Metabolite Profiles in Various Plant Organs of Justicia gendarussa Burm.f. and Its in Vitro Cultures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. Preparation of Extracts and Quality Control (QC) Samples
2.3. Instrumentation
2.4. Data Processing
2.5. Analytical Method Validation
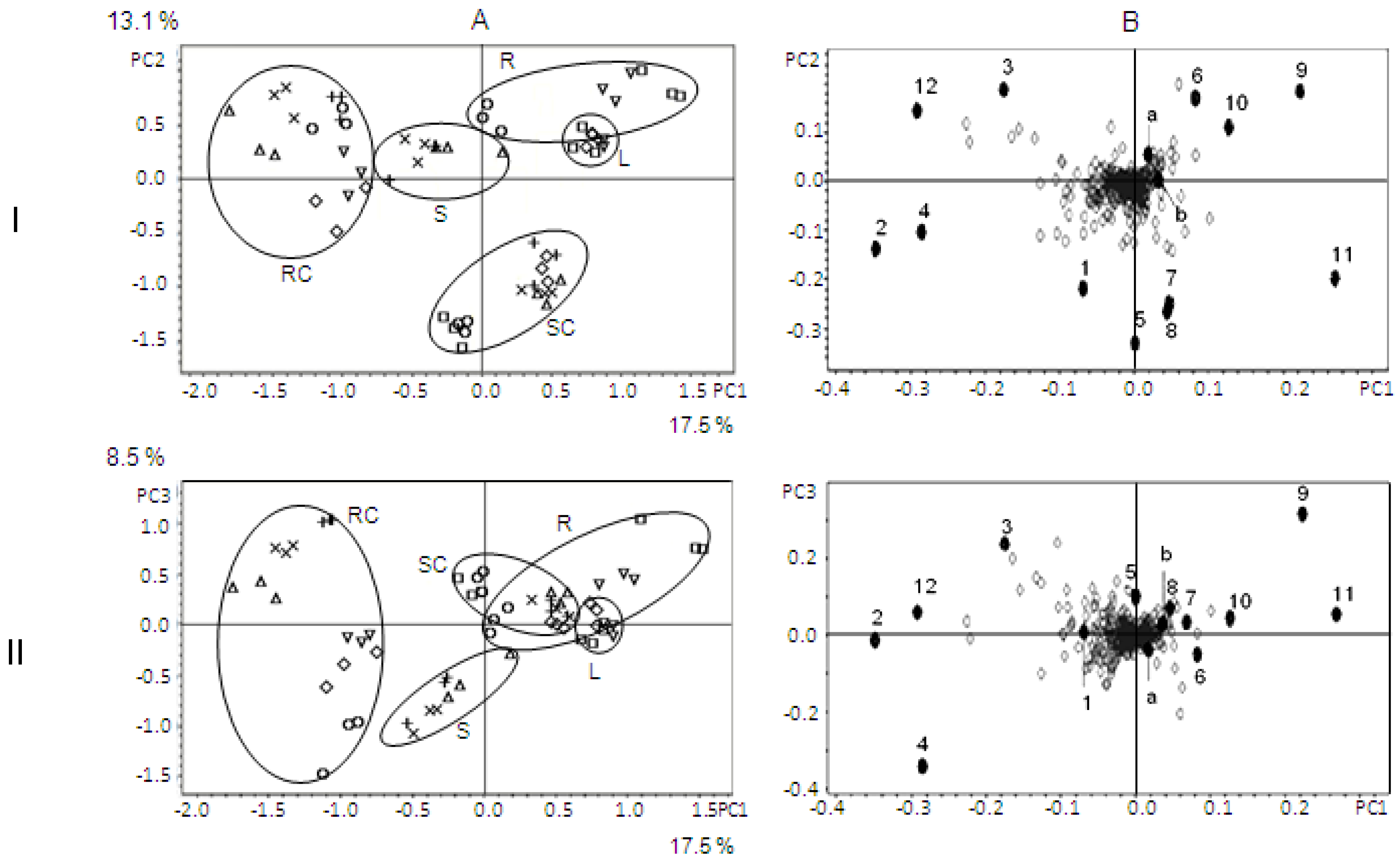
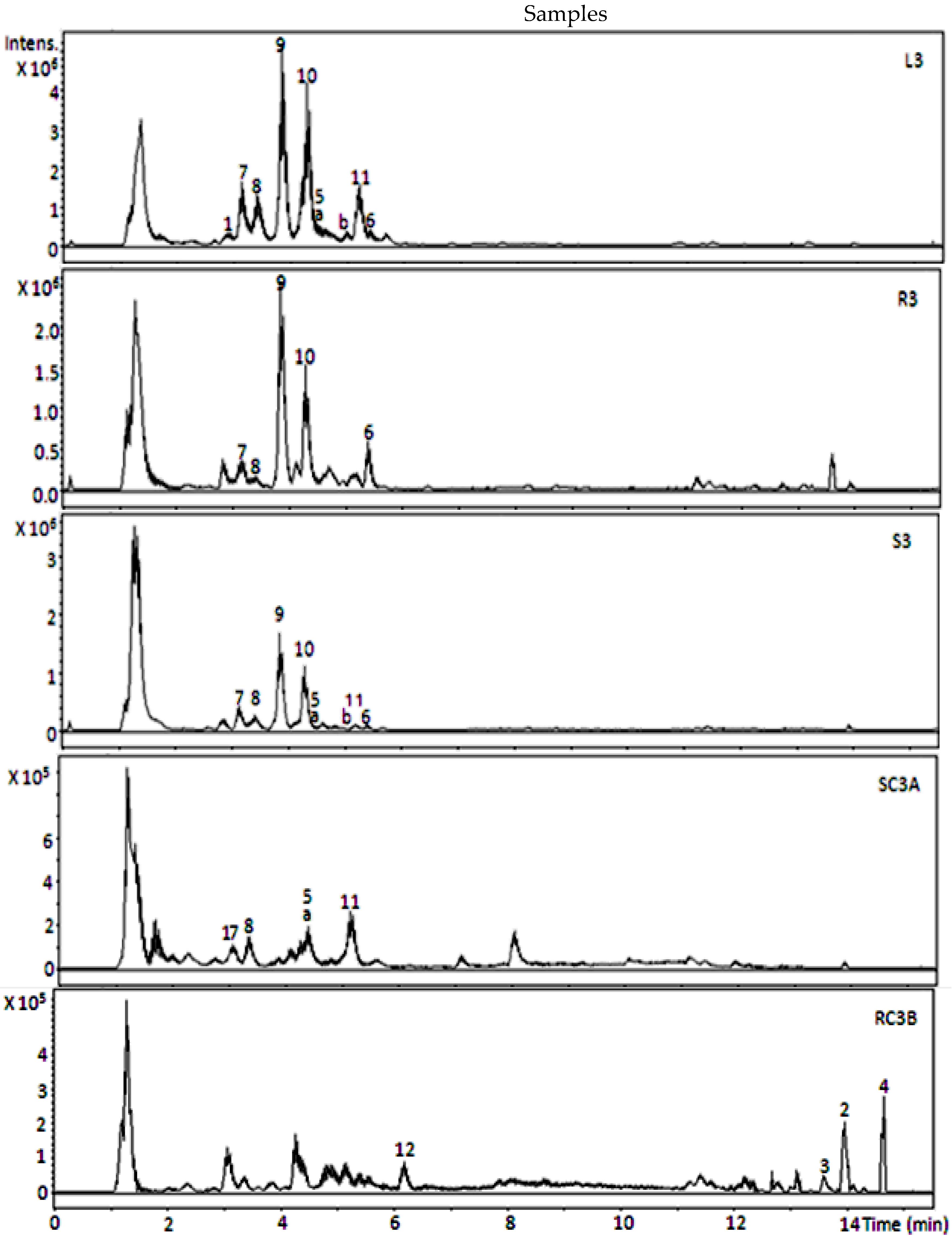
3. Results and Discussion
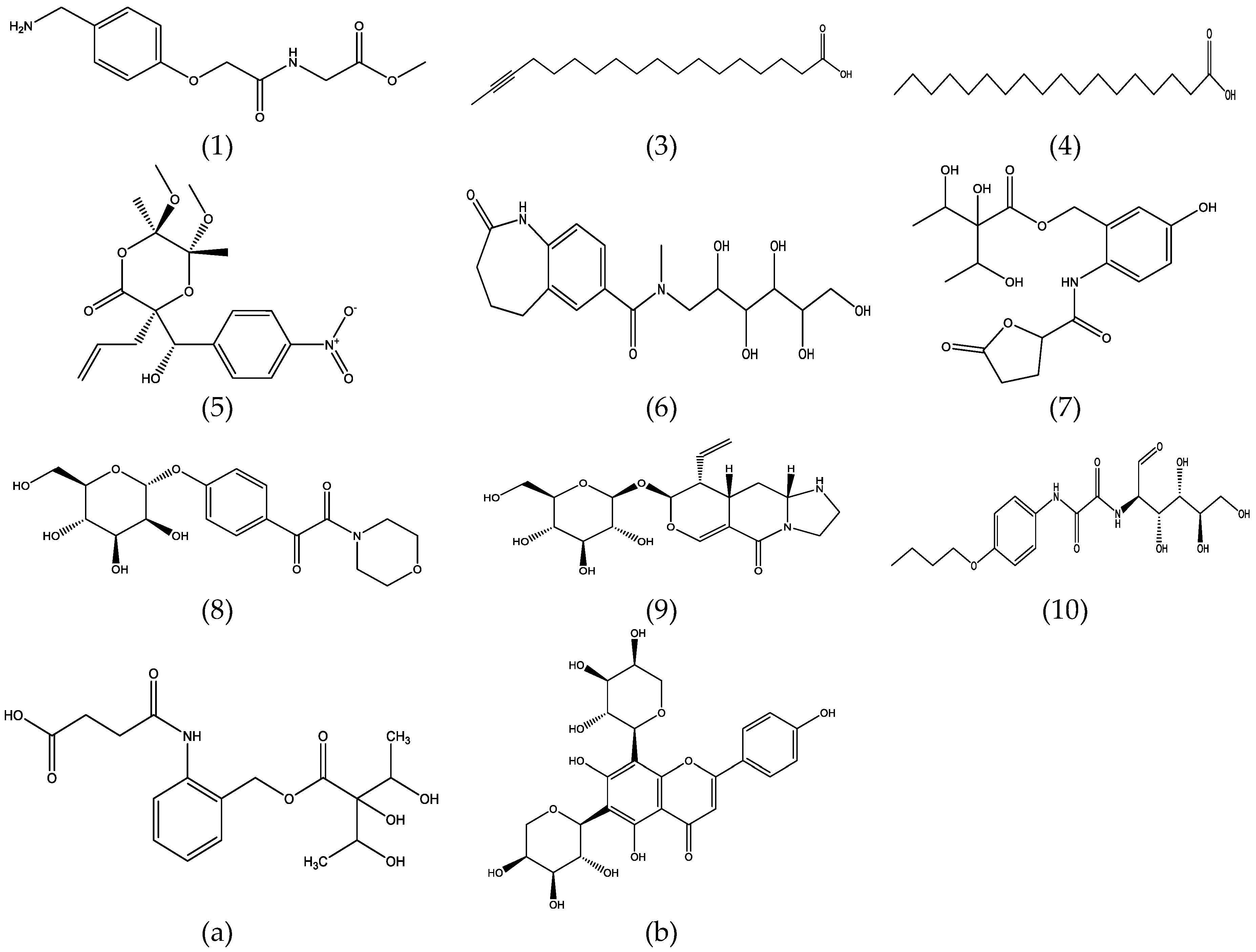
Identification of Metabolites
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ayob, Z.; Bohari, S.P.; Samad, A.A.; Jamil, S. Cytotoxic Activities against Breast Cancer Cells of Local Justicia gendarussa Crude Extracts. Evid-Based Complement. Altern. Med. 2014. [Google Scholar] [CrossRef] [PubMed]
- Arokiyaraj, S.; Perinbam, K.; Agastian, P.; Balaraju, K. Immunosuppresive Effect of Medicinal Plants of Kolli Hills on Mitogen-Stimulated Proliferation of The Human Peripheral Blood Mononuclear Cells in Vitro. Indian J. Pharmacol. 2007, 39, 180–183. [Google Scholar]
- Jothimanivannan, C.; Kumar, R.S.; Subramanian, N. Anti-Inflammatory and Analgesic Activities of Ethanol Extract of Aerial Parts of Justicia gendarussa Burm. Int. J. Biol. Chem. 2010, 6, 278–283. [Google Scholar]
- Krishna, K.L.; Mruthunjaya, K.; Patel, J.A. Antioxidant and Hepatoprotective Activity of Leaf Extract of Justicia gendarussa Burm. Int. J. Biol. Chem. 2009, 3, 99–110. [Google Scholar] [CrossRef]
- Krishna, K.L.; Mruthunjaya, K.; Patel, J.A. Antioxidant and Hepatoprotective Potential of Stem Methanolic Extract of Justicia gendarussa Burm. Int. J. Pharmacol. 2010, 6, 72–80. [Google Scholar] [CrossRef]
- Subramanian, N.; Jothimanivannan, C.; Kumar, R.S.; Kameshwaran, S. Evaluation of Anti-anxiety Activity of Justicia gendarussa Burm. Pharmacologia 2013, 4, 404–407. [Google Scholar]
- Sivasakthi, A.; Vijayalakshmi, M. An in Vitro Study of Antibactericidal Activity of Some Secondary Metabolites Rich Fraction from The Leaves of Justicia gendarussa. Int. J. Ethnomed. Pharmacol. Res. 2014, 2, 44–50. [Google Scholar]
- Nirmalraj, S.; Ravikumar, M.; Mahendrakumar, M.; Bharath, B.; Perinbam, K. Antibacterial and Anti-Inflammatory Activity of Justicia gendarussa Burm. F. Leaves. J. Plant Sci. 2015, 10, 70–74. [Google Scholar]
- Periyanayagam, K.; Umamaheswari, B.; Suseela, L.; Padmini, M.; Ismail, M. Evaluation of Antiangiogenic Effect of the Leaves of Justicia gendarussa (Burm. f) (Acanthaceae) by Chrio Allontoic Membrane Method. Am. J. Infect. Dis. 2009, 5, 180–182. [Google Scholar] [CrossRef]
- Sharma, K.K.; Saikia, R.; Kotoky, J.; Kalita, J.C.; Devi, R. Antifungal activity of Solanum melongena L., Lawsoniainermis L. and Justicia gendarussa B. against Dermatophytes. Int. J. PharmTech Res. 2011, 3, 1635–1640. [Google Scholar]
- Mpiana, P.T.; Bokota, M.T.; Ndjele, M.B.; Mudogo, V.; Tshibangu, D.S.; Ngbolua, K.N.; Atibu, E.K.; Kwembe, J.T.; Makelele, L.K. Antisickling Activity of Three Species of Justicia from Kisangani (D.R. Congo): J. tenella, J. gendarussa and J. insularis. Int. J. Biol. Chem. Sci. 2010, 4, 1953–1961. [Google Scholar] [CrossRef]
- Saha, M.R.; Debnath, P.C.; Rahman, A.; Islam, A.U. Evaluation of in vitro anthelmintic activities of leaf and stem extracts of Justicia gendarussa. Bangladesh J. Pharmacol. 2012, 7, 50–53. [Google Scholar] [CrossRef]
- Senthilkumar, N.; Varma, P.; Gurusubramanian, G. Larvicidal and Adulticidal Activities of Some Medicinal Plants against the Malarial Vector Anopheles stephensi (Liston). Parasitol. Res. 2009, 104, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Woradulayapinij, W.; Soonthornchareonnon, N.; Wiwat, C. In vitro HIV type 1 reverse transcriptase inhibitory activities of Thai medicinal plants and Canna indica L. rhizomes. J. Ethnopharmacol. 2005, 101, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.S.; Zaveri, M.N. Trypsin and Protein Denaturation Inhibitory Activity of Different Fractionation and Isolated Compound of Leaf and Root of Justicia Gendarussa. IJPSR 2014, 5, 5564–5571. [Google Scholar]
- Prajogo, B.E. Aktivitas Antifertilitas Flavonoid Daun Justicia gendarussa Burm.f.: Penelitian Eksperimental Pencegahan Penetrasi Spermatozoa Mencit dalam Proses Fertilisasi in Vitro (in Indonesian). Ph.D. Thesis, Airlangga University, Surabaya, Indonesia, 2002. [Google Scholar]
- Prajogo, B.E.; Widjiati, T.M. Pengaruh Fraksi Polifenol Gendarussa vulgaris Nees pada Penurunan Aktivitas Hialuronidase Spermatozoa Mencit melalui Uji Fertilitas in Vitro (in Indonesian). Laporan Penelitian Ilmu Pengetahuan Dasar, Lembaga Penelitian Universitas Airlangga, Surabaya, Indonesia, 2002. [Google Scholar]
- Prajogo, B.E.; Ifadotunnikmah, F.; Febriyanti, A.P.; Jusak, N. Efek Fase Air Daun Gandarusa (Justicia gendarussa Burm.f.) pada Fungsi Hati dan Fungsi Ginjal Kelinci Jantan (Uji Toksisitas Fase Air Daun Gandarusa sebagai Bahan Kontrasepsi Pria) (in Indonesian). Vet. Med. 2008, 1, 79. [Google Scholar]
- Prajogo, B.E.; Juliaan, F.; Hinting, A.; Pramesti, M.P.; Anggraeni, M.; Radjaram, A.; Musta’ina. Laporan Pelaksanaan Uji Klinik Fase I; (in Indonesian). Universitas Airlangga dan Badan Koordinasi Keluarga Berencana Nasional: Surabaya, Indonesia, 2008. [Google Scholar]
- Prajogo, B.E.; Juliaan, F.; Hinting, A.; Pramesti, M.P.; Anggraeni, M.; Radjaram, A.; Musta’ina. Laporan Pelaksanaan Uji Klinik Fase II; (in Indonesian). Universitas Airlangga dan Badan Koordinasi Keluarga Berencana Nasional: Surabaya, Indonesia, 2009. [Google Scholar]
- Prajogo, B.E.; Juliaan, F.; Hinting, A.; Pramesti, M.P.; Anggraeni, M.; Radjaram, A.; Musta’ina. Laporan Pelaksanaan Uji Klinik Fase III; (in Indonesian). Universitas Airlangga dan Badan Koordinasi Keluarga Berencana Nasional: Surabaya, Indonesia, 2011. [Google Scholar]
- Satapathy, A.K.; Gunasekaran, G.; Sahoo, S.C.; Amit, K.; Rodrigues, P.V. Corrosion Inhibition by Justicia gendarussa Plant Extract in Hydrochloric Acid Solution. Corros. Sci. 2009, 51, 2848–2856. [Google Scholar] [CrossRef]
- Shikha, P.; Latha, P.G.; Suja, S.R.; Anuja, G.I.; Shyamal, S.; Shine, V.J.; Sini, S.; Kumar, N.M.; Rajasekaran, S. Anti-inflammatory and antinociceptive activity of Justicia gendarussa Burm. f. Leaves. Indian J. Nat. Prod. Resour. 2010, 1, 456–461. [Google Scholar]
- Kiren, Y.; Deguchi, J.; Hirasawa, Y.; Morita, H.; Prajogo, B.E. Justidrusamides A-D, New 2-Aminobenzyl Alcohol Derivatives from Justicia gendarussa. J. Nat. Med. 2014, 68, 754–758. [Google Scholar] [CrossRef] [PubMed]
- Ningsih, I.Y.; Purwanti, D.I.; Wongso, S.; Prajogo, B.E.; Indrayanto, G. Metabolite Profiling of Justicia gendarussa Burm. f. Leaves Using UPLC-UHR-QTOF-MS. Sci. Pharm. 2015, 83, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Indrayanto, G.; Setiawan, B.; Cholies, N. Differential Diosgenin Accumulation in Costus speciosus and its Tissue Cultures. Planta Medica Lett. 1994, 60, 483–484. [Google Scholar] [CrossRef] [PubMed]
- Bhagya, N.; Chandrashekar, K.R. In Vitro Production of Bioactive Compounds from Stem and Leaf Explants of Justicia gendarussa Burm. f. Asian J. Pharm. Clin. Res. 2013, 6, 100–105. [Google Scholar]
- Ma, X.; Gang, D.R. Metabolic Profiling of Turmeric (Curcuma longa L.) Plants Derived from in Vitro Micropropagation and Conventional Greenhouse Cultivation. J. Agric. Food Chem. 2006, 54, 9573–9583. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.M.; Wang, R.; Li, X.D.; Jiang, Y.M.; Peng, F.; Xia, B. Alkaloid accumulation in different parts and ages of Lycoris chinensis. Z. Naturforschung C. 2010, 65, 458–462. [Google Scholar] [CrossRef]
- Butkienė, R.; Būdienė, J.; Judžentienė, A. Variation of Secondary Metabolites (Essential Oils) in Various Plant Organs of Juniperus communis L. Wild Growing in Lithuania. Balt. For. 2015, 21, 59–64. [Google Scholar]
- Want, E.J.; Wilson, I.D.; Gika, H.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Holmes, E.; Nicholson, J.K. Global Metabolic Profiling Procedures for Urine Using UPLC-MS. Nat. Protoc. 2010, 5, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- MetFrag. Available online: http://msbi.ipb-halle.de/MetFrag/ (accessed on 26 August–5 September 2015).
- Metlin. Available online: https://metlin.scripps.edu/index.php (accessed on 26 August–5 September 2015).
- MassBank. Available online: http://www.massbank.jp/index.html (accessed on 26 August–5 September 2015).
- Ponnamma, S.U.; Manjunath, K. GC-MS Analysis of Phytocomponents in The Methanolic Extract of Justicia wynaadensis (Nees) T. Anders. Int. J. Pharm. Bio. Sci. 2012, 3, 570–576. [Google Scholar]
- Lu, S.; Zhang, G. Alkaloids from Gendarussa vulgaris Nees. Nat. Prod. Res. 2008, 22, 1610–1613. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, A.K.; Dastidar, P.P.; Pakrashi, S.C. Simple Aromatic Amines from Justicia gendarussa: 13C NMR Spectra of The Bases and Their Analogues. Tetrahedron 1982, 18, 1797–1802. [Google Scholar] [CrossRef]
| Samples | Codes |
|---|---|
| Leaves | L1, L2, L3 |
| Roots | R1, R2, R3 |
| Stems | S1, S2, S3 |
| Shoot Cultures a | SC1A, SC2A, SC3A |
| Root Cultures b | RC1B, RC2B, RC3B |
| Root Cultures c | RC1C, RC2C, RC3C |
| Metabolites | RT (min) | Ions | Measured m/z | Score (Err[mDa]; mSigma) Ions a | Probable Elemental Formulas a,b |
|---|---|---|---|---|---|
| HRMS Ions | |||||
| (m/z calc.) a | |||||
| 1 | 3.09 | [M − H]− | 251.1039 | 77 | C12H16N2O4 |
| (251.1037) | (0.1;2.8) | ||||
| 2 | 13.94 | [M − H]− | 255.2326 | 88 | C16H32O2 |
| (255.2330) | (−0.3;1.7) | ||||
| 3 | 13.64 | [M − H]− | 279.2339 | 63 | C18H32O2 |
| (279.2330) | (1.0;12.9) | ||||
| 4 | 14.66 | [M − H]− | 283.2638 | 85 | C18H36O2 |
| (283.2643) | (−0.4;0.7) | ||||
| 5 | 4.43 | [M − H]− | 380.1339 | 75 | C18H23NO8 |
| (380.1351) | (−1.2;8.7) | ||||
| 6 | 5.42 | [M − H]− | 381.1668 | 100 | C18H26N2O7 |
| (381.1667) | (−0.1;5.6) | ||||
| [2M − H]− | 763.3405 | 96 | C36H52N4O14 | ||
| (763.3407) | (−0.3;31.8) | ||||
| 7 | 3.09 | [M − H]− | 396.1291 | 84 | C18H23NO9 |
| (396.1300) | (−0.9;7.7) | ||||
| 8 | 3.36 | [M − H]− | 396.1299 | 100 | C18H23NO9 |
| (396.1300) | (0.1;6.4) | ||||
| 9 | 3.84 | [M − H]− | 397.1611 | 89 | C18H26N2O8 |
| (397.1616) | (−0.5;2.4) | ||||
| 10 | 4.29 | [M − H]− | 397.1614 | 100 | C18H26N2O8 |
| (397.1616) | (0.3;1.9) | ||||
| [2M − H]− | 795.3290 | 48 | C36H52N4O16 | ||
| (795.3306) | (1.5;16.3) | ||||
| 11 | 5.18 | [M − H]− | 533.1311 | 100 | C26H22N4O9 |
| (533.1314) | (−0.3;6.3) | ||||
| 12 | 6.17 | [M − H]− | 651.2301 | 100 | C31H40O15 |
| (651.2294) | (0.7;10.0) | ||||
| a | 4.26 | [M − H]− | 368.1357 | 69 | C17H23NO8 |
| (368.1351) | (0.6;5.9) | ||||
| [2M − H]− | 737.2779 | 100 | C34H46N2O16 | ||
| (737.2775) | (0.4;2.1) | ||||
| b | 5.01 | [M − H]− | 533.1313 | 67 | C25H26O13 |
| (533.1301) | (−1.3;5.0) |
| Metabolites | Score (Chem Spider a/Pub Chem b) | Explained c and MS/MS Fragment Ions d | Measured m/z HRMS Fragment Ions | Score(Err[mDa]; mSigma) Fragment Ions d | Proposed Metabolites | Metabolite IDs and References |
|---|---|---|---|---|---|---|
| (m/z calc.) d | ||||||
| 1 | 0.991/1.0 | [C11H12NO4]− | 222.0778 | 55 | Methyl N-{[4-(aminomethyl)phenoxy]acetyl} glycinate c | ID 11862597 a; CID 16777361 b; |
| (222.0772) | (0.6;21.8) | |||||
| [C5H8NO4]− | 146.0463 | 75 | ||||
| (146.0459) | (0.4;10.8) | |||||
| [C5H6NO3]− | 128.0355 | 100 | ||||
| (128.0353) | (0.1;1.8) | |||||
| [C7H8NO]− | 122.0612 | 66 | ||||
| (122.0611) | (0.1;4.3) | |||||
| [C7H6NO]− | 120.0451 | 59 | ||||
| (120.0455) | (−0.4;3.8) | |||||
| [C3H5O2]− | 73.0294 | 63 | ||||
| (73.0295) | (−0.1;6.4) | |||||
| 2 | -/- | [C11H15O]− | 163.1125 | 43 | Unknown g | |
| (163.1128) | (0.3;21.8) | |||||
| 3 | 1.0/1.0 | [C15H29O2]− | 241.2177 | 45 | 16-Octadecynoic acid c,e | ID 4472124 a; CID 5312699 b; ID 74231 e |
| (241.2173) | (−0.4;11.1) | |||||
| [C14H27O2]− | 227.2024 | 24 | ||||
| (227.2017) | (0.7;33.5) | |||||
| 4 | 0.911/0.911 | [C16H31O2]− | 255.2326 | 42 | Stearic acid c,e | ID 5091 a; CID 164708 b; ID 189 e; C01530 f |
| (255.2330) | (0.4;17.2) | |||||
| 5 | 1.0/0.835 | [C12H10NO4]− | 232.0611 | 69 | (3S,5R,6R)-3-Allyl-3-[(S)-hydroxy(4-nitrophenyl) methyl]-5,6-dimethoxy-5,6-dimethyl-1,4-dioxan-2-one c | ID 9328344 a; CID 11153236 b |
| (232.0615) | (0.4;13.5) | |||||
| [C11H10NO3]− | 204.0654 | 26 | ||||
| (204.0666) | (1.2;36.7) | |||||
| [C6H11O4]− | 147.0658 | 51 | ||||
| (147.0663) | (−0.5;6.0) | |||||
| 6 | 0.968/- | [C6H11O5]− | 163.0609 | 64 | 1-Deoxy-1-{methyl[(2-oxo-2,3,4,5-tetrahydro-1H-1-benzazepin-7-yl) carbonyl] amino}hexitol c | ID 34743441 a |
| (163.0612) | (0.3;6.2) | |||||
| [C6H11O4]− | 147.0663 | 75 | ||||
| (147.0663) | (0.0;4.9) | |||||
| 7 | 1.0/- | [C6H11O5]− | 163.0610 | 89 | 1,5-Dideoxy-3-C-{[(5-hydroxy-2-{[(5-oxotetra hydro-2-furanyl) carbonyl] amino}benzyl) oxy]carbonyl} pentitol c | ID 29814435 a [25] |
| (163.0612) | (0.1;1.5) | |||||
| 8 | 0.802/- | [C6H11O5]− | 163.0611 | 85 | 4-[4-Morpholinyl(oxo)acetyl] phenyl α-d-manno pyranoside c | ID 32768629 a [25] |
| (163.0612) | (−0.1;2.5) | |||||
| [C4H5O3]− | 101.0241 | 50 | ||||
| (101.0244) | (0.3;5.0) | |||||
| 9 | 0.841/- | [C6H11O5]− | 163.0612 | 84 | (8S,9R,9aS, 10aR)-5-Oxo-9-vinyl-1,2,3,8,9,9a,10,10a-octahydro-5H-imidazo[1,2-a] pyrano [4,3-d] pyridin-8-yl β-d-glucopyranoside c | ID 26570736 a |
| (163.0612) | (−0.0;5.2) | |||||
| [C5H9O5]− | 149.0452 | 77 | ||||
| (149.0455) | (0.3;3.5) | |||||
| [C4H7O4]− | 119.0349 | 35 | ||||
| (119.0350) | (0.1;26.5) | |||||
| [C4H5O3]− | 101.0246 | 84 | ||||
| (101.0244) | (0.2;1.4) | |||||
| 10 | 0.885/0.97 | [C6H11O5]− | 163.0612 | 86 | 2-({[(4-Butoxyphenyl)amino](oxo)acetyl}amino)-2-deoxy-d-glucose c | ID 21249273 a; CID 24838413 b |
| (163.0612) | (−0.0;3.0) | |||||
| [C4H7O4]− | 119.0350 | 86 | ||||
| (119.0350) | (−0.1;2.9) | |||||
| [C4H5O3]− | 101.0244 | 52 | ||||
| (101.0244) | (0.1;6.4) | |||||
| 11 | -/- | [C6H11O4]− | 147.0663 | 96 | Unknown g | |
| (147.0663) | (0.0;1.7) | |||||
| 12 | -/- | [C27H32O7]− | 468.2158 | 66 | Unknown g | |
| (468.2154) | (0.4;20.1) | |||||
| a | 1.0/1.0 | [C6H11O5]− | 163,0618 | 64 | 3-C-[({2-[(3-Carboxypropanoyl)amino]benzyl}oxy)carbonyl]-1,5-dideoxy-L-arabinitol(Justidrusamide A/B) c | ID 22943323 a; CID 38352741b; [24,25] |
| (163,0612) | (0.6;5.9) | |||||
| [C4H5O3]− | 101,0246 | 81 | ||||
| (101,0244) | (−0.2;2.5) | |||||
| b | 0.969/1.0 | [C9H5O2]− | 145,0301 | 52 | 6,8-Di-C-alpha-l-arabino pyranosylapigenin c,e | ID 26504074 a; CID 10918510 b; ID 48669 e; [16,25] |
| (145,0295) | (0.6;24.1) | |||||
| [C4H5O3]− | 101.0249 | 50 | ||||
| (101.0244) | (−0.5;8.8) | |||||
| 89.0249 | 52 | |||||
| (89.0244) | (−0.5;6.7) |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Indrayoni, P.; Purwanti, D.I.; Wongso, S.; Prajogo, B.E.W.; Indrayanto, G. Metabolite Profiles in Various Plant Organs of Justicia gendarussa Burm.f. and Its in Vitro Cultures. Sci. Pharm. 2016, 84, 555-566. https://doi.org/10.3390/scipharm84030555
Indrayoni P, Purwanti DI, Wongso S, Prajogo BEW, Indrayanto G. Metabolite Profiles in Various Plant Organs of Justicia gendarussa Burm.f. and Its in Vitro Cultures. Scientia Pharmaceutica. 2016; 84(3):555-566. https://doi.org/10.3390/scipharm84030555
Chicago/Turabian StyleIndrayoni, Putu, Diah Intan Purwanti, Suwidji Wongso, Bambang E.W. Prajogo, and Gunawan Indrayanto. 2016. "Metabolite Profiles in Various Plant Organs of Justicia gendarussa Burm.f. and Its in Vitro Cultures" Scientia Pharmaceutica 84, no. 3: 555-566. https://doi.org/10.3390/scipharm84030555





