Arming Technology in Yeast—Novel Strategy for Whole-cell Biocatalyst and Protein Engineering
Abstract
:1. Introduction
2. Cell Surface Display System in Yeast
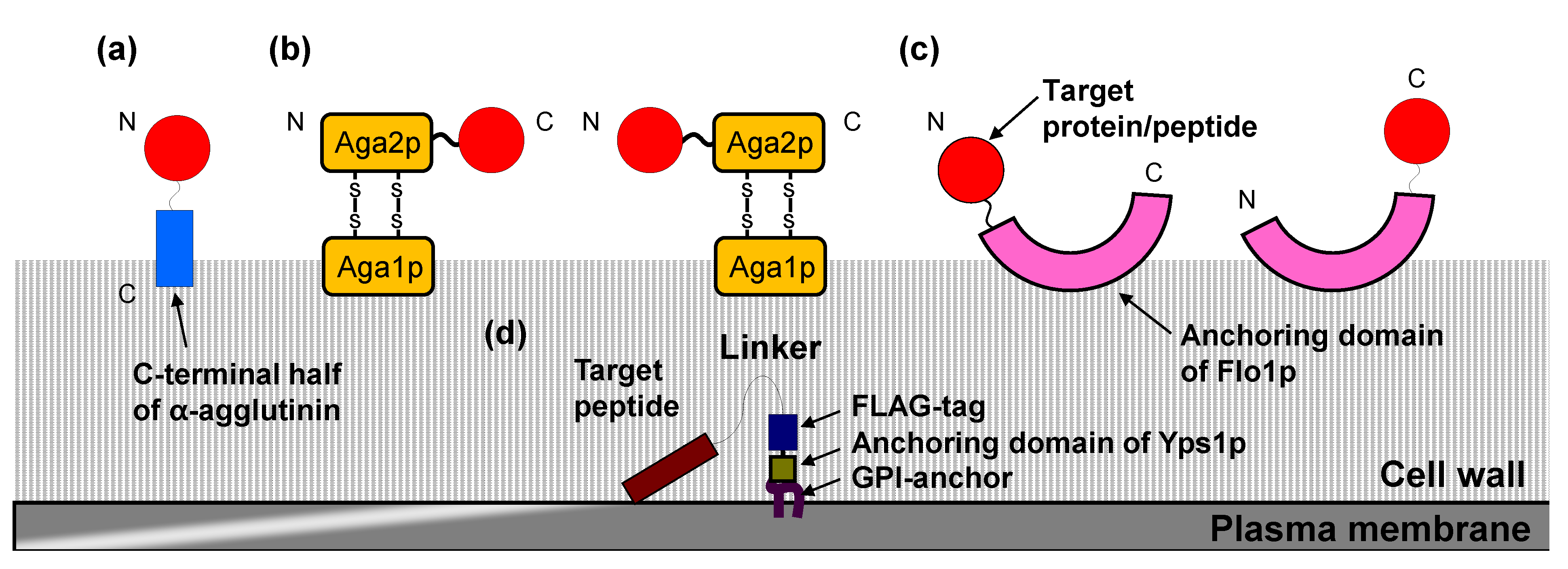
2.1. Cell Surface Display System in S. cerevisiae
2.2. Cell Surface Display System in P. pastoris
2.3. Cell Surface Display System in Y. lipolytica
2.4. Membrane Display System in S. cerevisiae
3. Whole-Cell Biocatalyst by Arming Technology
3.1. Biofuel Production
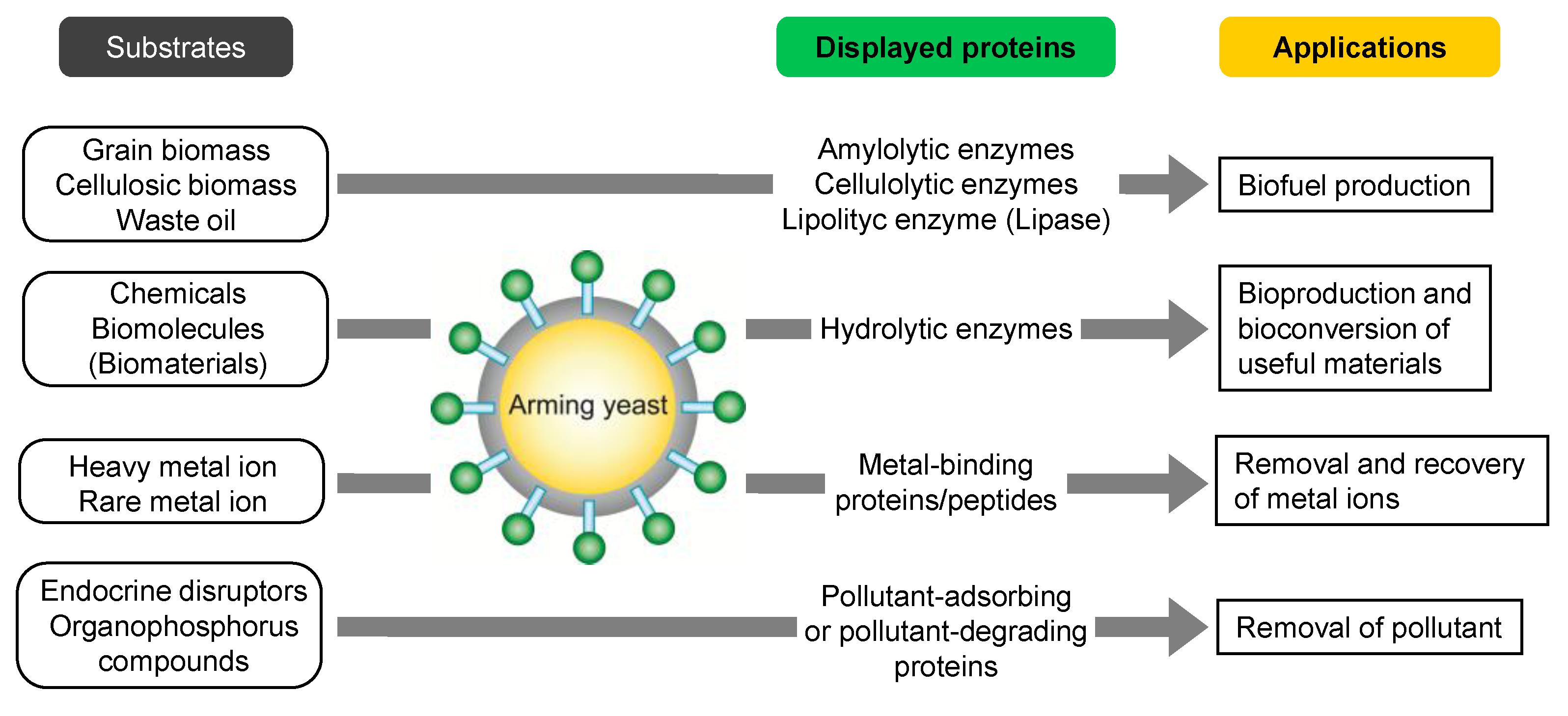
3.2. Bioproduction of Chemicals
3.3. Bioadsorption
3.4. Bioremediation
3.5. Other Applications
4. Protein Engineering and Directed Evolution by Arming Technology
4.1. Engineering of Enzymes
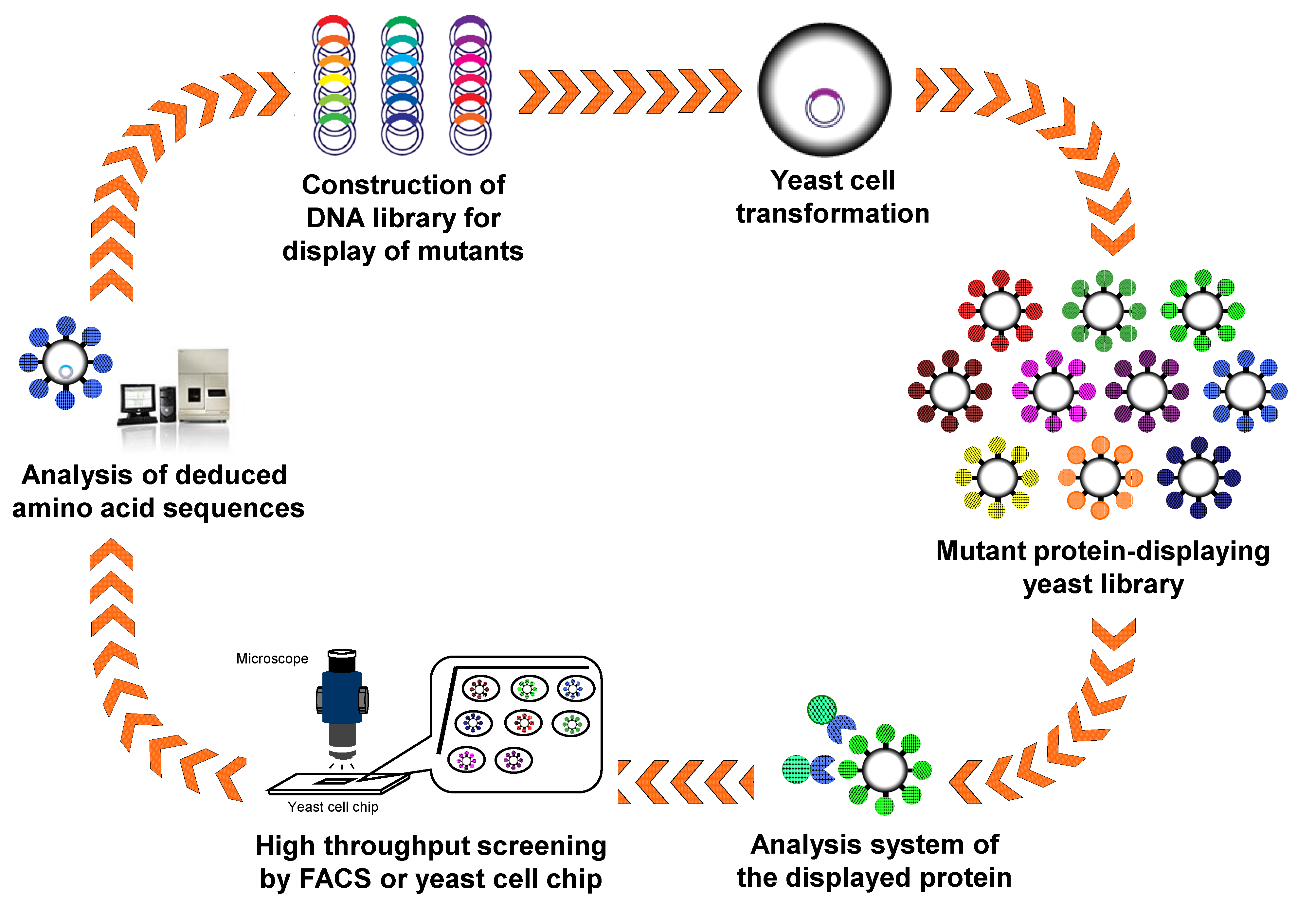
4.2. Antibody Engineering
4.3. Ligand Screening for Activation of GPCRs
5. Conclusions
Conflicts of Interest
References
- Anonymous. Arming yeast with cell-surface catalysts. Chem. Eng. News 1997, 75, 32. [Google Scholar]
- Ueda, M.; Tanaka, A. Genetic immobilization of proteins on the yeast cell surface. Biotechnol. Adv. 2000, 18, 121–140. [Google Scholar] [CrossRef]
- Ueda, M.; Tanaka, A. Cell surface engineering of yeast: Construction of arming yeast with biocatalyst. J. Biosci. Bioeng. 2000, 90, 125–136. [Google Scholar]
- Su, G.D.; Zhang, X.; Lin, Y. Surface display of active lipase in Pichia pastoris using Sed1 as an anchor protein. Biotechnol. Lett. 2010, 32, 1131–1136. [Google Scholar] [CrossRef]
- Chen, Y.P.; Hwang, I.E.; Lin, C.J.; Wang, H.J.; Tseng, C.P. Enhancing the stability of xylanase from Cellulomonas fimi by cell-surface display on Escherichia coli. J. Appl. Microbiol. 2012, 112, 455–463. [Google Scholar] [CrossRef]
- Aoki, W.; Yoshino, Y.; Morisaka, H.; Tsunetomo, K.; Koyo, H.; Kamiya, S.; Kawata, N.; Kuroda, K.; Ueda, M. High-throughput screening of improved protease inhibitors using a yeast cell surface display system and a yeast cell chip. J. Biosci. Bioeng. 2011, 111, 16–18. [Google Scholar] [CrossRef]
- Boder, E.T.; Wittrup, K.D. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 1997, 15, 553–557. [Google Scholar] [CrossRef]
- Chen, W.; Georgiou, G. Cell-Surface display of heterologous proteins: From high-throughput screening to environmental applications. Biotechnol. Bioeng. 2002, 79, 496–503. [Google Scholar] [CrossRef]
- Fukuda, T.; Kato-Murai, M.; Kadonosono, T.; Sahara, H.; Hata, Y.; Suye, S.; Ueda, M. Enhancement of substrate recognition ability by combinatorial mutation of β-glucosidase displayed on the yeast cell surface. Appl. Microbiol. Biotechnol. 2007, 76, 1027–1033. [Google Scholar] [CrossRef]
- Yeung, Y.A.; Wittrup, K.D. Quantitative screening of yeast surface-displayed polypeptide libraries by magnetic bead capture. Biotechnol. Prog. 2002, 18, 212–220. [Google Scholar] [CrossRef]
- Georgiou, G.; Poetschke, H.L.; Stathopoulos, C.; Francisco, J.A. Practical applications of engineering gram-negative bacterial cell surfaces. Trends Biotechnol. 1993, 11, 6–10. [Google Scholar] [CrossRef]
- Kuroda, K.; Ueda, M. Engineering of microorganisms towards recovery of rare metal ions. Appl. Microbiol. Biotechnol. 2010, 87, 53–60. [Google Scholar] [CrossRef]
- Kuroda, K.; Ueda, M. Molecular design of the microbial cell surface toward the recovery of metal ions. Curr. Opin. Biotechnol. 2011, 22, 427–433. [Google Scholar] [CrossRef]
- Kuroda, K.; Ueda, M. Cell surface engineering of yeast for applications in white biotechnology. Biotechnol. Lett. 2011, 33, 1–9. [Google Scholar]
- Samuelson, P.; Gunneriusson, E.; Nygren, P.A.; Ståhl, S. Display of proteins on bacteria. J. Biotechnol. 2002, 96, 129–154. [Google Scholar] [CrossRef]
- Ståhl, S.; Uhlén, M. Bacterial surface display: Trends and progress. Trends Biotechnol. 1997, 15, 185–192. [Google Scholar] [CrossRef]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar]
- Smith, G.P. Surface presentation of protein epitopes using bacteriophage expression systems. Curr. Opin. Biotechnol. 1991, 2, 668–673. [Google Scholar] [CrossRef]
- Tabuchi, S.; Ito, J.; Adachi, T.; Ishida, H.; Hata, Y.; Okazaki, F.; Tanaka, T.; Ogino, C.; Kondo, A. Display of both N- and C-terminal target fusion proteins on the Aspergillus oryzae cell surface using a chitin-binding module. Appl. Microbiol. Biotechnol. 2010, 87, 1783–1789. [Google Scholar] [CrossRef]
- Adachi, T.; Ito, J.; Kawata, K.; Kaya, M.; Ishida, H.; Sahara, H.; Hata, Y.; Ogino, C.; Fukuda, H.; Kondo, A. Construction of an Aspergillus oryzae cell-surface display system using a putative GPI-anchored protein. Appl. Microbiol. Biotechnol. 2008, 81, 711–719. [Google Scholar] [CrossRef]
- Breinig, F.; Schmitt, M.J. Spacer-elongated cell wall fusion proteins improve cell surface expression in the yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2002, 58, 637–644. [Google Scholar] [CrossRef]
- Washida, M.; Takahashi, S.; Ueda, M.; Tanaka, A. Spacer-mediated display of active lipase on the yeast cell surface. Appl. Microbiol. Biotechnol. 2001, 56, 681–686. [Google Scholar] [CrossRef]
- Lipke, P.N.; Kurjan, J. Sexual agglutination in budding yeasts: Structure, function, and regulation of adhesion glycoproteins. Microbiol. Rev. 1992, 56, 180–194. [Google Scholar]
- Kuroda, K.; Ueda, M. Generation of arming yeasts with active proeins and peptides via cell surface display system—Cell surface engineering, bio-arming technology. Methods Mol. Biol. 2013, in press. [Google Scholar]
- Sato, N.; Matsumoto, T.; Ueda, M.; Tanaka, A.; Fukuda, H.; Kondo, A. Long anchor using Flo1 protein enhances reactivity of cell surface-displayed glucoamylase to polymer substrates. Appl. Microbiol. Biotechnol. 2002, 60, 469–474. [Google Scholar] [CrossRef]
- Van der Vaart, J.M.; te Biesebeke, R.; Chapman, J.W.; Toschka, H.Y.; Klis, F.M.; Verrips, C.T. Comparison of cell wall proteins of Saccharomyces cerevisiae as anchors for cell surface expression of heterologous proteins. Appl. Environ. Microbiol. 1997, 63, 615–620. [Google Scholar]
- Wang, Z.; Mathias, A.; Stavrou, S.; Neville, D.M., Jr. A new yeast display vector permitting free scFv amino termini can augment ligand binding affinities. Protein Eng. Des. Sel. 2005, 18, 337–343. [Google Scholar] [CrossRef]
- Matsumoto, T.; Fukuda, H.; Ueda, M.; Tanaka, A.; Kondo, A. Construction of yeast strains with high cell surface lipase activity by using novel display systems based on the Flo1p flocculation functional domain. Appl. Environ. Microbiol. 2002, 68, 4517–4522. [Google Scholar] [CrossRef]
- Abe, H.; Ohba, M.; Shimma, Y.; Jigami, Y. Yeast cells harboring human α-1,3-fucosyltransferase at the cell surface engineered using Pir, a cell wall-anchored protein. FEMS Yeast Res. 2004, 4, 417–425. [Google Scholar] [CrossRef]
- Kuroda, K.; Matsui, K.; Higuchi, S.; Kotaka, A.; Sahara, H.; Hata, Y.; Ueda, M. Enhancement of display efficiency in yeast display system by vector engineering and gene disruption. Appl. Microbiol. Biotechnol. 2009, 82, 713–719. [Google Scholar] [CrossRef]
- Wentz, A.E.; Shusta, E.V. A novel high-throughput screen reveals yeast genes that increase secretion of heterologous proteins. Appl. Environ. Microbiol. 2007, 73, 1189–1198. [Google Scholar] [CrossRef]
- Tanino, T.; Fukuda, H.; Kondo, A. Construction of a Pichia pastoris cell-surface display system using Flo1p anchor system. Biotechnol. Prog. 2006, 22, 989–993. [Google Scholar] [CrossRef]
- Wang, Q.; Li, L.; Chen, M.; Qi, Q.; Wang, P.G. Construction of a novel system for cell surface display of heterologous proteins on Pichia pastoris. Biotechnol. Lett. 2007, 29, 1561–1566. [Google Scholar] [CrossRef]
- Wang, Q.; Li, L.; Chen, M.; Qi, Q.; Wang, P.G. Construction of a novel Pichia pastoris cell-surface display system based on the cell wall protein Pir1. Curr. Microbiol. 2008, 56, 352–357. [Google Scholar] [CrossRef]
- Yuzbasheva, E.Y.; Yuzbashev, T.V.; Laptev, I.A.; Konstantinova, T.K.; Sineoky, S.P. Efficient cell surface display of Lip2 lipase using C-domains of glycosylphosphatidylinositol-anchored cell wall proteins of Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2011, 91, 645–654. [Google Scholar] [CrossRef]
- Hara, K.; Ono, T.; Kuroda, K.; Ueda, M. Membrane-displayed peptide ligand activates the pheromone response pathway in Saccharomyces cerevisiae. J. Biochem. 2012, 151, 551–557. [Google Scholar] [CrossRef]
- Hara, K.; Shigemori, T.; Kuroda, K.; Ueda, M. Membrane-displayed somatostatin activates somatostatin receptor subtype-2 heterologously produced in Saccharomyces cerevisiae. AMB Express 2012, 2, e63. [Google Scholar] [CrossRef]
- Lynd, L.R.; van Zyl, W.H.; McBride, J.E.; Laser, M. Consolidated bioprocessing of cellulosic biomass: An update. Curr. Opin. Biotechnol. 2005, 16, 577–583. [Google Scholar] [CrossRef]
- Murai, T.; Ueda, M.; Yamamura, M.; Atomi, H.; Shibasaki, Y.; Kamasawa, N.; Osumi, M.; Amachi, T.; Tanaka, A. Construction of a starch-utilizing yeast by cell surface engineering. Appl. Environ. Microbiol. 1997, 63, 1362–1366. [Google Scholar]
- Murai, T.; Ueda, M.; Shibasaki, Y.; Kamasawa, N.; Osumi, M.; Imanaka, T.; Tanaka, A. Development of an arming yeast strain for efficient utilization of starch by co-display of sequential amylolytic enzymes on the cell surface. Appl. Microbiol. Biotechnol. 1999, 51, 65–70. [Google Scholar] [CrossRef]
- Shigechi, H.; Koh, J.; Fujita, Y.; Matsumoto, T.; Bito, Y.; Ueda, M.; Satoh, E.; Fukuda, H.; Kondo, A. Direct production of ethanol from raw corn starch via fermentation by use of a novel surface-engineered yeast strain codisplaying glucoamylase and α-amylase. Appl. Environ. Microbiol. 2004, 70, 5037–5040. [Google Scholar] [CrossRef]
- Murai, T.; Ueda, M.; Kawaguchi, T.; Arai, M.; Tanaka, A. Assimilation of cellooligosaccharides by a cell surface-engineered yeast expressing β-glucosidase and carboxymethylcellulase from Aspergillus aculeatus. Appl. Environ. Microbiol. 1998, 64, 4857–4861. [Google Scholar]
- Fujita, Y.; Ito, J.; Ueda, M.; Fukuda, H.; Kondo, A. Synergistic saccharification, and direct fermentation to ethanol, of amorphous cellulose by use of an engineered yeast strain codisplaying three types of cellulolytic enzyme. Appl. Environ. Microbiol. 2004, 70, 1207–1212. [Google Scholar] [CrossRef]
- Kotaka, A.; Bando, H.; Kaya, M.; Kato-Murai, M.; Kuroda, K.; Sahara, H.; Hata, Y.; Kondo, A.; Ueda, M. Direct ethanol production from barley β-glucan by sake yeast displaying Aspergillus oryzae β-glucosidase and endoglucanase. J. Biosci. Bioeng. 2008, 105, 622–627. [Google Scholar] [CrossRef]
- Ito, J.; Kosugi, A.; Tanaka, T.; Kuroda, K.; Shibasaki, S.; Ogino, C.; Ueda, M.; Fukuda, H.; Doi, R.H.; Kondo, A. Regulation of the display ratio of enzymes on the Saccharomyces cerevisiae cell surface by the immunoglobulin G and cellulosomal enzyme binding domains. Appl. Environ. Microbiol. 2009, 75, 4149–4154. [Google Scholar] [CrossRef]
- Yamada, R.; Taniguchi, N.; Tanaka, T.; Ogino, C.; Fukuda, H.; Kondo, A. Cocktail δ-integration: A novel method to construct cellulolytic enzyme expression ratio-optimized yeast strains. Microb. Cell Fact. 2010, 9, e32. [Google Scholar] [CrossRef]
- Nakanishi, A.; Bae, J.; Kuroda, K.; Ueda, M. Construction of a novel selection system for endoglucanases exhibiting carbohydrate-binding modules optimized for biomass using yeast cell-surface engineering. AMB Express 2012, 2, e56. [Google Scholar] [CrossRef]
- Tsai, S.L.; Oh, J.; Singh, S.; Chen, R.; Chen, W. Functional assembly of minicellulosomes on the Saccharomyces cerevisiae cell surface for cellulose hydrolysis and ethanol production. Appl. Environ. Microbiol. 2009, 75, 6087–6093. [Google Scholar] [CrossRef]
- Goyal, G.; Tsai, S.L.; Madan, B.; DaSilva, N.A.; Chen, W. Simultaneous cell growth and ethanol production from cellulose by an engineered yeast consortium displaying a functional mini-cellulosome. Microb. Cell Fact. 2011, 10, e89. [Google Scholar] [CrossRef]
- Katahira, S.; Fujita, Y.; Mizuike, A.; Fukuda, H.; Kondo, A. Construction of a xylan-fermenting yeast strain through codisplay of xylanolytic enzymes on the surface of xylose-utilizing Saccharomyces cerevisiae cells. Appl. Environ. Microbiol. 2004, 70, 5407–5414. [Google Scholar] [CrossRef]
- Katahira, S.; Mizuike, A.; Fukuda, H.; Kondo, A. Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose- and cellooligosaccharide-assimilating yeast strain. Appl. Microbiol. Biotechnol. 2006, 72, 1136–1143. [Google Scholar] [CrossRef]
- Ota, M.; Sakuragi, H.; Morisaka, H.; Kuroda, K.; Miyake, H.; Tamaru, Y.; Ueda, M. Display of Clostridium cellulovorans xylose isomerase on the cell surface of Saccharomyces cerevisiae and its direct application to xylose fermentation. Biotechnol. Prog. 2013, 29, 346–351. [Google Scholar] [CrossRef]
- Nakanishi, A.; Bae, J.G.; Fukai, K.; Tokumoto, N.; Kuroda, K.; Ogawa, J.; Nakatani, M.; Shimizu, S.; Ueda, M. Effect of pretreatment of hydrothermally processed rice straw with laccase-displaying yeast on ethanol fermentation. Appl. Microbiol. Biotechnol. 2012, 94, 939–948. [Google Scholar] [CrossRef]
- Shiraga, S.; Kawakami, M.; Ishiguro, M.; Ueda, M. Enhanced reactivity of Rhizopus oryzae lipase displayed on yeast cell surfaces in organic solvents: Potential as a whole-cell biocatalyst in organic solvents. Appl. Environ. Microbiol. 2005, 71, 4335–4338. [Google Scholar] [CrossRef]
- Nakamura, Y.; Matsumoto, T.; Nomoto, F.; Ueda, M.; Fukuda, H.; Kondo, A. Enhancement of activity of lipase-displaying yeast cells and their application to optical resolution of (R,S)-1-benzyloxy-3-chloro-2-propyl monosuccinate. Biotechnol. Prog. 2006, 22, 998–1002. [Google Scholar] [CrossRef]
- Kato, M.; Fuchimoto, J.; Tanino, T.; Kondo, A.; Fukuda, H.; Ueda, M. Preparation of a whole-cell biocatalyst of mutated Candida antarctica lipase B (mCALB) by a yeast molecular display system and its practical properties. Appl. Microbiol. Biotechnol. 2007, 75, 549–555. [Google Scholar] [CrossRef]
- Inaba, C.; Maekawa, K.; Morisaka, H.; Kuroda, K.; Ueda, M. Efficient synthesis of enantiomeric ethyl lactate by Candida antarctica lipase B (CALB)-displaying yeasts. Appl. Microbiol. Biotechnol. 2009, 83, 859–864. [Google Scholar] [CrossRef]
- Tanino, T.; Aoki, T.; Chung, W.Y.; Watanabe, Y.; Ogino, C.; Fukuda, H.; Kondo, A. Improvement of a Candida antarctica lipase B-displaying yeast whole-cell biocatalyst and its application to the polyester synthesis reaction. Appl. Microbiol. Biotechnol. 2009, 82, 59–66. [Google Scholar] [CrossRef]
- Tanino, T.; Ohno, T.; Aoki, T.; Fukuda, H.; Kondo, A. Development of yeast cells displaying Candida antarctica lipase B and their application to ester synthesis reaction. Appl. Microbiol. Biotechnol. 2007, 75, 1319–1325. [Google Scholar] [CrossRef]
- Han, S.Y.; Pan, Z.Y.; Huang, D.F.; Ueda, M.; Wang, X.N.; Lin, Y. Highly efficient synthesis of ethyl hexanoate catalyzed by CALB-displaying Saccharomyces cerevisiae whole-cells in non-aqueous phase. J. Mol. Catal. B 2009, 59, 168–172. [Google Scholar] [CrossRef]
- Su, G.D.; Huang, D.F.; Han, S.Y.; Zheng, S.P.; Lin, Y. Display of Candida antarctica lipase B on Pichia pastoris and its application to flavor ester synthesis. Appl. Microbiol. Biotechnol. 2010, 86, 1493–1501. [Google Scholar] [CrossRef]
- Jiang, Z.; Gao, B.; Ren, R.; Tao, X.; Ma, Y.; Wei, D. Efficient display of active lipase LipB52 with a Pichia pastoris cell surface display system and comparison with the LipB52 displayed on Saccharomyces cerevisiae cell surface. BMC Biotechnol. 2008, 8, e4. [Google Scholar]
- Jiang, Z.B.; Song, H.T.; Gupta, N.; Ma, L.X.; Wu, Z.B. Cell surface display of functionally active lipases from Yarrowia lipolytica in Pichia pastoris. Protein Expr. Purif. 2007, 56, 35–39. [Google Scholar] [CrossRef]
- Kaya, M.; Ito, J.; Kotaka, A.; Matsumura, K.; Bando, H.; Sahara, H.; Ogino, C.; Shibasaki, S.; Kuroda, K.; Ueda, M.; et al. Isoflavone aglycones production from isoflavone glycosides by display of β-glucosidase from Aspergillus oryzae on yeast cell surface. Appl. Microbiol. Biotechnol. 2008, 79, 51–60. [Google Scholar] [CrossRef]
- Inaba, C.; Higuchi, S.; Morisaka, H.; Kuroda, K.; Ueda, M. Synthesis of functional dipeptide carnosine from nonprotected amino acids using carnosinase-displaying yeast cells. Appl. Microbiol. Biotechnol. 2010, 86, 1895–1902. [Google Scholar] [CrossRef]
- Fukuda, T.; Isogawa, D.; Takagi, M.; Kato-Murai, M.; Kimoto, H.; Kusaoke, H.; Ueda, M.; Suye, S. Yeast cell-surface expression of chitosanase from Paenibacillus fukuinensis. Biosci. Biotechnol. Biochem. 2007, 71, 2845–2847. [Google Scholar] [CrossRef]
- Liu, G.; Yue, L.; Chi, Z.; Yu, W.; Chi, Z.; Madzak, C. The surface display of the alginate lyase on the cells of Yarrowia lipolytica for hydrolysis of alginate. Mar. Biotechnol. (NY) 2009, 11, 619–626. [Google Scholar]
- Kotrba, P.; Ruml, T. Surface display of metal fixation motifs of bacterial P1-type ATPases specifically promotes biosorption of Pb2+ by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2010, 76, 2615–2622. [Google Scholar] [CrossRef]
- Kuroda, K.; Shibasaki, S.; Ueda, M.; Tanaka, A. Cell surface-engineered yeast displaying a histidine oligopeptide (hexa-His) has enhanced adsorption of and tolerance to heavy metal ions. Appl. Microbiol. Biotechnol. 2001, 57, 697–701. [Google Scholar] [CrossRef]
- Kuroda, K.; Ueda, M. Bioadsorption of cadmium ion by cell surface-engineered yeasts displaying metallothionein and hexa-His. Appl. Microbiol. Biotechnol. 2003, 63, 182–186. [Google Scholar] [CrossRef]
- Kuroda, K.; Ueda, M. Effective display of metallothionein tandem repeats on the bioadsorption of cadmium ion. Appl. Microbiol. Biotechnol. 2006, 70, 458–463. [Google Scholar] [CrossRef]
- Grunden, A.M.; Ray, R.M.; Rosentel, J.K.; Healy, F.G.; Shanmugam, K.T. Repression of the Escherichia coli modABCD (molybdate transport) operon by ModE. J. Bacteriol. 1996, 178, 735–744. [Google Scholar]
- Nishitani, T.; Shimada, M.; Kuroda, K.; Ueda, M. Molecular design of yeast cell surface for adsorption and recovery of molybdenum, one of rare metals. Appl. Microbiol. Biotechnol. 2010, 86, 641–648. [Google Scholar] [CrossRef]
- Kuroda, K.; Nishitani, T.; Ueda, M. Specific adsorption of tungstate by cell surface display of the newly designed ModE mutant. Appl. Microbiol. Biotechnol. 2012, 96, 153–159. [Google Scholar]
- Yasui, M.; Shibasaki, S.; Kuroda, K.; Ueda, M.; Kawada, N.; Nishikawa, J.; Nishihara, T.; Tanaka, A. An arming yeast with the ability to entrap fluorescent 17β-estradiol on the cell surface. Appl. Microbiol. Biotechnol. 2002, 59, 329–331. [Google Scholar] [CrossRef]
- Fukuda, T.; Tsuchiyama, K.; Makishima, H.; Takayama, K.; Mulchandani, A.; Kuroda, K.; Ueda, M.; Suye, S. Improvement in organophosphorus hydrolase activity of cell surface-engineered yeast strain using Flo1p anchor system. Biotechnol. Lett. 2010, 32, 655–659. [Google Scholar] [CrossRef]
- Takayama, K.; Suye, S.; Kuroda, K.; Ueda, M.; Kitaguchi, T.; Tsuchiyama, K.; Fukuda, T.; Chen, W.; Mulchandani, A. Surface display of organophosphorus hydrolase on Saccharomyces cerevisiae. Biotechnol. Prog. 2006, 22, 939–943. [Google Scholar] [CrossRef]
- Fukuda, T.; Tsuchiya, K.; Makishima, H.; Tsuchiyama, K.; Mulchandani, A.; Kuroda, K.; Ueda, M.; Suye, S. Organophosphorus compound detection on a cell chip with yeast coexpressing hydrolase and eGFP. Biotechnol. J. 2010, 5, 515–519. [Google Scholar] [CrossRef]
- Takayama, K.; Suye, S.; Tanaka, Y.; Mulchandani, A.; Kuroda, K.; Ueda, M. Estimation of enzyme kinetic parameters of cell surface-displayed organophosphorus hydrolase and construction of a biosensing system for organophosphorus compounds. Anal. Sci. 2011, 27, 823–826. [Google Scholar] [CrossRef]
- Shibasaki, S.; Ueda, M.; Ye, K.; Shimizu, K.; Kamasawa, N.; Osumi, M.; Tanaka, A. Creation of cell surface-engineered yeast that display different fluorescent proteins in response to the glucose concentration. Appl. Microbiol. Biotechnol. 2001, 57, 528–533. [Google Scholar] [CrossRef]
- Shibasaki, S.; Ninomiya, Y.; Ueda, M.; Iwahashi, M.; Katsuragi, T.; Tani, Y.; Harashima, S.; Tanaka, A. Intelligent yeast strains with the ability to self-monitor the concentrations of intra- and extracellular phosphate or ammonium ion by emission of fluorescence from the cell surface. Appl. Microbiol. Biotechnol. 2001, 57, 702–707. [Google Scholar]
- Shibasaki, S.; Tanaka, A.; Ueda, M. Development of combinatorial bioengineering using yeast cell surface display—Order-made design of cell and protein for bio-monitoring. Biosens. Bioelectron. 2003, 19, 123–130. [Google Scholar] [CrossRef]
- Tamaru, Y.; Ohtsuka, M.; Kato, K.; Manabe, S.; Kuroda, K.; Sanada, M.; Ueda, M. Application of the arming system for the expression of the 380R antigen from red sea bream iridovirus (RSIV) on the surface of yeast cells: A first step for the development of an oral vaccine. Biotechnol. Prog. 2006, 22, 949–953. [Google Scholar] [CrossRef]
- Wasilenko, J.L.; Sarmento, L.; Spatz, S.; Pantin-Jackwood, M. Cell surface display of highly pathogenic avian influenza virus hemagglutinin on the surface of Pichia pastoris cells using α-agglutinin for production of oral vaccines. Biotechnol. Prog. 2010, 26, 542–547. [Google Scholar]
- Lin, Y.; Shiraga, S.; Tsumuraya, T.; Matsumoto, T.; Kondo, A.; Fujii, I.; Ueda, M. Comparison of two forms of catalytic antibody displayed on yeast-cell surface. J. Mol. Catal. B 2004, 28, 241–246. [Google Scholar] [CrossRef]
- Lin, Y.; Tsumuraya, T.; Wakabayashi, T.; Shiraga, S.; Fujii, I.; Kondo, A.; Ueda, M. Display of a functional hetero-oligomeric catalytic antibody on the yeast cell surface. Appl. Microbiol. Biotechnol. 2003, 62, 226–232. [Google Scholar] [CrossRef]
- Okochi, N.; Kato-Murai, M.; Kadonosono, T.; Ueda, M. Design of a serine protease-like catalytic triad on an antibody light chain displayed on the yeast cell surface. Appl. Microbiol. Biotechnol. 2007, 77, 597–603. [Google Scholar] [CrossRef]
- Nakamura, Y.; Shibasaki, S.; Ueda, M.; Tanaka, A.; Fukuda, H.; Kondo, A. Development of novel whole-cell immunoadsorbents by yeast surface display of the IgG-binding domain. Appl. Microbiol. Biotechnol. 2001, 57, 500–505. [Google Scholar] [CrossRef]
- Matsui, K.; Kuroda, K.; Ueda, M. Creation of a novel peptide endowing yeasts with acid tolerance using yeast cell-surface engineering. Appl. Microbiol. Biotechnol. 2009, 82, 105–113. [Google Scholar] [CrossRef]
- Zou, W.; Ueda, M.; Tanaka, A. Screening of a molecule endowing Saccharomyces cerevisiae with n-nonane-tolerance from a combinatorial random protein library. Appl. Microbiol. Biotechnol. 2002, 58, 806–812. [Google Scholar] [CrossRef]
- Zou, W.; Ueda, M.; Yamanaka, H.; Tanaka, A. Construction of a combinatorial protein library displayed on yeast cell surface using DNA random priming method. J. Biosci. Bioeng. 2001, 92, 393–396. [Google Scholar]
- Andreu, C.; del Olmo, M. Yeast arming by the Aga2p system: Effect of growth conditions in galactose on the efficiency of the display and influence of expressing leucine-containing peptides. Appl. Microbiol. Biotechnol. 2013. [Google Scholar] [CrossRef]
- Shiraga, S.; Ishiguro, M.; Fukami, H.; Nakao, M.; Ueda, M. Creation of Rhizopus oryzae lipase having a unique oxyanion hole by combinatorial mutagenesis in the lid domain. Appl. Microbiol. Biotechnol. 2005, 68, 779–785. [Google Scholar] [CrossRef]
- Kadonosono, T.; Kato-Murai, M.; Ueda, M. Alteration of substrate specificity of rat neurolysin from matrix metalloproteinase-2/9-type to -3-type specificity by comprehensive mutation. Protein Eng. Des. Sel. 2008, 21, 507–513. [Google Scholar] [CrossRef]
- Fushimi, T.; Miura, N.; Shintani, H.; Tsunoda, H.; Kuroda, K.; Ueda, M. Mutant firefly luciferases with improved specific activity and dATP discrimination constructed by yeast cell surface engineering. Appl. Microbiol. Biotechnol. 2013, 97, 4003–4011. [Google Scholar]
- Van den Beucken, T.; Pieters, H.; Steukers, M.; van der Vaart, M.; Ladner, R.C.; Hoogenboom, H.R.; Hufton, S.E. Affinity maturation of Fab antibody fragments by fluorescent-activated cell sorting of yeast-displayed libraries. FEBS Lett. 2003, 546, 288–294. [Google Scholar] [CrossRef]
- Rajpal, A.; Beyaz, N.; Haber, L.; Cappuccilli, G.; Yee, H.; Bhatt, R.R.; Takeuchi, T.; Lerner, R.A.; Crea, R. A general method for greatly improving the affinity of antibodies by using combinatorial libraries. Proc. Natl. Acad. Sci. USA 2005, 102, 8466–8471. [Google Scholar]
- Graff, C.P.; Chester, K.; Begent, R.; Wittrup, K.D. Directed evolution of an anti-carcinoembryonic antigen scFv with a 4-day monovalent dissociation half-time at 37 °C. Protein Eng. Des. Sel. 2004, 17, 293–304. [Google Scholar] [CrossRef]
- Razai, A.; Garcia-Rodriguez, C.; Lou, J.; Geren, I.N.; Forsyth, C.M.; Robles, Y.; Tsai, R.; Smith, T.J.; Smith, L.A.; Siegel, R.W.; et al. Molecular evolution of antibody affinity for sensitive detection of botulinum neurotoxin type A. J. Mol. Biol. 2005, 351, 158–169. [Google Scholar] [CrossRef]
- VanAntwerp, J.J.; Wittrup, K.D. Thermodynamic characterization of affinity maturation: The D1.3 antibody and a higher-affinity mutant. J. Mol. Recognit. 1998, 11, 10–13. [Google Scholar] [CrossRef]
- Wang, Z.; Kim, G.B.; Woo, J.H.; Liu, Y.Y.; Mathias, A.; Stavrou, S.; Neville, D.M., Jr. Improvement of a recombinant anti-monkey anti-CD3 diphtheria toxin based immunotoxin by yeast display affinity maturation of the scFv. Bioconjug. Chem. 2007, 18, 947–955. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kuroda, K.; Ueda, M. Arming Technology in Yeast—Novel Strategy for Whole-cell Biocatalyst and Protein Engineering. Biomolecules 2013, 3, 632-650. https://doi.org/10.3390/biom3030632
Kuroda K, Ueda M. Arming Technology in Yeast—Novel Strategy for Whole-cell Biocatalyst and Protein Engineering. Biomolecules. 2013; 3(3):632-650. https://doi.org/10.3390/biom3030632
Chicago/Turabian StyleKuroda, Kouichi, and Mitsuyoshi Ueda. 2013. "Arming Technology in Yeast—Novel Strategy for Whole-cell Biocatalyst and Protein Engineering" Biomolecules 3, no. 3: 632-650. https://doi.org/10.3390/biom3030632



