Regulation of Transcription Elongation and Termination
Abstract
:1. Introduction
2. Pausing
3. Intrinsic Termination
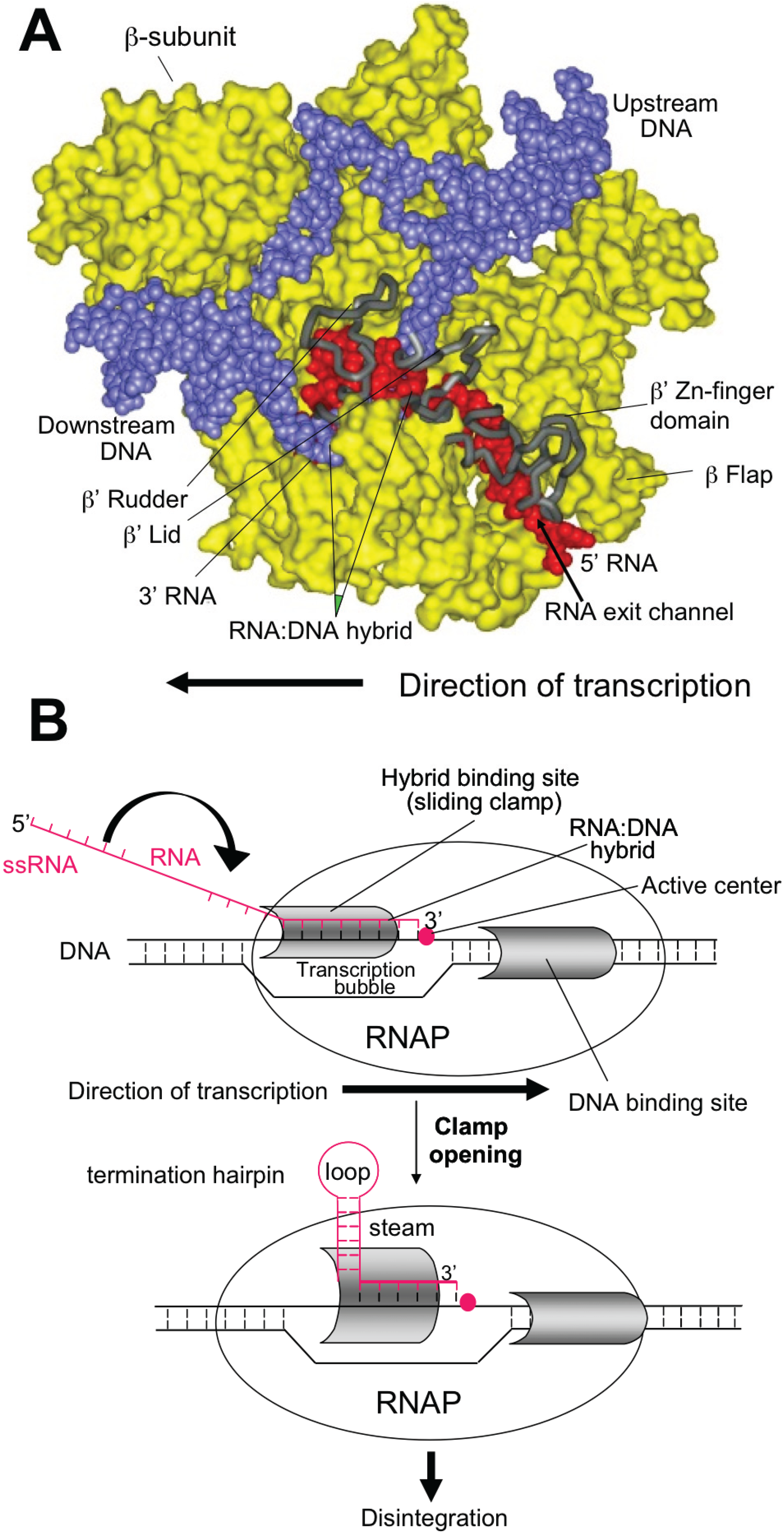
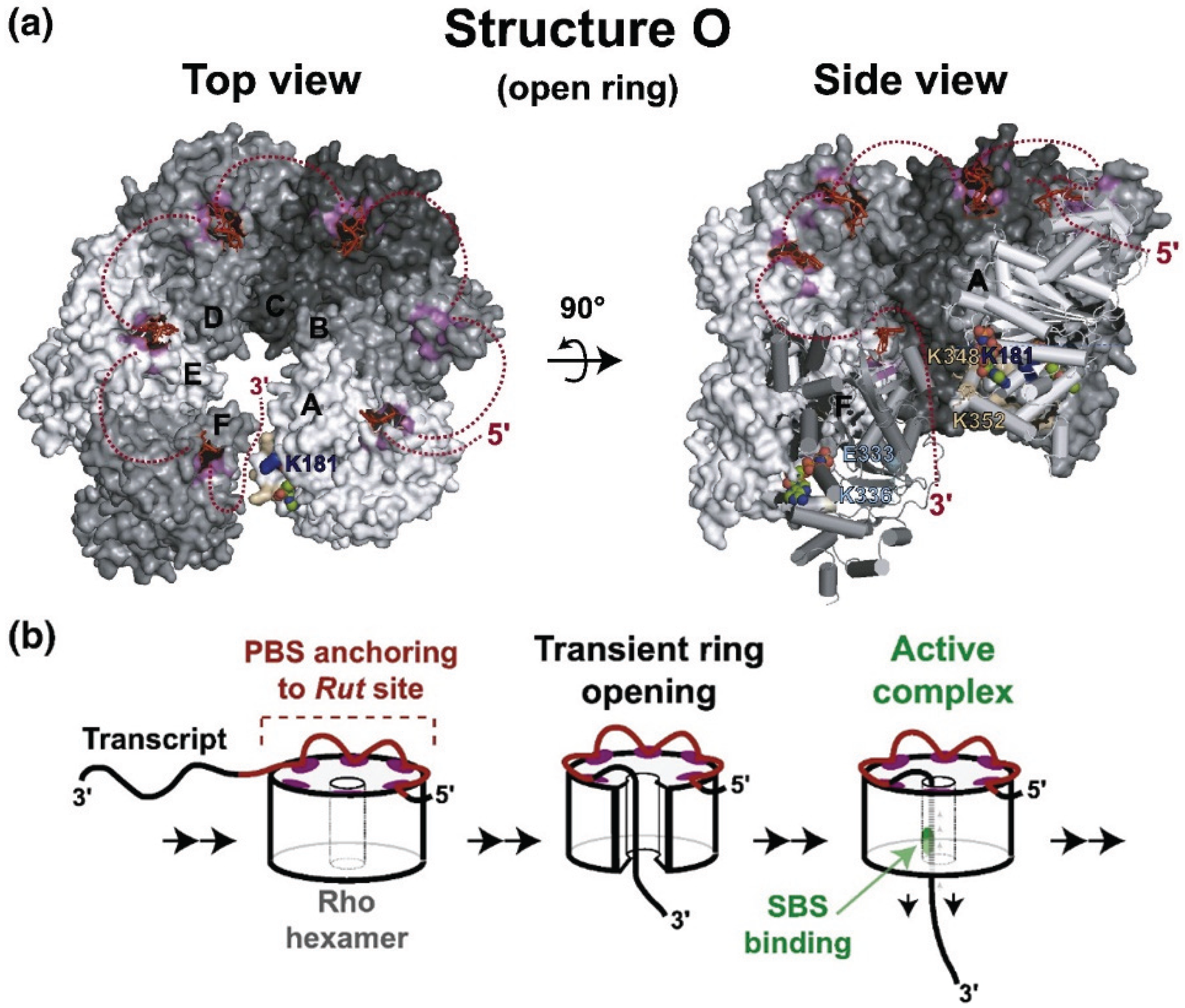
4. Rho-Dependent Termination
5. NusG, NusA and DksA
5.1. NusG
5.2. NusA
5.3. DksA
6. RNA-Binding Phage-Encoded Proteins that Affect Transcription Elongation
6.1. λ N Antitermination
6.2. HK022 Nun-Mediated Transcription Arrest

7. Conclusions
Acknowledgments
Conflicts of Interest
References
- Mejia, Y.X.; Nudler, E.; Bustamante, C. Trigger loop folding determines transcription rate of Escherichia coli’s RNA polymerase. Proc. Natl. Acad. Sci. USA 2015, 112, 743–774. [Google Scholar] [CrossRef] [PubMed]
- Kireeva, M.L.; Kashlev, M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc. Natl. Acad. Sci. USA 2009, 106, 8900–8905. [Google Scholar] [CrossRef] [PubMed]
- Bochkareva, A.; Yuzenkova, Y.; Tadigotla, V.R.; Zenkin, N. Factor-independent transcription pausing caused by recognition of the RNA-DNA hybrid sequence. EMBO J. 2012, 31, 630–639. [Google Scholar] [CrossRef] [PubMed]
- Vvedenskaya, I.O.; Vahedian-Movahed, H.; Bird, J.G.; Knoblauch, J.G.; Goldman, S.R.; Zhang, Y.; Ebright, R.H.; Nickels, B.E. Transcription. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science 2014, 344, 1285–1289. [Google Scholar] [CrossRef] [PubMed]
- Larson, M.H.; Mooney, R.A.; Peters, J.M.; Windgassen, T.; Nayak, D.; Gross, C.A.; Block, S.M.; Greenleaf, W.J.; Landick, R.; Weissman, J.S. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 2014, 344, 1042–1047. [Google Scholar] [CrossRef] [PubMed]
- Purdue, S.A.; Roberts, J.W. σ70-dependent transcription pausing in Escherichia coli. J. Mol. Biol. 2011, 412, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Zhilina, E.; Esyunina, D.; Brodolin, K.; Kulbachinskiy, A. Structural transitions in the transcription elongation complexes of bacterial RNA polymerase during σ-dependent pausing. Nucleic Acids Res. 2012, 40, 3078–3091. [Google Scholar] [PubMed]
- Wade, J.T.; Struhl, K. Association of RNA polymerase with transcribed regions in Escherichia coli. Proc. Natl. Acad. Sci. USA 2004, 101, 17777–17782. [Google Scholar] [CrossRef] [PubMed]
- Nedialkov, Y.A.; Opron, K.; Assaf, F.; Artsimovitch, I.; Kireeva, M.L.; Kashlev, M.; Cukier, R.I.; Nudler, E.; Burton, Z.F. The RNA polymerase bridge helix YFI motif in catalysis, fidelity and translocation. Biochim. Biophys. Acta 2013, 1829, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.W. Termination factor for RNA synthesis. Nature 1969, 224, 1168–1174. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.M.; Mooney, R.A.; Kuan, P.F.; Rowland, J.L.; Keles, S.; Landick, R. Rho directs widespread termination of intragenic and stable RNA transcription. Proc. Natl. Acad. Sci. USA 2009, 106, 15406–15411. [Google Scholar] [CrossRef] [PubMed]
- Opperman, T.; Richardson, J.P. Phylogenetic analysis of sequences from diverse bacteria with homology to the Escherichia coli Rho gene. J. Bacteriol. 1994, 176, 5033–5043. [Google Scholar] [PubMed]
- Lowery-Goldhammer, C.; Richardson, J.P. An RNA-dependent nucleoside triphosphate phosphohydrolase (ATPase) associated with Rho termination factor. Proc. Natl. Acad. Sci. USA 1974, 71, 2003–2007. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.A.; Dombroski, A.J.; Platt, T. Transcription termination factor Rho is an RNA-DNA helicase. Cell 1987, 48, 945–952. [Google Scholar] [CrossRef]
- Skordalakes, E.; Berger, J.M. Structure of the Rho transcription terminator: Mechanism of mRNA recognition and helicase loading. Cell 2003, 114, 135–146. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Shigesada, K.; Hirano, M.; Imai, M. Autogenous regulation of the gene for transcription termination factor Rho in Escherichia coli: Localization and function of its attenuators. J. Bacteriol. 1986, 166, 945–958. [Google Scholar] [PubMed]
- Chen, C.Y.; Richardson, J.P. Sequence elements essential for Rho-dependent transcription termination at λ tR1. J. Biol. Chem. 1987, 262, 11292–11299. [Google Scholar] [PubMed]
- Steinmetz, E.J.; Platt, T. Evidence supporting a tethered tracking model for helicase activity of Escherichia coli Rho factor. Proc. Natl. Acad. Sci. USA 1994, 91, 1401–1405. [Google Scholar] [CrossRef] [PubMed]
- Koslover, D.J.; Fazal, F.M.; Mooney, R.A.; Landick, R.; Block, S.M. Binding and translocation of termination factor Rho studied at the single-molecule level. J. Mol. Biol. 2012, 423, 664–676. [Google Scholar] [CrossRef] [PubMed]
- Hart, C.M.; Roberts, J.W. Rho-dependent transcription termination. Characterization of the requirement for cytidine in the nascent transcript. J. Biol. Chem. 1991, 266, 24140–24148. [Google Scholar] [PubMed]
- Richardson, J.P. Activation of Rho protein ATPase requires simultaneous interaction at two kinds of nucleic acid-binding sites. J. Biol. Chem. 1982, 257, 5760–5766. [Google Scholar] [PubMed]
- Skordalakes, E.; Berger, J.M. Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell 2006, 127, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Rabhi, M.; Gocheva, V.; Jacquinot, F.; Lee, A.; Margeat, E.; Boudvillain, M. Mutagenesis-based evidence for an asymmetric configuration of the ring-shaped transcription termination factor Rho. J. Mol. Biol. 2011, 405, 497–518. [Google Scholar] [CrossRef] [PubMed]
- Epshtein, V.; Dutta, D.; Wade, J.; Nudler, E. An allosteric mechanism of Rho-dependent transcription termination. Nature 2010, 463, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Roberts, J.W. Role of DNA bubble rewinding in enzymatic transcription termination. Proc. Natl. Acad. Sci. USA 2006, 103, 4870–4875. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-Y.; Platt, T. Transcriptional arrest of yeast RNA polymerase II by Escherichia coli Rho protein in vitro. Proc. Natl. Acad. Sci. USA 1993, 90, 6606–6610. [Google Scholar] [CrossRef] [PubMed]
- Alifano, P.; Rivellini, F.; Limauro, D. A consensus motif common to all Rho-dependent prokaryotic transcription terminators. Cell 1991, 64, 553–563. [Google Scholar] [CrossRef]
- Zalatan, F.; Platt, T. Effects of decreased cytosine content on Rho interaction with the Rho-dependent terminator trp T' in Escherichia coli. J. Biol. Chem. 1992, 267, 19082–19088. [Google Scholar] [PubMed]
- Morgan, W.D.; Bear, D.G.; von Hippel, P.H. Rho-dependent termination of transcription. I. Identification and characterization of termination sites for transcription from the bacteriophage λ PR promoter. J. Biol. Chem. 1983, 258, 9553–9564. [Google Scholar] [PubMed]
- Sullivan, S.L.; Gottesman, M.E. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell 1992, 68, 989–994. [Google Scholar] [CrossRef]
- Li, J.; Mason, S.W.; Greenblatt, J. Elongation factor NusG interacts with termination factor Rho to regulate termination and antitermination of transcription. Genes Dev. 1993, 7, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Burns, C.M.; Richardson, L.V.; Richardson, J.P. Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J. Mol. Biol. 1998, 278, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Nehrke, K.W.; Zalatan, F.; Platt, T. NusG alters Rho-dependent termination of transcription in vitro independent of kinetic coupling. Gene Expr. 1993, 3, 119–133. [Google Scholar] [PubMed]
- Washburn, R.S.; Jin, D.J.; Stitt, B.L. The mechanism of early transcription termination by Rho026. J. Mol. Biol. 1996, 260, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Nehrke, K.W.; Platt, T. A quaternary transcription termination complex. Reciprocal stabilization by Rho factor and NusG protein. J. Mol. Biol. 1994, 243, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Mooney, R.A.; Davis, S.E.; Mooney, J.M.; Rowland, J.L.; Ansari, A.Z.; Landick, R. Regulator trafficking on bacterial transcription units in vivo. Mol. Cell. 2009, 33, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Kalyani, B.S.; Muteeb, G.; Qayyum, M.Z.; Sen, J. Interaction with the nascent RNA is a prerequisite for the recruitment of Rho to the transcription elongation complex in vitro. J. Mol. Biol. 2011, 413, 548–560. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, C.J.; Washburn, R.S.; Tadigotla, V.R.; Brown, L.M.; Gottesman, M.E.; Nudler, E. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science 2008, 320, 935–938. [Google Scholar] [CrossRef] [PubMed]
- Washburn, R.S.; Gottesman, M.E. Transcription termination maintains chromosome integrity. Proc. Natl. Acad. Sci. USA 2011, 108, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Washburn, R.S.; Hashem, Y.; Sun, M.; Shen, B.; Harvey, S.; Frank, J.; Gottesman, M.E. NusG and RelA couple and tmRNA uncouples transcription with translation in E. coli. Unpublished work. 2015. [Google Scholar]
- Harinarayanan, R.; Gowrishankar, J. Host factor titration by chromosomal R-loops as a mechanism for runaway plasmid replication in transcription termination-defective mutants of Escherichia coli. J. Mol. Biol. 2003, 332, 31–46. [Google Scholar] [CrossRef]
- Leela, J.K.; Syeda, A.H.; Anupama, K.; Gowrishankar, J. Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc. Natl. Acad. Sci. USA 2013, 110, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.; van Baarsel, J.A.; Washburn, R.S.; Gottesman, M.E.; Miller, J.H. Single-gene deletion mutants of Escherichia coli with altered sensitivity to bicyclomycin, an inhibitor of transcription termination factor Rho. J. Bacteriol. 2011, 193, 2229–2235. [Google Scholar] [CrossRef] [PubMed]
- Nichols, R.J.; Sen, S.; Choo, Y.J.; Beltrao, P.; Zietek, M.; Chaba, R.; Lee, S.; Kazmierczak, K.M.; Lee, K.J.; Wong, A.; et al. Phenotypic landscape of a bacterial cell. Cell 2011, 144, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Shatalin, K.; Epshtein, V.; Gottesman, M.E.; Nudler, E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell 2011, 146, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.M.; Mooney, R.A.; Grass, J.A.; Jessen, E.D.; Tran, F.; Landick, R. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012, 26, 2621–2633. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.J.; Burgess, R.R.; Richardson, J.P.; Gross, C.A. Termination efficiency at Rho-dependent terminators depends on kinetic coupling between RNA polymerase and Rho. Proc. Natl. Acad. Sci. USA 1992, 89, 1453–1457. [Google Scholar] [CrossRef] [PubMed]
- Navarre, W.W.; Porwollik, S.; Wang, Y.; McClelland, M.; Rosen, H.; Libby, S.J.; Fang, F.C. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 2006, 313, 236–238. [Google Scholar] [CrossRef] [PubMed]
- Mooney, R.A.; Schweimer, K.; Rösch, P.; Gottesman, M.; Landick, R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J. Mol. Biol. 2009, 391, 341–358. [Google Scholar] [CrossRef] [PubMed]
- Svetlov, V.; Nudler, E. Clamping the clamp of RNA polymerase. EMBO J. 2011, 30, 1190–1191. [Google Scholar] [CrossRef] [PubMed]
- Herbert, K.M.; Zhou, J.; Mooney, R.A.; la Porta, A.; Landick, R.; Block, S.M. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J. Mol. Biol. 2010, 399, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Burmann, B.M.; Schweimer, K.; Luo, X.; Wahl, M.C.; Stitt, B.L.; Gottesman, M.E.; Rösch, P. A NusE:NusG complex links transcription and translation. Science 2010, 328, 501–504. [Google Scholar] [CrossRef] [PubMed]
- Burmann, B.M.; Knauer, S.H.; Sevostyanova, A.; Schweimer, K.; Mooney, R.A.; Landick, R.; Artsimovitch, I.; Rösch, P. An helix to barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell 2012, 150, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Balada, J.-M.; Zellars, M.; Squires, C.; Squires, C.L. In vivo effect of NusB and NusG on rRNA transcription antitermination. J. Bacteriol. 2004, 186, 1304–1310. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, S.; Siryaporn, A.; Goulian, M.; Weisshaar, J.C. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol. Microbiol. 2012, 85, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Yakhnin, A.V.; Babitzke, P. NusG/Spt5: Are there common functions of this ubiquitous transcription elongation factor? Curr. Opin. Microbiol. 2014, 18, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Tomar, S.K.; Knauer, S.H.; Nandymazumdar, M.; Rösch, P.; Artsimovitch, I. Interdomain contacts control folding of transcription factor RfaH. Nucleic Acids Res. 2013, 41, 10077–10085. [Google Scholar] [CrossRef] [PubMed]
- Nudler, E.; Gottesman, M.E. Transcription termination and anti-termination in E. coli. Genes Cells 2002, 7, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Kolb, K.E.; Hein, P.P.; Landick, R. Antisense oligonucleotide-stimulated transcriptional pausing reveals RNA exit channel specificity of RNA polymerase and mechanistic contributions of NusA and RfaH. J. Biol. Chem. 2014, 289, 1151–1163. [Google Scholar] [CrossRef] [PubMed]
- Gusarov, I.; Nudler, E. Control of intrinsic transcription termination by N and NusA: The basic mechanisms. Cell 2001, 107, 437–449. [Google Scholar] [CrossRef]
- Ha, K.S.; Tiulokhonev, I.; Vassylyev, D.G.; Landick, R. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J. Mol. Biol. 2010, 401, 708–725. [Google Scholar] [CrossRef] [PubMed]
- Schweimer, K.; Prasch, S.; Sujatha, P.S.; Bubunenko, M.; Gottesman, M.E.; Rösch, P. NusA interaction with the α subunit of E. coli RNA polymerase is via the UP element site and releases autoinhibition. Structure 2011, 19, 945–954. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lewis, P.J. The interaction between RNA polymerase and the elongation factor NusA. RNA Biol. 2010, 7, 272–275. [Google Scholar] [CrossRef] [PubMed]
- Washburn, R.S.; Gottesman, M.E.; Columbia University Medical Center, New York, NY, USA. Unpublished work. 2015.
- Ross, W.; Vrentas, C.E.; Sanchez-Vazquez, P.; Gaal, T.; Gourse, R.L. The magic spot: A ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell 2013, 50, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lennon, C.W.; Ross, W.; Gourse, R.L. Role of the coiled-coil tip of Escherichia coli DksA in promoter control. J. Mol. Biol. 2012, 416, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Vinella, D.; Potrykus, K.; Murphy, H.; Cashel, M. Effects on growth by changes of the balance between GreA, GreB, and DksA suggest mutual competition and functional redundancy in Escherichia coli. J. Bacteriol. 2012, 194, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, E.; Lee, J.; Ozerova, M.; Semenova, E.; Datsenko, K.; Wanner, B.L.; Severinov, K.; Borukhov, S. Analysis of promoter targets for Escherichia coli transcription elongation factor GreA in vivo and in vitro. J. Bacteriol. 2007, 189, 8772–8785. [Google Scholar] [CrossRef] [PubMed]
- Furman, R.; Sevostyanova, A.; Artsimovitch, I. Transcription initiation factor DksA has diverse effects on RNA chain elongation. Nucleic Acids Res. 2012, 40, 3392–3402. [Google Scholar] [CrossRef] [PubMed]
- Trautinger, B.W.; Jaktaji, R.P.; Rusakova, E.; Lloyd, R.G. RNA polymerase modulators and DNA repair activities resolve conflicts between replication and transcription. Mol. Cell 2005, 19, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Tehranchi, A.K.; Blankenschien, M.D.; Zhang, Y.; Halliday, J.A.; Srivatsan, A.; Peng, J.; Herman, C.; Wang, J.D. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell 2010, 141, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mooney, R.A.; Grass, J.A.; Sivaramakrishnan, P.; Herman, C.; Landick, R.; Wang, J.D. DksA guards elongating RNA polymerase against ribosome-stalling-induced arrest. Mol. Cell 2014, 53, 766–778. [Google Scholar] [CrossRef] [PubMed]
- Satory, D.; Halliday, J.A.; Sivaramakrishnan, P.; Lua, R.C.; Herman, C. Characterization of a novel RNA polymerase mutant that alters DksA activity. J. Bacteriol. 2013, 195, 4187–4194. [Google Scholar] [CrossRef] [PubMed]
- Parks, A.R.; Court, C.; Lubkowska, L.; Jin, D.J.; Kashlev, M.; Court, D.L. Bacteriophage λ N protein inhibits transcription slippage by Escherichia coli RNA polymerase. Nucleic Acids Res. 2014, 42, 5823–5829. [Google Scholar] [CrossRef] [PubMed]
- Washburn, R.S.; Wang, Y.; Gottesman, M.E. Role of E. coli transcription-repair coupling factor Mfd in Nun-mediated transcription termination. J. Mol. Biol. 2003, 329, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Hung, S.C.; Gottesman, M.E. Phage HK022 Nun protein arrests transcription on phage λ DNA in vitro and competes with the phage lambda N antitermination protein. J. Mol. Biol. 1995, 247, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Uc-Mass, A.; Khodursky, A.; Brown, L.; Gottesman, M.E. Overexpression of phage HK022 Nun protein is toxic for Escherichia coli. J. Mol. Biol. 2008, 380, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Hung, S.C.; Stuart, A.C.; Palmer, A.G., 3rd; Garcia-Mena, J.; Das, A.; Gottesman, M.E. Interaction between the phage HK022 Nun protein and the nut RNA of phage λ. Proc. Natl. Acad. Sci. USA 1995, 92, 12131–12135. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, C.L.; Kireeva, M.L.; Lubkowska, L.; Kashlev, M.; Gottesman, M. Coliphage HK022 Nun protein inhibits RNA polymerase translocation. Proc. Natl. Acad. Sci. USA 2014, 111, E2368–E2375. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, C.L.; Gottesman, M.E. Bacteriophage HK022 Nun protein arrests transcription by blocking lateral mobility of RNA polymerase during transcription elongation. Bacteriophage 2014. [Google Scholar] [CrossRef] [PubMed]
- Rees, W.A.; Weitzel, S.E.; Yager, T.D.; Das, A.; von Hippel, P.H. Bacteriophage λ N protein alone can induce transcription antitermination in vitro. Proc. Natl. Acad. Sci. USA 1996, 93, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Watnick, R.S.; Gottesman, M.E. Escherichia coli NusA is required for efficient RNA binding by phage HK022 nun protein. Proc. Natl. Acad. Sci. USA 1998, 95, 1546–1551. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Washburn, R.S.; Gottesman, M.E. Regulation of Transcription Elongation and Termination. Biomolecules 2015, 5, 1063-1078. https://doi.org/10.3390/biom5021063
Washburn RS, Gottesman ME. Regulation of Transcription Elongation and Termination. Biomolecules. 2015; 5(2):1063-1078. https://doi.org/10.3390/biom5021063
Chicago/Turabian StyleWashburn, Robert S., and Max E. Gottesman. 2015. "Regulation of Transcription Elongation and Termination" Biomolecules 5, no. 2: 1063-1078. https://doi.org/10.3390/biom5021063




