M.; Pearlman, A.; Wang, G.; Vega Y Saenz de Miera, E.C.; Medicherla, R.; Christos, P.J.; Shapiro, R.; et al. Integrative genomics identifies molecular alterations that challenge the linear model of melanoma progression. Cancer Res. 2011, 71, 2561–2571. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Peng, Y.; Tobin, D.J. A new 12-gene diagnostic biomarker signature of melanoma revealed by integrated microarray analysis. Peer J. 2013, 1, e49. [Google Scholar] [CrossRef] [PubMed]
- Lohr, J.G.; Stojanov, P.; Carter, S.L.; Cruz-Gordillo, P.; Lawrence, M.S.; Auclair, D.; Sougnez, C.; Knoechel, B.; Gould, J.; Saksena, G.; et al. Widespread genetic heterogeneity in multiple myeloma: Implications for targeted therapy. Cancer Cell 2014, 25, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.A.; Lawrence, M.S.; Keats, J.J.; Cibulskis, K.; Sougnez, C.; Schinzel, A.C.; Harview, C.L.; Brunet, J.P.; Ahmann, G.J.; Adli, M.; et al. Initial genome sequencing and analysis of multiple myeloma. Nature 2011, 471, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.A.; Wardell, C.P.; Melchor, L.; Hulkki, S.; Potter, N.E.; Johnson, D.C.; Fenwick, K.; Kozarewa, I.; Gonzalez, D.; Lord, C.J.; et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood 2012, 120, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Weißbach, S.; Langer, C.; Puppe, B.; Nedeva, T.; Bach, E.; Kull, M.; Bargou, R.; Einsele, H.; Rosenwald, A.; Knop, S.; et al. The molecular spectrum and clinical impact of DIS3 mutations in multiple myeloma. Br. J. Haematol. 2015, 169, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Leich, E.; Weißbach, S.; Klein, H.U.; Grieb, T.; Pischimarov, J.; Stühmer, T.; Chatterjee, M.; Steinbrunn, T.; Langer, C.; Eilers, M.; et al. Multiple myeloma is affected by multiple and heterogeneous somatic mutations in adhesion- and receptor tyrosine kinase signaling molecules. Blood Cancer J. 2013, 3, e102. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.; Deutscher, M.P. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001, 29, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Tomecki, R.; Kristiansen, M.S.; Lykke-Andersen, S.; Chlebowski, A.; Larsen, K.M.; Szczesny, R.J.; Drazkowska, K.; Pastula, A.; Andersen, J.S.; Stepien, P.P.; et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010, 29, 2342–2357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frazão, C.; McVey, C.E.; Amblar, M.; Barbas, A.; Vonrhein, C.; Arraiano, C.M.; Carrondo, M.A. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature 2006, 443, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, A.; Tomecki, R.; Dziembowski, A.; Séraphin, B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 2008, 456, 993–996. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, D.; Reis, F.P.; Johnson, S.J.; Arraiano, C.M.; van Hoof, A. The CR3 motif of Rrp44p is important for interaction with the core exosome and exosome function. Nucleic Acids Res. 2012, 40, 9298–9307. [Google Scholar] [CrossRef] [PubMed]
- Staals, R.H.; Bronkhorst, A.W.; Schilders, G.; Slomovic, S.; Schuster, G.; Heck, A.J.; Raijmakers, R.; Pruijn, G.J. Dis3-like 1: A novel exoribonuclease associated with the human exosome. EMBO J. 2010, 29, 2358–2367. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Murphy, C.; Sieburth, L.E. Conserved RNaseII domain protein functions in cytoplasmic mRNA decay and suppresses Arabidopsis decapping mutant phenotypes. Proc. Natl. Acad. Sci. USA 2010, 107, 15981–15985. [Google Scholar] [CrossRef] [PubMed]
- Kumakura, N.; Otsuki, H.; Tsuzuki, M.; Takeda, A.; Watanabe, Y. Arabidopsis AtRRP44A is the functional homolog of Rrp44/Dis3, an exosome component, is essential for viability and is required for RNA processing and degradation. PLoS ONE 2013, 8, e79219. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.M.; Triboulet, R.; Thornton, J.E.; Gregory, R.I. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 2013, 497, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, D.; Tsanova, B.; Barbas, A.; Reis, F.P.; Dastidar, E.G.; Sanchez-Rotunno, M.; Arraiano, C.M.; van Hoof, A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat. Struct. Mol. Biol. 2009, 16, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Lorentzen, E.; Basquin, J.; Tomecki, R.; Dziembowski, A.; Conti, E. Structure of the active subunit of the yeast exosome core, Rrp44: Diverse modes of substrate recruitment in the RNase II nuclease family. Mol. Cell 2008, 29, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Dziembowski, A.; Lorentzen, E.; Conti, E.; Seraphin, B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007, 14, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Greimann, J.C.; Lima, C.D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 2006, 127, 1223–1237. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Bratkowski, M.A.; Ding, F.; Ke, A.; Ha, T. Elastic coupling between RNA degradation and unwinding by an exoribonuclease. Science 2012, 336, 1726–1729. [Google Scholar] [CrossRef] [PubMed]
- Makino, D.L.; Baumgartner, M.; Conti, E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature 2013, 495, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Graham, A.C.; Davis, S.M.; Andrulis, E.D. Interdependent nucleocytoplasmic trafficking and interactions of Dis3 with Rrp6, the core exosome and importin-alpha3. Traffic 2009, 10, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Symmons, M.F.; Luisi, B.F. Through ancient rings thread programming strings. Structure 2009, 17, 1429–1431. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.; Leung, E.; Brown, J.; Tollervey, D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009, 37, 1127–1140. [Google Scholar] [CrossRef] [PubMed]
- Drazkowska, K.; Tomecki, R.; Stodus, K.; Kowalska, K.; Czarnocki-Cieciura, M.; Dziembowski, A. The RNA exosome complex central channel controls both exonuclease and endonuclease Dis3 activities in vivo and in vitro. Nucleic Acids Res. 2013, 41, 3845–3858. [Google Scholar] [CrossRef] [PubMed]
- Halbach, F.; Reichelt, P.; Rode, M.; Conti, E. The yeast ski complex: Crystal structure and RNA channeling to the exosome complex. Cell 2013, 154, 814–826. [Google Scholar] [CrossRef] [PubMed]
- LaCava, J.; Houseley, J.; Saveanu, C.; Petfalski, E.; Thompson, E.; Jacquier, A.; Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 2005, 121, 713–724. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.; Kudla, G.; Wlotzka, W.; Tuck, A.; Tollervey, D. Transcriptome-wide analysis of exosome targets. Mol. Cell 2012, 48, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P. Exosome substrate targeting: The long and short of it. Biochem. Soc. Trans. 2014, 42, 1129–1134. [Google Scholar] [CrossRef] [PubMed]
- Briggs, M.W.; Burkard, K.T.; Butler, J.S. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3' end formation. J. Biol. Chem. 1998, 273, 13255–13263. [Google Scholar] [CrossRef] [PubMed]
- Graham, A.C.; Kiss, D.L.; Andrulis, E.D. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol. Biol. Cell 2006, 17, 1399–1409. [Google Scholar] [CrossRef] [PubMed]
- Shiomi, T.; Fukushima, K.; Suzuki, N.; Nakashima, N.; Noguchi, E.; Nishimoto, T. Human Dis3p, which binds to either GTP- or GDP-Ran, complements Saccharomyces cerevisiae Dis3. J. Biochem. 1998, 123, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Mamolen, M.; Smith, A.; Andrulis, E.D. Drosophila melanogaster Dis3 N-terminal domains are required for ribonuclease activities, nuclear localization and exosome interactions. Nucleic Acids Res. 2010, 38, 5507–5517. [Google Scholar] [CrossRef] [PubMed]
- Chlebowski, A.; Lubas, M.; Jensen, T.H.; Dziembowski, A. RNA decay machines: The exosome. Biochim. Biophys. Acta 2013, 1829, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Garneau, N.L.; Wilusz, J.; Wilusz, C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007, 8, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Newbury, S.F. Control of mRNA stability in eukaryotes. Biochem. Soc. Trans. 2006, 34, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Andrulis, E.D.; Werner, J.; Nazarian, A.; Erdjument-Bromage, H.; Tempst, P.; Lis, J.T. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 2002, 420, 837–841. [Google Scholar] [CrossRef] [PubMed]
- Lejeune, F.; Li, X.; Maquat, L.E. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 2003, 12, 675–687. [Google Scholar] [CrossRef]
- Frischmeyer, P.A.; van Hoof, A.; O’Donnell, K.; Guerrerio, A.L.; Parker, R.; Dietz, H.C. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 2002, 295, 2258–2261. [Google Scholar] [CrossRef] [PubMed]
- Van Hoof, A.; Frischmeyer, P.A.; Dietz, H.C.; Parker, R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 2002, 295, 2262–2264. [Google Scholar] [CrossRef] [PubMed]
- Doma, M.K.; Parker, R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 2006, 440, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Kashima, I.; Yamashita, A.; Izumi, N.; Kataoka, N.; Morishita, R.; Hoshino, S.; Ohno, M.; Dreyfuss, G.; Ohno, S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006, 20, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Kiss, D.L.; Andrulis, E.D. Genome-wide analysis reveals distinct substrate specificities of Rrp6, Dis3, and core exosome subunits. RNA 2010, 16, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Bakheet, T.; Williams, B.R.; Khabar, K.S. ARED 3.0: The large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006, 34, D111–D114. [Google Scholar] [CrossRef] [PubMed]
- Halees, A.S.; Hitti, E.; Al-Saif, M.; Mahmoud, L.; Vlasova-St Louis, I.A.; Beisang, D.J.; Bohjanen, P.R.; Khabar, K. Global assessment of GU-rich regulatory content and function in the human transcriptome. RNA Biol. 2011, 8, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Rehwinkel, J.; Behm-Ansmant, I.; Gatfield, D.; Izaurralde, E. A crucial role for GW182 and the DCP1: DCP2 decapping complex in miRNA-mediated gene silencing. RNA 2005, 11, 1640–1647. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.S. The yin and yang of the exosome. Trends Cell Biol. 2002, 12, 90–96. [Google Scholar] [CrossRef]
- Mitchell, P.; Petfalski, E.; Tollervey, D. The 3' end of yeast 5.8 S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 1996, 10, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Kadaba, S.; Krueger, A.; Trice, T.; Krecic, A.M.; Hinnebusch, A.G.; Anderson, J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004, 18, 1227–1240. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.; Anderson, J.T.; Tollervey, D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol. Cell 2007, 27, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Gudipati, R.K.; Xu, Z.; Lebreton, A.; Séraphin, B.; Steinmetz, L.M.; Jacquier, A.; Libri, D. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol. Cell 2012, 48, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Hata, A.; Lieberman, J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci. Signal. 2015. [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Triboulet, R.; Chang, H.M.; Lapierre, R.J.; Gregory, R.I. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA 2009, 15, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Pedersen, J.S.; Kwon, S.C.; Belair, C.D.; Kim, Y.K.; Yeom, K.H.; Yang, W.Y.; Haussler, D.; Blelloch, R.; Kim, V.N. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 2009, 136, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Towler, B.P.; Jones, C.I.; Viegas, S.C.; Apura, P.; Waldron, J.A.; Smalley, S.K.; Arraiano, C.M.; Newbury, S.F. The 3'-5' exoribonuclease Dis3 regulates the expression of specific microRNAs in Drosophila wing imaginal discs. RNA Biol. 2015. [CrossRef]
- Flynt, A.S.; Greimann, J.C.; Chung, W.J.; Lima, C.D.; Lai, E.C. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol. Cell 2010, 38, 900–907. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zheng, Q.; Vrettos, N.; Maragkakis, M.; Alexiou, P.; Gregory, B.D.; Mourelatos, Z. A MicroRNA precursor surveillance system in quality control of MicroRNA synthesis. Mol. Cell 2014, 55, 868–879. [Google Scholar] [CrossRef] [PubMed]
- Preker, P.; Nielsen, J.; Kammler, S.; Lykke-Andersen, S.; Christensen, M.S.; Mapendano, C.K.; Schierup, M.H.; Jensen, T.H. RNA exosome depletion reveals transcription upstream of active human promoters. Science 2008, 322, 1851–1854. [Google Scholar] [CrossRef] [PubMed]
- Preker, P.; Almvig, K.; Christensen, M.S.; Valen, E.; Mapendano, C.K.; Sandelin, A.; Jensen, T.H. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011, 39, 7179–7193. [Google Scholar] [CrossRef] [PubMed]
- Tomecki, R.; Drazkowska, K.; Kucinski, I.; Stodus, K.; Szczesny, R.J.; Gruchota, J.; Owczarek, E.P.; Kalisiak, K.; Dziembowski, A. Multiple myeloma-associated hDIS3 mutations cause perturbations in cellular RNA metabolism and suggest hDIS3 PIN domain as a potential drug target. Nucleic Acids Res. 2014, 42, 1270–1290. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.; Hyman, A. Microtubule cytoskeleton: No longer an also Ran. Curr. Biol. 1999, 9, R704–R707. [Google Scholar] [CrossRef]
- Sazer, S.; Dasso, M. The ran decathlon: Multiple roles of Ran. J. Cell Sci. 2000, 113, 1111–1118. [Google Scholar] [PubMed]
- Carazo-Salas, R.E.; Guarguaglini, G.; Gruss, O.J.; Segref, A.; Karsenti, E.; Mattaj, I.W. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 1999, 400, 178–181. [Google Scholar] [PubMed]
- Clarke, P.R.; Zhang, C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat. Rev. Mol. Cell Biol. 2008, 9, 464–477. [Google Scholar] [CrossRef] [PubMed]
- Arnaoutov, A.; Dasso, M. The Ran GTPase regulates kinetochore function. Dev. Cell 2003, 5, 99–111. [Google Scholar] [CrossRef]
- Suzuki, N.; Noguchi, E.; Nakashima, N.; Oki, M.; Ohba, T.; Tartakoff, A.; Ohishi, M.; Nishimoto, T. The Saccharomyces cerevisiae small GTPase, Gsp1p/Ran, is involved in 3' processing of 7 S-to-5.8 S rRNA and in degradation of the excised 5'-A0 fragment of 35 S pre-rRNA, both of which are carried out by the exosome. Genetics 2001, 158, 613–625. [Google Scholar] [PubMed]
- Maiato, H.; DeLuca, J.; Salmon, E.D.; Earnshaw, W.C. The dynamic kinetochore-microtubule interface. J. Cell Sci. 2004, 117, 5461–5477. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, T.; Chen, S.; Hitomi, M.; Jacobs, E.; Kumagai, C.; Liang, S.; Schneiter, R.; Singleton, D.; Wisniewska, J.; Tartakoff, A.M. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J. Cell Biol. 1994, 126, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Vasiljeva, L.; Kim, M.; Terzi, N.; Soares, L.M.; Buratowski, S. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol. Cell 2008, 29, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.W.; Stevenson, A.L.; Kearsey, S.E.; Watt, S.; Bähler, J. Global role for polyadenylation-assisted nuclear RNA degradation in posttranscriptional gene silencing. Mol. Cell Biol. 2008, 28, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Grewal, S.I.; Elgin, S.C. Heterochromatin: New possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 2002, 12, 178–187. [Google Scholar] [CrossRef]
- Buhler, M.; Haas, W.; Gygi, S.P.; Moazed, D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 2007, 129, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Camps, J.; Pitt, J.J.; Emons, G.; Hummon, A.B.; Case, C.M.; Grade, M.; Jones, T.L.; Nguyen, Q.T.; Ghadimi, B.M.; Beissbarth, T.; et al. Genetic amplification of the NOTCH modulator LNX2 upregulates the WNT/β-catenin pathway in colorectal cancer. Cancer Res. 2013, 73, 2003–2013. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Ley, T.J.; Larson, D.E.; Miller, C.A.; Koboldt, D.C.; Welch, J.S.; Ritchey, J.K.; Young, M.A.; Lamprecht, T.; McLellan, M.D.; et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012, 481, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Welch, J.S.; Ley, T.J.; Link, D.C.; Miller, C.A.; Larson, D.E.; Koboldt, D.C.; Wartman, L.D.; Lamprecht, T.L.; Liu, F.; Xia, J.; et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012, 150, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Kyle, R.A.; Rajkumar, S.V. Multiple myeloma. N. Engl. J. Med. 2004, 351, 1860–1873. [Google Scholar] [CrossRef] [PubMed]
- Laubach, J.; Richardson, P.; Anderson, K. Multiple myeloma. Annu. Rev. Med. 2011, 62, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Sternberg, M.J. Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Shlush, L.I.; Hershkovitz, D. Clonal evolution models of tumor heterogeneity. Am. Soc. Clin. Oncol. Educ. Book 2015, 35, e662–e665. [Google Scholar] [CrossRef] [PubMed]
- Greaves, M.; Maley, C.C. Clonal evolution in cancer. Nature 2012, 481, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Melchor, L.; Brioli, A.; Wardell, C.P.; Murison, A.; Potter, N.E.; Kaiser, M.F.; Fryer, R.A.; Johnson, D.C.; Begum, D.B.; Hulkki Wilson, S.; et al. Single-cell genetic analysis reveals the composition of initiating clones and phylogenetic patterns of branching and parallel evolution in myeloma. Leukemia 2014, 28, 1705–1715. [Google Scholar] [CrossRef] [PubMed]
- Morgan, G.J.; Walker, B.A.; Davies, F.E. The genetic architecture of multiple myeloma. Nat. Rev. Cancer 2012, 12, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Boyd, K.D.; Pawlyn, C.; Morgan, G.J.; Davies, F.E. Understanding the molecular biology of myeloma and its therapeutic implications. Expert Rev. Hematol. 2012, 5, 603–617. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, G.; Ghobrial, I.M. Biological and clinical implications of clonal heterogeneity and clonal evolution in multiple myeloma. Curr. Cancer Ther. Rev. 2014, 10, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Kuchta, K.; Knizewski, L.; Wyrwicz, L.S.; Rychlewski, L.; Ginalski, K. Comprehensive classification of nucleotidyltransferase fold proteins: Identification of novel families and their representatives in human. Nucleic Acids Res. 2009, 37, 7701–7714. [Google Scholar] [CrossRef] [PubMed]
- Meng, T. The Molecular Cloning and Characterization of Fam46c RNA Stability Factor. Ph.D. Thesis, Harvard University, Cambridge, MA, USA, 2010. [Google Scholar]
- Drach, J.; Schuster, J.; Nowotny, H.; Angerler, J.; Rosenthal, F.; Fiegl, M.; Rothermundt, C.; Gsur, A.; Jäger, U.; Heinz, R. Multiple myeloma: High incidence of chromosomal aneuploidy as detected by interphase fluorescence in situ hybridization. Cancer Res. 1995, 55, 3854–3859. [Google Scholar] [PubMed]
- Diaz-Rodriguez, E.; Alvarez-Fernandez, S.; Chen, X.; Paiva, B.; Lopez-Perez, R.; Garcia-Hernandez, J.L.; San Miguel, J.F.; Pandiella, A. Deficient spindle assembly checkpoint in multiple myeloma. PLoS ONE 2011, 6, e27583. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robinson, S.R.; Oliver, A.W.; Chevassut, T.J.; Newbury, S.F. The 3' to 5' Exoribonuclease DIS3: From Structure and Mechanisms to Biological Functions and Role in Human Disease. Biomolecules 2015, 5, 1515-1539. https://doi.org/10.3390/biom5031515
Robinson SR, Oliver AW, Chevassut TJ, Newbury SF. The 3' to 5' Exoribonuclease DIS3: From Structure and Mechanisms to Biological Functions and Role in Human Disease. Biomolecules. 2015; 5(3):1515-1539. https://doi.org/10.3390/biom5031515
Chicago/Turabian StyleRobinson, Sophie R., Antony W. Oliver, Timothy J. Chevassut, and Sarah F. Newbury. 2015. "The 3' to 5' Exoribonuclease DIS3: From Structure and Mechanisms to Biological Functions and Role in Human Disease" Biomolecules 5, no. 3: 1515-1539. https://doi.org/10.3390/biom5031515