Towards the Fecal Metabolome Derived from Moderate Red Wine Intake
Abstract
:1. Introduction
2. Experimental Section
2.1. Chemicals
2.2. Red Wine
2.3. Human Intervention Study Design
2.4. Preparation of Fecal Solutions
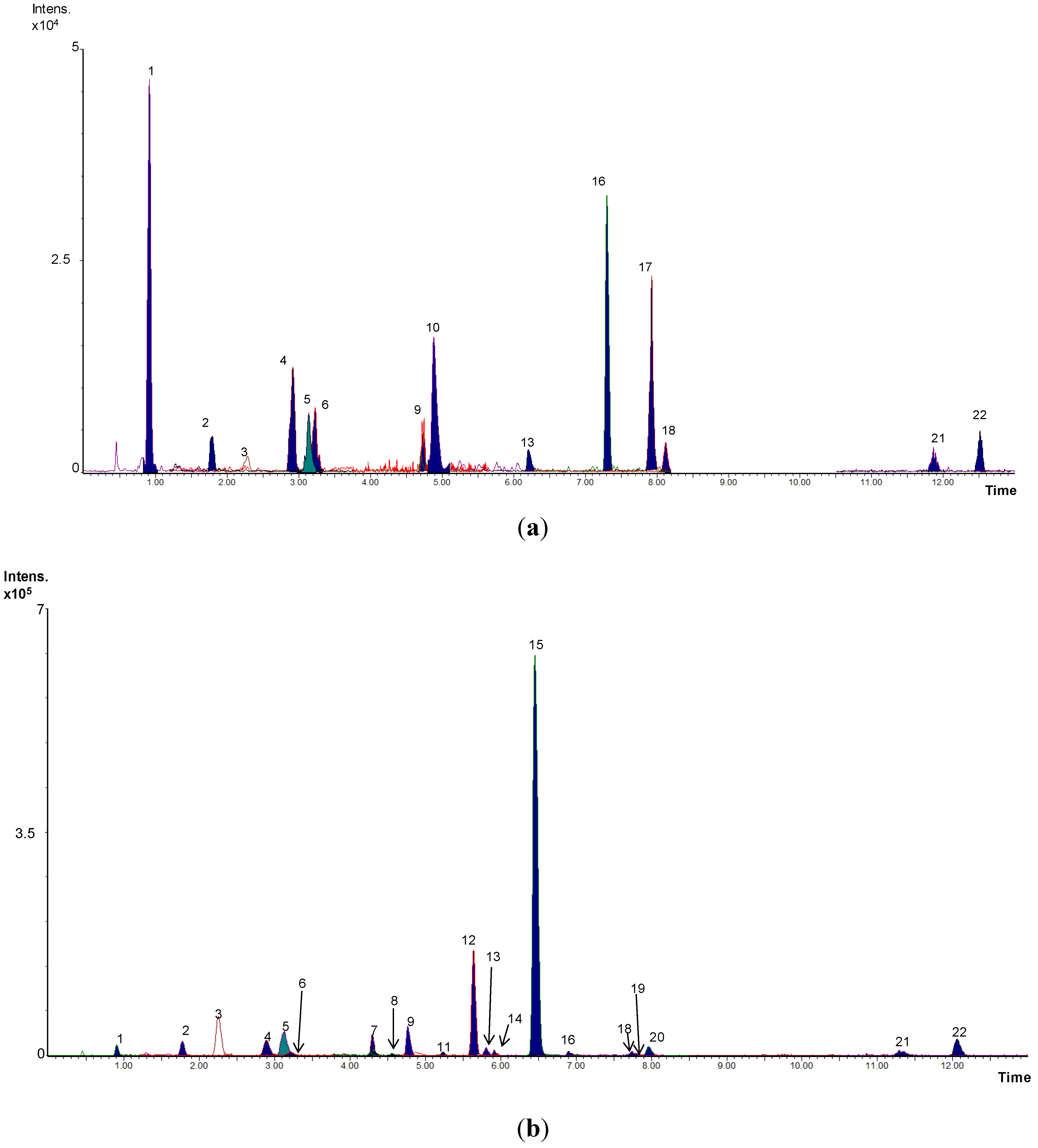
2.5. Targeted Analysis of Phenolic Metabolites
2.6. Non-Targeted Analysis: Metabolomic Study and Metabolite Identification
2.7. Statistical Analysis
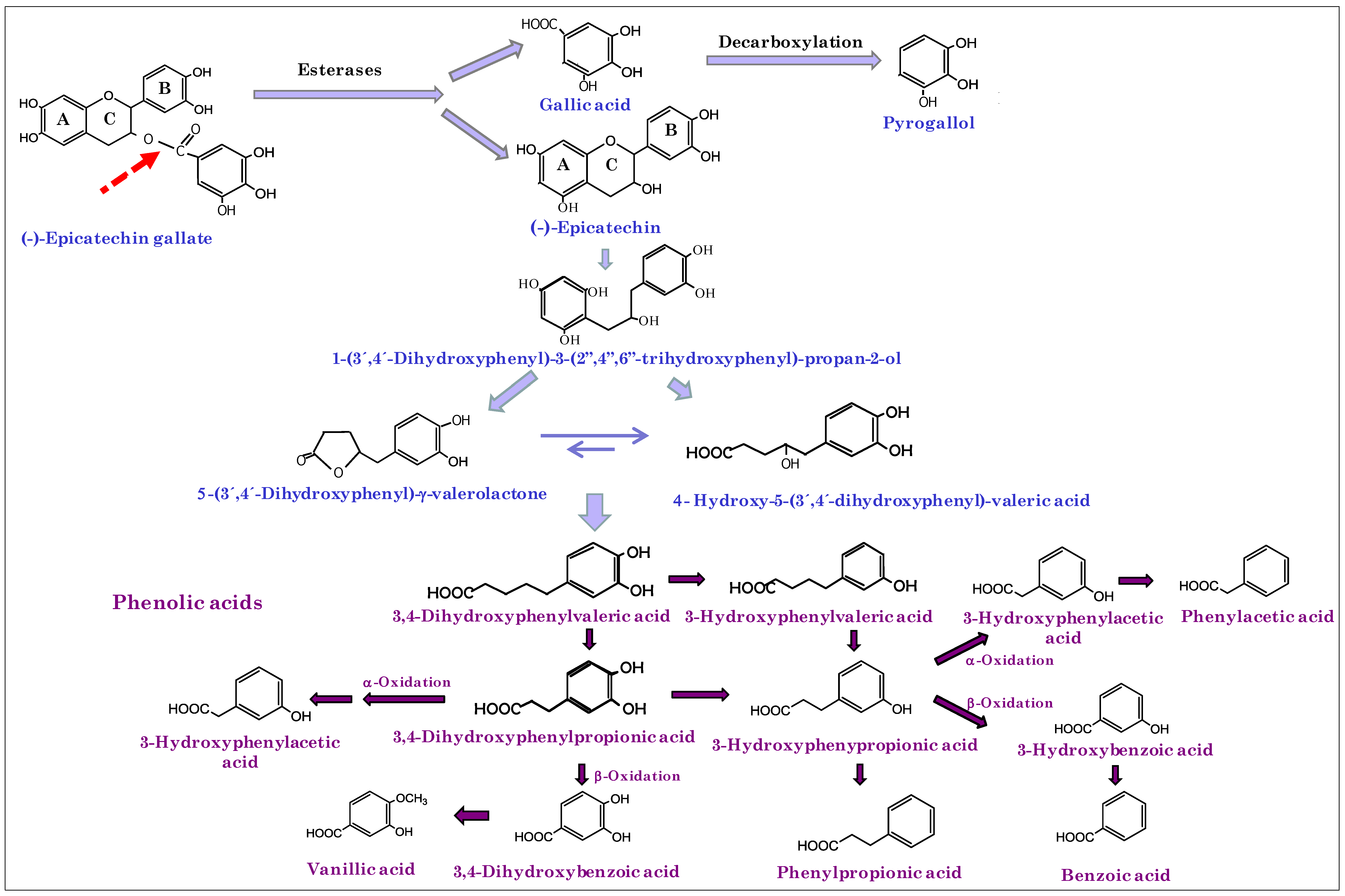
3. Results and Discussion
| Compound | Analysis | Effect of Wine Intake (↑, ↓) | |
|---|---|---|---|
| Targeted | Non-Targeted | ||
| Group I: Wine compounds | |||
| 2-Hydroxyglutaric acid a | + | ↑ | |
| 2-Methylbutyric acid | + | ↑ | |
| 2,3-Pentanedione | + | ↑ | |
| Diethylmalonate | + | ↑ | |
| 2-Phenethyl butyrate | + | ↑ | |
| 2-Phenylethyl hexanoate | + | ↑ | |
| Group II: Phenolic metabolites | |||
| Benzoic acids | |||
| 3,5-Dihydroxybenzoic acid | + | ↑ | |
| Protocatechuic acid | + | ↑ | |
| 3-O-Methylgallic acid | + | ↑ | |
| Vanillic acid | + | ↑ | |
| Syringic acid | + | ↑ | |
| Benzoic acid b | + | ↑ | |
| Phenylacetic acids | |||
| 3-Hydroxyphenylacetic acid | + | ↑ | |
| Phenylpropionic acids | |||
| 3-Phenylpropionic acid | + | ↑ | |
| 3-(3’-Hydroxyphenyl) propionic acid b | + | ↓ | |
| Valeric acids | |||
| 4-Hydroxy-5-(3’-hydroxyphenyl) valeric acid | + | ↑ | |
| 4-Hydroxy-5-(phenyl) valeric acid | + | + | ↑ |
| 4-Hydroxy-5-(3’,4’-dihydroxyphenyl) valeric acid | + | ↑ | |
| Valerolactones | |||
| 5-(3’,4’-Dihydroxyphenyl)- γ-valerolactone | + | ↑ | |
| 5-(3’-Hydroxyphenyl)-γ-valerolactone | + | ↑ | |
| Group III: Endogenous metabolites | |||
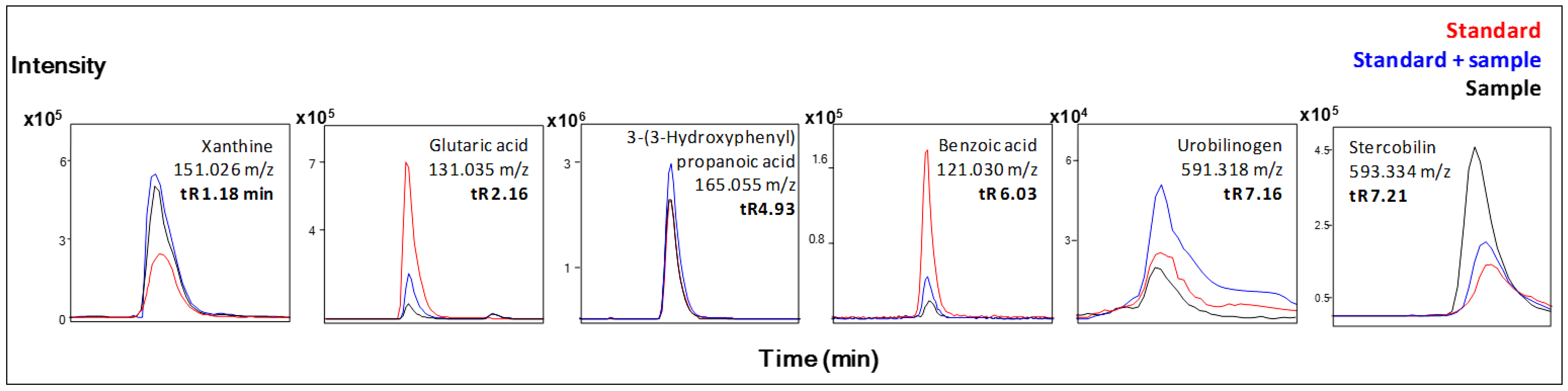
| Xanthine b | + | ↓ | |
| Tricarballylic acid | + | ↓ | |
| Glutaric acid b | + | ↑ | |
| Urobilinogen b | + | ↑ | |
| Stercobilin b | + | ↑ | |
| Docosahexaenoic acid methyl ester | + | ↑ | |
| Deoxycholic acid | + | ↑ | |
| Sulfolithocholic acid | + | ↓ | |
| Cholesterol sulfate | + | ↑ | |
 : 25th–75th percentiles;
: 25th–75th percentiles;  : non-outlier range; ○: outliers;
: non-outlier range; ○: outliers;  : extremes) of the total phenolic metabolites content in the intervention group (n = 33).
: extremes) of the total phenolic metabolites content in the intervention group (n = 33).
 : 25th–75th percentiles;
: 25th–75th percentiles;  : non-outlier range; ○: outliers;
: non-outlier range; ○: outliers;  : extremes) of the total phenolic metabolites content in the intervention group (n = 33).
: extremes) of the total phenolic metabolites content in the intervention group (n = 33).
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pozo-Bayón, M.A.; Monagas, M.; Bartolomé, B.; Moreno-Arribas, M.V. Wine features related to safety and consumer’health: An integrated perspective. Crit. Rev. Food Sci. Nutr. 2012, 52, 31–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arranz, S.; Chiva-Blanch, G.; Valderas-Martínez, P.; Medina-Remón, A.; Lamuela-Raventós, R.M.; Estruch, R. Wine, Beer, Alcohol and Polyphenols on Cardiovascular Disease and Cancer. Nutrients 2012, 4, 759–781. [Google Scholar] [CrossRef] [PubMed]
- Queipo-Ortuño, M.I.; Boto-Ordóñez, M.; Murri, M.; Gómez-Zumaquero, J.M.; Clemente-Postigo, M.; Estruch, R.; Cardona Díaz, F.; Andrés-Lacueva, C.; Tinahones, F.J. Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. Am. J. Clin. Nutr. 2012, 95, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Aura, A. Microbial metabolism of dietary phenolic compounds in the colon. Phytochem. Rev. 2008, 7, 407–429. [Google Scholar] [CrossRef]
- Selma, M.V.; Espín, J.C.; Tomás-Barberán, F.A. Interaction between phenolics and gut microbiota: Role in human health. J. Agric. Food Chem. 2009, 57, 6485–6501. [Google Scholar] [CrossRef] [PubMed]
- Monagas, M.; Urpi-Sarda, M.; Sánchez-Patán, F.; Llorach, R.; Garrido, I.; Gómez-Cordovés, C.; Andrés-Lacueva, C.; Bartolome, B. Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food Funct. 2010, 1, 233–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotches-Ribalta, M.; Urpi-Sarda, M.; Llorach, R.; Boto-Ordonez, M.; Jauregui, O.; Chiva-Blanch, G.; Perez-Garcia, L.; Jaeger, W.; Guillen, M.; Corella, D.; et al. Gut and microbial resveratrol metabolite profiling after moderate long-term consumption of red wine versus dealcoholized red wine in humans by an optimized ultra-high-pressure liquid chromatography tandem mass spectrometry method. J. Chomatogr. A 2012, 120, 105–113. [Google Scholar] [CrossRef]
- Jacobs, D.M.; Fuhrmann, J.C.; van Dorsten, F.A.; Rein, D.; Peters, S.; van Velzen, E.J.J.; Hollebrands, B.; Draijer, R.; van Duynhoven, J. Impact of short-term intake of red wine and grape polyphenol extract on the human metabolome. J. Agric. Food Chem. 2012, 60, 3078–3085. [Google Scholar] [CrossRef] [PubMed]
- Van Dorsten, F.A.; Peters, S.; Gross, G.; Gómez-Roldán, V.; Klinkenberg, M.; de Vos, R.C.; Vaughan, E.E.; van Duynhoven, J.; Possemiers, S.; van de Wiele, T.; et al. Gut microbial metabolism of polyphenols from black tea and red wine/grape juice is source-specific and colon-region dependent. J. Agric. Food Chem. 2012, 60, 11331–11342. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Patán, F.; Cueva, C.; Monagas, M.; Walton, G.E.; Gibson, G.R.; Quintanilla-López, J.E.; Lebrón-Aguilar, R.; Martín-Álvarez, P.J.; Moreno-Arribas, M.V.; Bartolomé, B. In vitro fermentation of a red wine extract by human gut microbiota: Changes in microbial groups and formation of phenolic metabolites. J. Agric. Food Chem. 2012, 60, 2136–2147. [Google Scholar] [CrossRef] [PubMed]
- Aura, A.M.; Mattila, I.; Hyotylainen, T.; Gopalacharyulu, P.; Cheynier, V.; Souquet, J.M.; Bes, M.; le Bourvellec, C.; Guyot, S.; Oresic, M. Characterization of microbial metabolism of Syrah grape products in an in vitro colon model using targeted and non-targeted analytical approaches. Eur. J. Nutr. 2013, 52, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Stalmach, A.; Edwards, C.A.; Wightman, J.D.; Crozier, A. Colonic catabolism of dietary phenolic and polyphenolic compounds from Concord grape juice. Food Funct. 2013, 4, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Girón, A.; Queipo-Ortuño, M.I.; Boto-Ordóñez, M.; Muñoz-González, I.; Sánchez-Patán, F.; Monagas, M.; Martín-Álvarez, P.J.; Murri, M.; Tinahones, F.J.; Andrés-Lacueva, C.; et al. Comparative study of microbial-derived phenolic metabolites in human feces after intake of gin, red wine, and dealcoholized red wine. J. Agric. Food Chem. 2013, 61, 3909–3915. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-González, I.; Jiménez-Girón, A.; Martín-Álvarez, P.J.; Bartolomé, B.; Moreno-Arribas, M.V. Profiling of Microbial-Derived Phenolic Metabolites in Human Feces after Moderate Red Wine Intake. J. Agric. Food Chem. 2013, 61, 9470–9479. [Google Scholar] [CrossRef] [PubMed]
- García-Cañas, V.; Simó, C.; León, C.; Cifuentes, A. Advances in Nutrigenomics research: Novel and future analytical approaches to investigate the biological activity of natural compounds and food functions. J. Pharmaceut. Biomed. 2010, 51, 290–304. [Google Scholar] [CrossRef] [Green Version]
- Llorach, R.; Garcia-Aloy, M.; Tulipani, S.; Vazquez-Fresno, R.; Andres-Lacueva, C. Nutrimetabolomic strategies to develop new biomarkers of intake and health effects. J. Agric. Food Chem. 2012, 60, 8797–8808. [Google Scholar] [CrossRef]
- Metabolomics Society. Metabolomics. Available online: http://metabolomicssociety.org/index.php/metabolomics (accessed on 24 July 2014).
- Pujos-Guillot, E.; Hubert, J.; Martin, J.F.; Lyan, B.; Quintana, M.; Claude, S.; Chabanas, B.; Rothwell, J.A.; Bennetau-Pelissero, C.; Scalbert, A.; et al. Mass spectrometry-based metabolomics for the discovery of biomarkers of fruit and vegetable intake: Citrus fruit as a case study. J. Proteome Res. 2013, 12, 1645–1659. [Google Scholar] [CrossRef] [PubMed]
- May, D.H.; Navarro, S.L.; Ruczinski, I.; Hogan, J.; Ogata, Y.; Schwarz, Y.; Levy, L.; Holzman, T.; McIntosh, M.W.; Lampe, J.W. Metabolomic profiling of urine: Response to a randomised, controlled feeding study of select fruits and vegetables, and application to an observational study. Br. J. Nutr. 2013, 110, 1760–1770. [Google Scholar] [CrossRef] [PubMed]
- Llorach, R.; Urpi-Sarda, M.; Tulipani, S.; Garcia-Aloy, M.; Monagas, M.; Andres-Lacueva, C. Metabolomic fingerprint in patients at high risk of cardiovascular disease by cocoa intervention. Mol. Nutr. Food Res. 2013, 57, 962–973. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, M.; Engelsen, S.B.; Dragsted, L.O. LC-MS metabolomics top-down approach reveals new exposure and effect biomarkers of apple and apple-pectin intake. Metabolomics 2012, 8, 64–73. [Google Scholar] [CrossRef]
- Tulipani, S.; Llorach, R.; Jáuregui, O.; López-Uriarte, P.; Garcia-Aloy, M.; Bullo, M.; Salas-Salvadó, J.; Andrés-Lacueva, C. Metabolomics unveils urinary changes in subjects with metabolic syndrome following 12-week nut consumption. J. Proteome Res. 2011, 10, 5047–5058. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Fresno, R.; Llorach, R.; Alcaro, F.; Rodríguez, M.A.; Vinaixa, M.; Chiva-Blanch, G.; Estruch, R.; Correig, X.; Andrés-Lacueva, C. H-NMR-based metabolomic analysis of the effect of moderate wine consumption on subjects with cardiovascular risk factors. Electrophoresis 2012, 33, 2345–2354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xuan, H; Marco, M.L.; Slupsky, C.M. Emerging Aspects of Food and Nutrition on Gut Microbiota. J. Agric. Food Chem. 2013, 61, 9559–9574. [Google Scholar] [PubMed]
- Phua, L.C.; Koh, P.K.; Cheah, P.Y.; Ho, H.K.; Chan, E.C.Y. Global gas chromatography/time-of-flight mass spectrometry (GC/TOFMS)-based metabonomic profiling of lyophilized human feces. J. Chromatogr. B 2013, 937, 103–113. [Google Scholar] [CrossRef]
- Jacobs, D.M.; Deltimple, N.; van Velzen, E.; van Dorsten, F.A.; Bingham, M.; Vaughan, E.E.; van Duynhoven, J. (1)H NMR metabolite profiling of feces as a tool to assess the impact of nutrition on the human microbiome. NMR Biomed. 2008, 21, 615–626. [Google Scholar] [CrossRef] [PubMed]
- </i>Jiménez-Girón, A.; Ibáñez, C.; Cifuentes, A.; Simó, C.; Muñoz-González, I.; Martín-Álvarez, P.J.; Bartolomé, B.; Moreno-Arribas, M.V. Faecal metabolomic fingerprint after moderate consumption of red wine by healthy subjects. J. Proteome Res. 2014. [Google Scholar] [CrossRef]
- Sánchez-Patán, F.; Chioua, M.; Garrido, I.; Cueva, C.; Samadi, A.; Marco-Contelles, J.; Moreno-Arribas, M.V.; Bartolomé, B.; Monagas, M. Synthesis, analytical features, and biological relevance of 5-(3′,4′-Dihydroxyphenyl)-gamma-valerolactone, a microbial metabolite derived from the catabolism of dietary flavan-3-ols. J. Agric. Food. Chem. 2011, 59, 7083–7091. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Patán, F.; Monagas, M.; Moreno-Arribas, M.V.; Bartolomé, B. Determination of microbial phenolic acids in human faeces by UPLC-ESI-TQ MS. J. Agric. Food Chem. 2011, 59, 2241–2247. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 2010, 11, 395–405. [Google Scholar]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, 480–484. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2009, 37, 603–610. [Google Scholar] [CrossRef]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A Metabolite Mass Spectral Database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Boto-Ordóñez, M.; Urpi-Sarda, M.; Queipo-Ortuño, M.I.; Corella, D.; Tinahones, F.J.; Estruch, R.; Andres-Lacueva, C. Microbial metabolomic fingerprinting in urine after regular dealcoholized red wine consumption in humans. J. Agric. Food Chem. 2013, 61, 9166–9175. [Google Scholar] [CrossRef] [PubMed]
- Caccetta, R.A.A.; Croft, K.D.; Beilin, L.J.; Puddey, I.B. Ingestion of red wine significantly increases plasma phenolic acid concentrations but does not acutely affect ex vivo lipoprotein oxidizability. Am. J. Clin. Nutr. 2000, 71, 67–74. [Google Scholar] [PubMed]
- Truchado, P.; Larrosa, M.; García-Conesa, M.T.; Cerdá, B.; Vidal-Guevara, M.L.; Tomás-Barberán, F.A.; Espín, J.C. Strawberry processing does not affect the production and urinary excretion of urolithins, ellagic acid metabolites, in humans. J. Agric. Food Chem. 2012, 60, 5749–5754. [Google Scholar] [CrossRef] [PubMed]
- Bolca, S.; Possemiers, S.; Verstraete, W.; van de Wiele, T.; Huybrechts, I.; de Henauw, S.; Depypere, H.; Bracke, M. Microbial and dietary factors associated with the equol and 8-prenylnaringenin producer phenotypes. Acta Hort. 2009, 848, 235–246. [Google Scholar]
- Van Velzen, E.J.J.; Westerhuis, J.A.; van Duynhoven, J.P.M.; van Dorsten, F.A.; Hoefsloot, H.C.J.; Jacobs, D.M.; Smit, S.; Draijer, R.; Kroner, C.I.; Smilde, A.K. Multilevel Data Analysis of a Crossover Designed Human Nutritional Intervention Study. J. Proteome Res. 2008, 7, 4483–4491. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Pujos-Guillot, E.; Sebedio, J.L. Development of a Quantitative Metabolomic Approach to Study Clinical Human Fecal Water Metabolome Based on Trimethylsilylation Derivatization and GC/MS Analysis. Anal. Chem. 2010, 82, 6447–6456. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Patán, F.; Cueva, C.; Monagas, M.; Walton, G.E.; Gibson, G.R.; Martín-Álvarez, P.J.; Moreno-Arribas, M.V.; Bartolomé, B. Gut microbial catabolism of grape seed flavan-3-ols by human faecal microbiota. Targetted analysis of precursor compounds, intermediate metabolites and end-products. Food Chem. 2012, 131, 337–347. [Google Scholar] [CrossRef] [Green Version]
- Cueva, C.; Moreno-Arribas, M.V.; Martin-Alvarez, P.J.; Bills, G.; Vicente, M.F.; Basilio, A.; Rivas, C.L.; Requena, T.; Rodriguez, J.M.; Bartolome, B. Antimicrobial activity of phenolic acids against commensal, probiotic and pathogenic bacteria. Res. Microbiol. 2010, 161, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, S.L.; da Silva, A.; Honório, K.M.; Marangoni, S.; Toyama, M.H.; da Silva, A.B.F. The influence of electronic, steric and hydrophobic properties of flavonoid compounds in the inhibition of the Xanthine Oxidase. J. Mol. Struc-Theochem. 2004, 684, 1–7. [Google Scholar] [CrossRef]
- Acquaviva, R.; Russo, A.; Russo, A.M.; Campisi, A.; Sorrenti, V.; di Giacomo, C.; Barcellona, M.L.; Avitabile, M.; Vanella, A. Antioxidant Activity and Protective Effect on DNA Cleavage of Resveratrol. J. Food Sci. 2002, 67, 137–141. [Google Scholar] [CrossRef]
- Li, Y.; Cao, Z.; Zhu, H. Upregulation of endogenous antioxidants and phase 2 enzymes by the red wine polyphenol, resveratrol in cultured aortic smooth muscle cells leads to cytoprotection against oxidative and electrophilic stress. Pharmacol. Res. 2006, 53, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Wendel, U.; Bakkeren, J.; Dejong, J.; Bongaerts, G. Glutaric aciduria mediated by gut bacteria. J. Inherit. Metab. Dis. 1995, 18, 358–359. [Google Scholar] [CrossRef] [PubMed]
- Becker, N.; Kunath, J.; Loh, G.; Blaut, M. Human intestinal microbiota: Characterization of a simplified and stable gnotobiotic rat model. Gut Microbes 2011, 2, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Tzounis, X.; Rodriguez-Mateos, A.; Vulevic, J.; Gibson, G.R.; Kwik-Uribe, C.; Spencer, J.P.E. Prebiotic evaluation of cocoa-derived flavanols in healthy humans by using a randomized, controlled, doubleblind, crossover intervention study. Am. J. Clin. Nutr. 2011, 93, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Molan, A.L.; Liu, Z.; Kruger, M. The ability of blackcurrant extracts to positively modulate key markers of gastrointestinal function in rats. World J. Microbiol. Biotechnol. 2011, 26, 1735–1743. [Google Scholar] [CrossRef]
- Viveros, A.; Chamorro, S.; Pizarro, M.; Arija, I.; Centeno, C.; Brenes, A. Effects of dietary polyphenol-rich grape products on intestinal microflora and gut morphology in broiler chicks. Poult. Sci. 2011, 90, 566–578. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.P.J.; Montoliu, I.; Nagy, K.; Moco, S.; Collino, S.; Guy, P.; Redeuil, K.; Scherer, M.; Rezzi, S.; Kochhar, S. Specific dietary preferences are linked to differing gut microbial metabolic activity in response to dark chocolate intake. J. Proteome Res. 2012, 11, 6252–6263. [Google Scholar] [CrossRef] [PubMed]
- Massot-Cladera, M.; Pérez-Berezo, T.; Franch, A.; Castell, M.; Pérez-Cano, F.J. Cocoa modulatory effect on rat faecal microbiota and colonic crosstalk. Arch. Biochem. Biophys. 2012, 527, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Cueva, C.; Sánchez-Patán, F.; Monagas, M.; Watson, G.; Gibson, G.R.; Martín-Álvarez, P.J.; Bartolomé, B.; Moreno-Arribas, M.V. In vitro fermentation of grape seed flavan-3-ol fractions by human faecal microbiota: Changes in microbial groups and phenolic metabolites. FEMS Microbiol. Ecol. 2013, 83, 792–805. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez-Girón, A.; Muñoz-González, I.; Martínlvarez, P.J.; Moreno-Arribas, M.V.; Bartolomé, B. Towards the Fecal Metabolome Derived from Moderate Red Wine Intake. Metabolites 2014, 4, 1101-1118. https://doi.org/10.3390/metabo4041101
Jiménez-Girón A, Muñoz-González I, Martínlvarez PJ, Moreno-Arribas MV, Bartolomé B. Towards the Fecal Metabolome Derived from Moderate Red Wine Intake. Metabolites. 2014; 4(4):1101-1118. https://doi.org/10.3390/metabo4041101
Chicago/Turabian StyleJiménez-Girón, Ana, Irene Muñoz-González, Pedro J. Martínlvarez, María Victoria Moreno-Arribas, and Begoña Bartolomé. 2014. "Towards the Fecal Metabolome Derived from Moderate Red Wine Intake" Metabolites 4, no. 4: 1101-1118. https://doi.org/10.3390/metabo4041101





