STAT5-Interacting Proteins: A Synopsis of Proteins that Regulate STAT5 Activity
Abstract
:1. Introduction
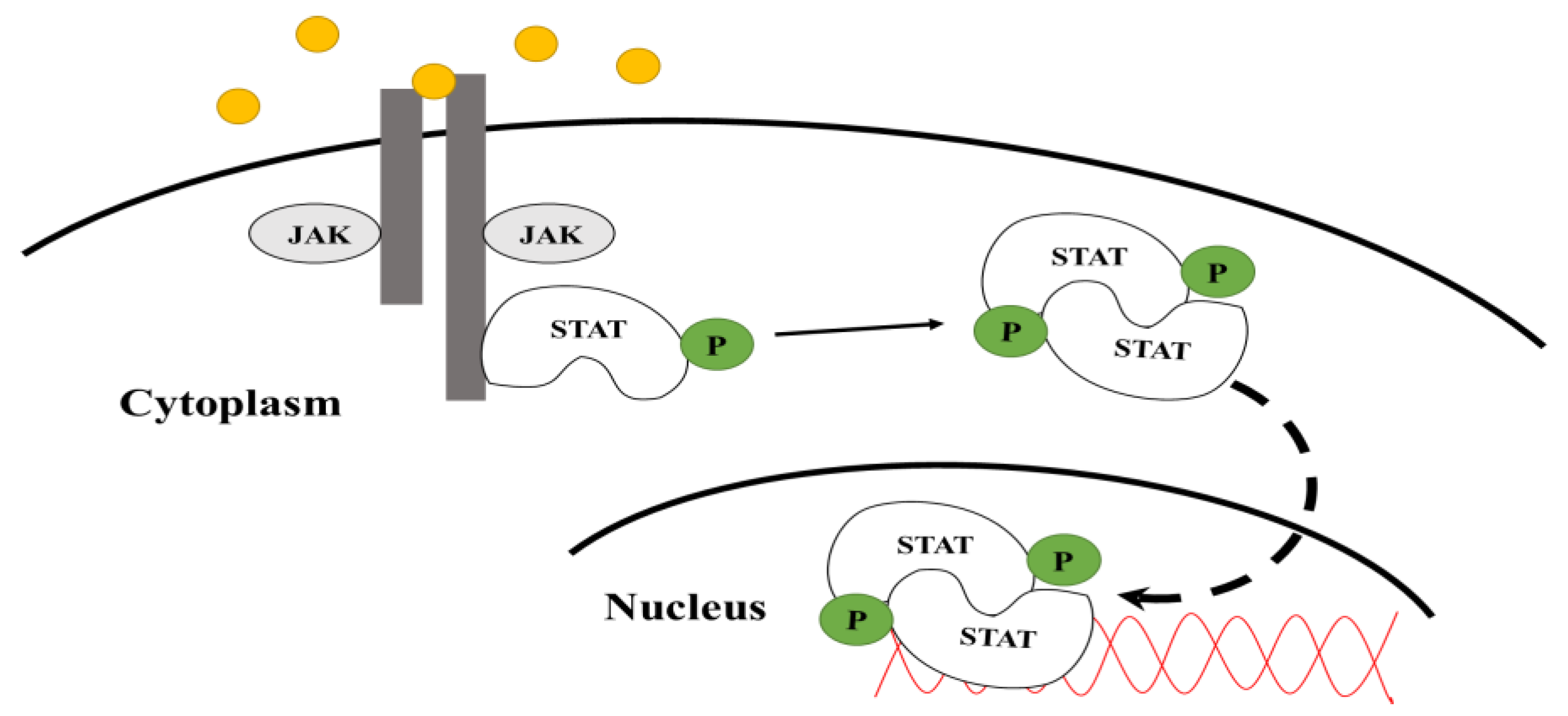
1.1. JAK-STAT Pathway
1.2. Signal Transducers and Activators of Transcription (STATs)
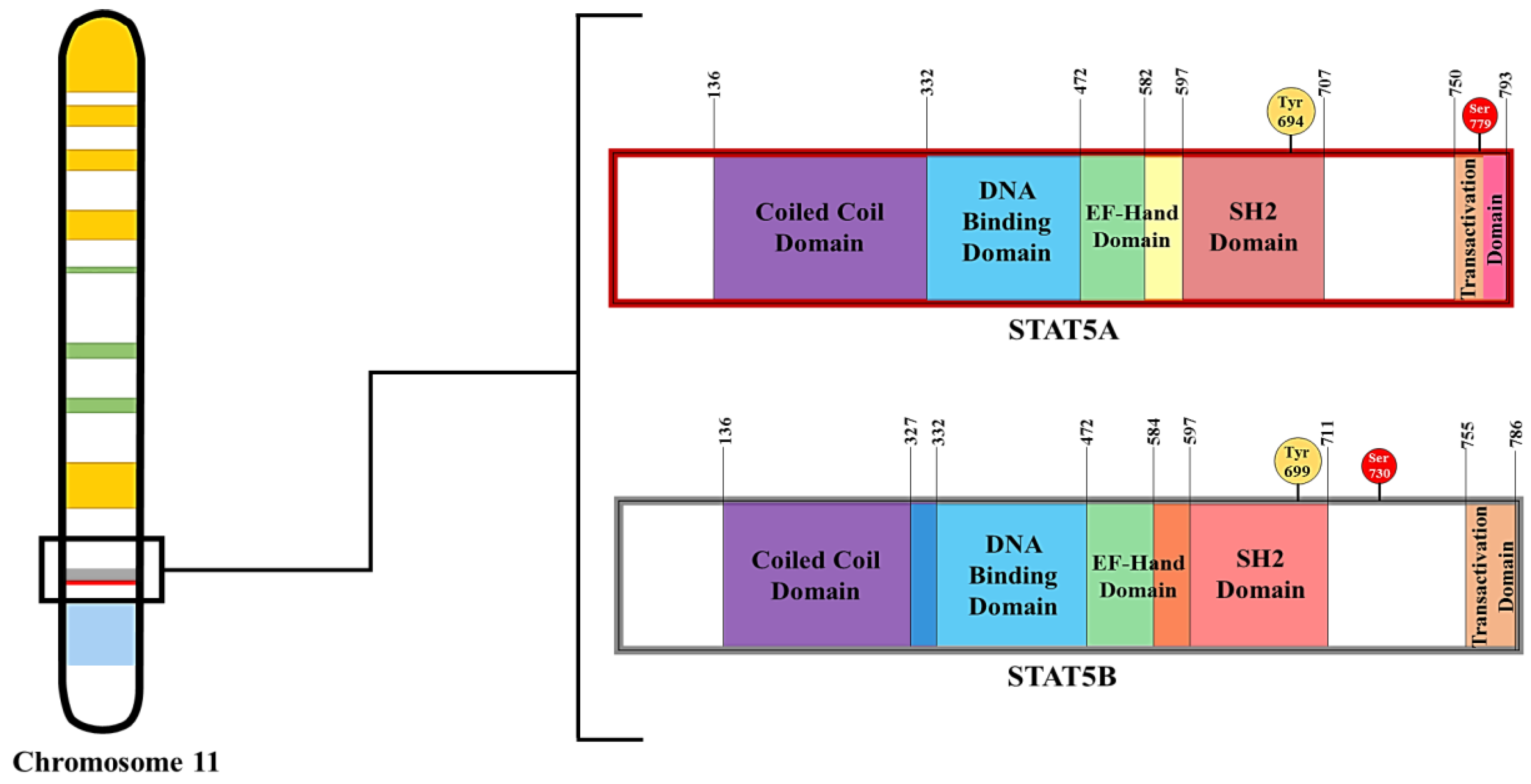
1.3. STAT5 Characteristics and Functions
1.4. Tools for Understanding the Function of STAT5 Proteins
2. STAT5 Interacting Proteins
2.1. Interactions with Cytokine/Hormone Receptors
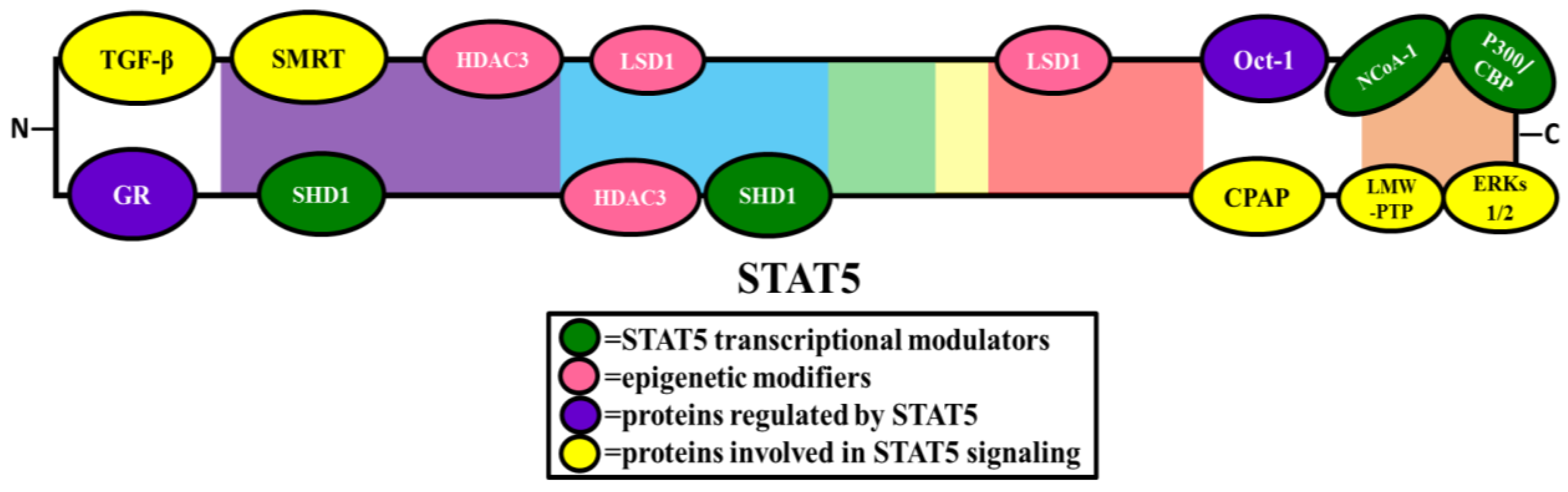
2.2. Transcriptional Modulators
2.2.1. Proteins That Enhance STAT5 Transcriptional Activity
2.2.2. Proteins that Repress STAT5 Transcriptional Activity
2.2.3. Role of STAT5 in Oct-1 Transcriptional Activity
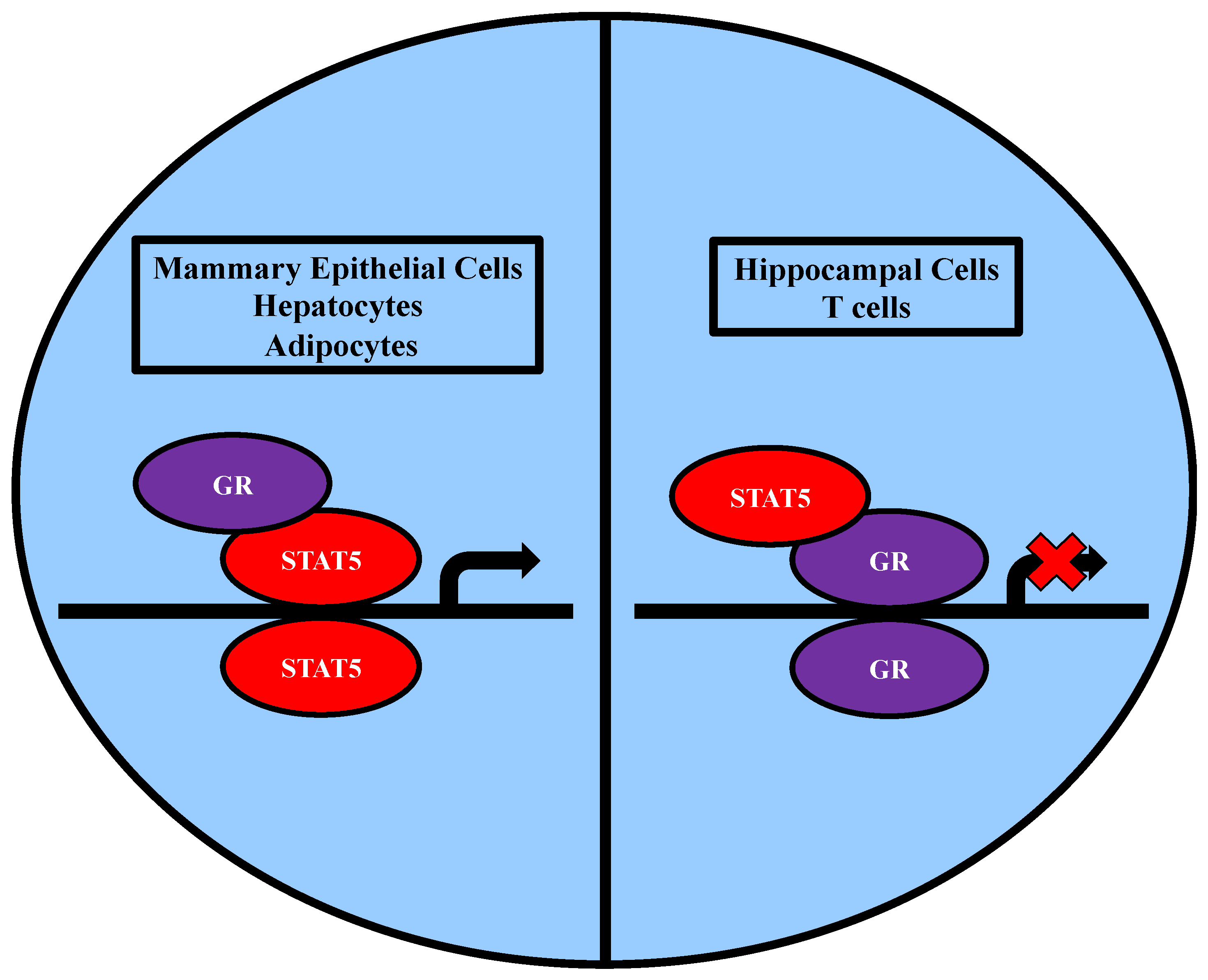
2.3. Association of STAT5 and Members of the Nuclear Receptor Family
2.3.1. Association with PR
2.3.2. Association with GR
2.4. Epigenetic Modifiers
2.5. Proteins Involved in STAT5 Signaling
2.5.1. Proteins That Modulate STAT5 Phosphorylation
2.5.2. Proteins That Modulate STAT5 Dephosphorylation
2.5.3. Proteins That Enhance STAT5 Signaling
2.5.4. Proteins That Repress STAT5 Signaling
3. Conclusions
Author Contributions
Conflicts of Interest
References
- Mullen, M.; Gonzalez-Perez, R. Leptin-Induced JAK/STAT Signaling and Cancer Growth. Vaccines 2016. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, K.; Saharinen, P.; Pesu, M.; Holt, V.E.T.; Silvennoinen, O.; O’Shea, J.J. The Janus kinases (Jaks). Genome Biol. 2004, 5, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, D.A.; McCoon, P.E.; Binari, R.; Gilman, M.; Perrimon, N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998, 12, 3252–3263. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef] [PubMed]
- Greenhalgh, C.J.; Hilton, D.J. Negative regulation of cytokine signaling. J. Leukoc. Biol. 2001, 70, 348–356. [Google Scholar] [PubMed]
- Szelag, M.; Piaszyk-Borychowska, A.; Plens-Galaska, M.; Wesoly, J.; Bluyssen, H.A.R. Targeted inhibition of STATs and IRFs as a potential treatment strategy in cardiovascular disease Atherosclerosis And inflAmmAtion. Oncotarget 2016, 7, 48788–48812. [Google Scholar] [PubMed]
- Richard, A.J.; Stephens, J.M. The role of JAK-STAT signaling in adipose tissue function. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Hogan, J.C.; Stephens, J.M. The Regulation of Fatty Acid Synthase by STAT5A. Diabetes 2005, 54, 1968–1975. [Google Scholar] [CrossRef] [PubMed]
- Floyd, Z.E.; Stephens, J.M. STAT5A promotes adipogenesis in nonprecursor cells and associates with the glucocorticoid receptor during adipocyte differentiation. Diabetes 2003, 52, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Seong, S.; Kim, J.H.; Kim, K.; Kim, I.; Jeong, B.; Nam, K.-I.; Kim, K.K.; Hennighausen, L.; Kim, N.; et al. STAT5 is a key transcription factor for IL-3-mediated inhibition of RANKL-induced osteoclastogenesis. Sci. Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Kaltenecker, D.; Mueller, K.M.; Benedikt, P.; Feiler, U.; Themanns, M.; Schlederer, M.; Kenner, L.; Schweiger, M.; Haemmerle, G.; Moriggl, R. Adipocyte STAT5 deficiency promotes adiposity and impairs lipid mobilisation in mice. Diabetologia 2017, 60, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Heltemes-Harris, L.M.; Farrar, M.A. The role of STAT5 in lymphocyte development and transformation. Curr. Opin. Immunol. 2012, 24, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Hennighausen, L.; Robinson, G.W. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 2008, 22, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Feldman, G.M.; Rosenthal, L.A.; Liu, X.; Hayes, M.P.; Wynshaw-Boris, A.; Leonard, W.J.; Hennighausen, L.; Finbloom, D.S. STAT5A-Deficient Mice Demonstrate a Defect in Granulocyte-Macrophage Colony-Stimulating Factor–Induced Proliferation and Gene Expression. Blood 1997, 90, 1768–1776. [Google Scholar] [PubMed]
- Mui, A.L.; Wakao, H.; Kinoshita, T.; Kitamura, T.; Miyajima, A. Suppression of interleukin-3-induced gene expression by a C-terminal truncated Stat5: Role of Stat5 in proliferation. EMBO J. 1996, 15, 2425–2433. [Google Scholar] [PubMed]
- Bachmann, J.; Raue, A.; Schilling, M.; Bohm, M.E.; Kreutz, C.; Kaschek, D.; Busch, H.; Gretz, N.; Lehmann, W.D.; Timmer, J.; et al. Division of labor by dual feedback regulators controls JAK2/STAT5 signaling over broad ligand range. Mol. Syst. Biol. 2011, 7, 516. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, M.; Ginter, T.; Brand, P.; Heinzel, T.; Krämer, O.H. Acetylation modulates the STAT signaling code. Cytokine Growth Factor Rev. 2012, 23, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Oshida, K.; Vasani, N.; Waxman, D.J.; Corton, J.C. Disruption of STAT5b-regulated sexual dimorphism of the liver transcriptome by diverse factors is a common event. PLoS ONE 2016, 11, e0148308. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Gallego, M.I.; Smith, G.H.; Robinson, G.W.; Hennighausen, L. Functional rescue of Stat5a-null mammary tissue through the activation of compensating signals including Stat5b. Cell Growth Differ. 1998, 9, 795–803. [Google Scholar] [PubMed]
- Wan, C.K.; Oh, J.; Li, P.; West, E.E.; Wong, E.A.; Andraski, A.B.; Spolski, R.; Yu, Z.X.; He, J.; Kelsall, B.L.; et al. The Cytokines IL-21 and GM-CSF Have Opposing Regulatory Roles in the Apoptosis of Conventional Dendritic Cells. Immunity 2013, 38, 514–527. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, D.; Na, R.; Feuermann, Y.; Pechhold, S.; Chen, W.; Robinson, G.W.; Hennighausen, L. Development of mammary luminal progenitor cells is controlled by the transcription factor STAT5A. Genes Dev. 2009, 23, 2382–2387. [Google Scholar] [CrossRef] [PubMed]
- Reichenstein, M.; Rauner, G.; Kfir, S.; Kisliouk, T.; Barash, I. Luminal STAT5 mediates H2AX promoter activity in distinct population of basal mammary epithelial cells. Oncotarget 2016, 7, 41781–41797. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.R.; Nelson, E.A.; Yeh, J.E.; Pinello, L.; Yuan, G.C.; Frank, D.A. STAT5 outcompetes STAT3 to regulate the expression of the oncogenic transcriptional modulator BCL6. Mol. Cell. Biol. 2013, 33, 2879–2890. [Google Scholar] [CrossRef] [PubMed]
- Iavnilovitch, E.; Cardiff, R.D.; Groner, B.; Barash, I. Deregulation of Stat5 expression and activation causes mammary tumors in transgenic mice. Int. J. Cancer 2004, 112, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Stewart, W.C.; Pearcy, L.A.; Floyd, Z.E.; Stephens, J.M. STAT5A expression in Swiss 3T3 cells promotes adipogenesis in vivo in an athymic mice model system. Obesity 2011, 19, 1731–1734. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Zhang, Y.; Liu, Y.; Chen, J.; Zong, C.; Yu, C.; Cui, S.; Gao, W.; Qin, D.; Sun, W.; et al. Signal transducer and activator of transcription 5B (STAT5B) modulates adipocyte differentiation via MOF. Cell. Signal. 2015, 27, 2434–2443. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, A.; Corl, B.A.; Jiang, H. Effect of growth hormone on the differentiation of bovine preadipocytes into adipocytes and the role of the signal transducer and activator of transcription 5b. J. Anim. Sci. 2014, 92, 1958–1967. [Google Scholar] [CrossRef] [PubMed]
- Tse, M.C.L.; Liu, X.; Yang, S.; Ye, K.; Chan, C.B. Fyn regulates adipogenesis by promoting PIKE-A/STAT5a interaction. Mol. Cell. Biol. 2013, 33, 1797–1808. [Google Scholar] [CrossRef] [PubMed]
- Schmitt-Ney, M.; Doppler, W.; Ball, R.K.; Groner, B. Beta-casein gene promoter activity is regulated by the hormone-mediated relief of transcriptional repression and a mammary-gland-specific nuclear factor. Mol. Cell. Biol. 1991, 11, 3745–3755. [Google Scholar] [CrossRef] [PubMed]
- Pezet, A.; Ferrag, F.; Kelly, P.A.; Edery, M. Tyrosine docking sites of the rat prolactin receptor required for association and activation of Stat5. J. Biol. Chem. 1997, 272, 25043–25050. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.C.; Wang, X.; Darus, C.J.; Kopchick, J.J. Growth hormone promotes the association of transcription factor STAT5 with the growth hormone receptor. J. Biol. Chem. 1996, 271, 19768–19773. [Google Scholar] [CrossRef] [PubMed]
- Chin, H.; Nakamura, N.; Kamiyama, R.; Miyasaka, N.; Ihle, J.N.; Miura, O. Physical and functional interactions between Stat5 and the tyrosine-phosphorylated receptors for erythropoietin and interleukin-3. Blood 1996, 88, 4415–4425. [Google Scholar] [PubMed]
- Pfitzner, E.; Jähne, R.; Wissler, M.; Stoecklin, E.; Groner, B. p300/CREB-binding protein enhances the prolactin-mediated transcriptional induction through direct interaction with the transactivation domain of Stat5, but does not participate in the Stat5-mediated suppression of the glucocorticoid response. Mol. Endocrinol. 1998, 12, 1582–1593. [Google Scholar] [CrossRef] [PubMed]
- Oñate, S.A.; Tsai, S.Y.; Tsai, M.J.; O’Malley, B.W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 1995, 270, 1354–1358. [Google Scholar] [PubMed]
- Yao, T.P.; Ku, G.; Zhou, N.; Scully, R.; Livingston, D.M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl. Acad. Sci. USA 1996, 93, 10626–10631. [Google Scholar] [CrossRef] [PubMed]
- Litterst, C.M.; Kliem, S.; Marilley, D.; Pfitzner, E. NCoA-1/SRC-1 Is an Essential Coactivator of STAT5 That Binds to the FDL Motif in the α-Helical Region of the STAT5 Transactivation Domain. J. Biol. Chem. 2003, 278, 45340–45351. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, H.; Brindle, P.K.; Handa, M.; Ihle, J.N. Functional interaction of STAT5 and nuclear receptor co-repressor SMRT: Implications in negative regulation of STAT5-dependent transcription. EMBO J. 2001, 20, 6836–6844. [Google Scholar] [CrossRef] [PubMed]
- Sefat-E-Khuda; Yoshida, M.; Xing, Y.; Shimasaki, T.; Takeya, M.; Kuwahara, K.; Sakaguchi, N. The Sac3 homologue shd1 is involved in mitotic progression in mammalian cells. J. Biol. Chem. 2004, 279, 46182–46190. [Google Scholar]
- Banks, A.S.; Li, J.; McKeag, L.; Hribal, M.L.; Kashiwada, M.; Accili, D.; Rothman, P.B. Deletion of SOCS7 leads to enhanced insulin action and enlarged islets of Langerhans. J. Clin. Investig. 2005, 115, 2462–2471. [Google Scholar] [CrossRef] [PubMed]
- Martens, N.; Uzan, G.; Wery, M.; Hooghe, R.; Hooghe-Peters, E.L.; Gertler, A. Suppressor of cytokine signaling 7 inhibits prolactin, growth hormone, and leptin signaling by interacting with STAT5 or STAT3 and attenuating their nuclear translocation. J. Biol. Chem. 2005, 280, 13817–13823. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, I.; Kitamura, T.; Wakao, H.; Tanaka, H.; Hashimoto, K.; Albanese, C.; Downward, J.; Pestell, R.G.; Kanakura, Y. Transcriptional regulation of the cyclin D1 promoter by STAT5: Its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 1999, 18, 1367–1377. [Google Scholar] [CrossRef] [PubMed]
- Sturm, R.A.; Das, G.; Herr, W. The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 1988, 2, 1582–1599. [Google Scholar] [CrossRef] [PubMed]
- Lydon, J.P.; De Mayo, F.J.; Funk, C.R.; Mani, S.K.; Hughes, A.R.; Montgomery, C.A.; Shyamala, G.; Conneely, O.M.; O’Malley, B.W. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 1995, 9, 2266–2278. [Google Scholar] [CrossRef] [PubMed]
- Brisken, C.; Park, S.; Vass, T.; Lydon, J.P.; O’Malley, B.W.; Weinberg, R.A. A paracrine role for the epithelial progesterone receptor in mammary gland development. Proc. Natl. Acad. Sci. USA 1998, 95, 5076–5081. [Google Scholar] [CrossRef] [PubMed]
- Silberstein, G.B.; Van Horn, K.; Shyamala, G.; Daniel, C.W. Progesterone receptors in the mouse mammary duct: Distribution and developmental regulation. Cell Growth Differ. 1996, 7, 945–952. [Google Scholar] [PubMed]
- Richer, J.K.; Lange, C.A.; Manning, N.G.; Owen, G.; Powell, R.; Horwitz, K.B. Convergence of progesterone with growth factor and cytokine signaling in breast cancer. Progesterone receptors regulate signal transducers and activators of transcription expression and activity. J. Biol. Chem. 1998, 273, 31317–31326. [Google Scholar] [CrossRef] [PubMed]
- Cerliani, J.P.; Guillardoy, T.; Giulianelli, S.; Vaque, J.P.; Gutkind, J.S.; Vanzulli, S.I.; Martins, R.; Zeitlin, E.; Lamb, C.A.; Lanari, C. Interaction between FGFR-2, STAT5, and progesterone receptors in breast cancer. Cancer Res. 2011, 71, 3720–3731. [Google Scholar] [CrossRef] [PubMed]
- Obr, A.E.; Grimm, S.L.; Bishop, K.A.; Pike, J.W.; Lydon, J.P.; Edwards, D.P. Progesterone receptor and Stat5 signaling cross talk through RANKL in mammary epithelial cells. Mol. Endocrinol. 2013, 27, 1808–1824. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.; Bresnick, J.; Rosewell, I.; Crafton, T.; Poulsom, R.; Stamp, G.; Dickson, C. Fibroblast growth factor receptor signalling has a role in lobuloalveolar development of the mammary gland. J. Cell Sci. 1997, 110, 1261–1268. [Google Scholar] [PubMed]
- Lu, P.; Ewald, A.J.; Martin, G.R.; Werb, Z. Genetic mosaic analysis reveals FGF receptor 2 function in terminal end buds during mammary gland branching morphogenesis. Dev. Biol. 2008, 321, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Planque, N. Nuclear trafficking of secreted factors and cell-surface receptors: New pathways to regulate cell proliferation and differentiation, and involvement in cancers. Cell Commun. Signal. 2006. [Google Scholar] [CrossRef] [PubMed]
- Stöcklin, E.; Wissler, M.; Gouilleux, F.; Groner, B. SL7 Functional interactions between Stat5 and the glucocorticoid receptor. Nature 1996, 383, 726–728. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Lichten, L.A.; Ryu, M.-S.; Liuzzi, J.P.; Wang, F.; Cousins, R.J. STAT5-glucocorticoid receptor interaction and MTF-1 regulate the expression of ZnT2 (Slc30a2) in pancreatic acinar cells. Proc. Natl. Acad. Sci. USA 2010, 107, 2818–2823. [Google Scholar] [CrossRef] [PubMed]
- Biola, A.; Lefebvre, P.; Perrin-Wolff, M.; Sturm, M.; Bertoglio, J.; Pallardy, M. Interleukin-2 inhibits glucocorticoid receptor transcriptional activity through a mechanism involving STAT5 (signal transducer and activator of transcription 5) but not AP-1. Mol. Endocrinol. 2001, 15, 1062–1076. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Pace, T.W.W.; Miller, A.H. Interferon-alpha inhibits glucocorticoid receptor-mediated gene transcription via STAT5 activation in mouse HT22 cells. Brain Behav. Immun. 2009, 23, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Engblom, D.; Kornfeld, J.W.; Schwake, L.; Tronche, F.; Reimann, A.; Beug, H.; Hennighausen, L.; Morigg, R.; Schütz, G. Direct glucocorticoid receptor-Stat5 interaction in hepatocytes controls body size and maturation-related gene expression. Genes Dev. 2007, 21, 1157–1162. [Google Scholar] [CrossRef] [PubMed]
- Baugh, J.E.; Floyd, Z.E.; Stephens, J.M. The modulation of STAT5A/GR complexes during fat cell differentiation and in mature adipocytes. Obesity 2007, 15, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Cella, N.; Groner, B.; Hynes, N.E. Characterization of Stat5a and Stat5b homodimers and heterodimers and their association with the glucocortiocoid receptor in mammary cells. Mol. Cell. Biol. 1998, 18, 1783–1792. [Google Scholar] [CrossRef] [PubMed]
- Stoecklin, E.; Wissler, M.; Moriggl, R.; Groner, B. Specific DNA binding of Stat5, but not of glucocorticoid receptor, is required for their functional cooperation in the regulation of gene transcription. Mol. Cell. Biol. 1997, 17, 6708–6716. [Google Scholar] [CrossRef] [PubMed]
- Tronche, F.; Opherk, C.; Moriggl, R.; Kellendonk, C.; Reimann, A.; Schwake, L.; Reichardt, H.M.; Stangl, K.; Gau, D.; Hoeflich, A.; et al. Glucocorticoid receptor function in hepatocytes is essential to promote postnatal body growth. Genes Dev. 2004, 18, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Fish, E.N.; Uddin, S.; Korkmaz, M.; Majchrzak, B.; Druker, B.J.; Platanias, L.C. Activation of a CrkL-Stat5 signaling complex by type I interferons. J. Biol. Chem. 1999, 274, 571–573. [Google Scholar] [CrossRef] [PubMed]
- Uddin, S.; Lekmine, F.; Sassano, A.; Rui, H.; Fish, E.N.; Platanias, L.C. Role of Stat5 in Type I interferon-signaling and transcriptional regulation. Biochem. Biophys. Res. Commun. 2003, 308, 325–330. [Google Scholar] [CrossRef]
- Nanou, A.; Toumpeki, C.; Lavigne, M.D.; Lazou, V.; Demmers, J.; Paparountas, T.; Thanos, D.; Katsantoni, E. The dual role of LSD1 and HDAC3 in STAT5-dependent transcription is determined by protein interactions, binding affinities, motifs and genomic positions. Nucleic Acids Res. 2016, 45, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Gall Trošelj, K.; Novak Kujundzic, R.; Ugarkovic, D. Polycomb repressive complex’s evolutionary conserved function: The role of EZH2 status and cellular background. Clin. Epigenetics 2016. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.R.; Yu, Y.N.; Ren, L.L.; Cui, Y.; Lu, Y.Y.; Chen, H.Y.; Ma, X.; Qin, W.X.; Cao, W.; Hong, J.; et al. Role of C9orf140 in the promotion of colorectal cancer progression and mechanisms of its upregulation via activation of STAT5, β-catenin and EZH2. Carcinogenesis 2014, 35, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.; Powers, S.E.; Maienschein-cline, M.; Bartom, E.T.; Keith, M. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 2012, 12, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Yoo, K.H.; Oh, S.; Kang, K.; Hensel, T.; Robinson, G.W.; Hennighausen, L. Loss of EZH2 results in precocious mammary gland development and activation of STAT5-dependent genes. Nucleic Acids Res. 2015, 43, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Gouilleux, F.; Wakao, H.; Mundt, M.; Groner, B. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J. 1994, 13, 4361–4369. [Google Scholar] [PubMed]
- Beuvink, I.; Hess, D.; Flotow, H.; Hofsteenge, J.; Groner, B.; Hynes, N.E. Stat5a serine phosphorylation. Serine 779 is constitutively phosphorylated in the mammary gland, and serine 725 phosphorylation influences prolactin-stimulated in vitro DNA binding activity. J. Biol. Chem. 2000, 275, 10247–10255. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Yamashita, H.; Rui, H.; Waxman, D.J. Serine phosphorylation of GH-activated signal transducer and activator of transcription 5a (STAT5a) and STAT5b: Impact on STAT5 transcriptional activity. Mol. Endocrinol. 2001, 15, 2157–2171. [Google Scholar] [CrossRef] [PubMed]
- Pircher, T.J.; Petersen, H.; Gustafsson, J.A.; Haldosén, L.A. Extracellular signal-regulated kinase (ERK) interacts with signal transducer and activator of transcription (STAT) 5a. Mol. Endocrinol. 1999, 13, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, H.; Xu, J.; Erwin, R.A.; Farrar, W.L.; Kirken, R.A.; Rui, H. Differential control of the phosphorylation state of proline-juxtaposed serine residues Ser725 of Stat5a and Ser730 of Stat5b in prolactin-sensitive cells. J. Biol. Chem. 1998, 273, 30218–30224. [Google Scholar] [CrossRef] [PubMed]
- David, M.; Chen, H.E.; Goelz, S.; Larner, A.C.; Neel, B.G. Differential Regulation of the Alpha/Beta Interferon-Stimulated Jak/Stat Pathway by the SH2 Domain-Containing Tyrosine Phosphatase SHPTP1. Mol. Cell. Biol. 1995, 15, 7050–7058. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Yu, D.H.; Feng, G.S. Shp-2 tyrosine phosphatase functions as a negative regulator of the interferon-stimulated Jak/STAT pathway. Mol. Cell. Biol. 1999, 19, 2416–2424. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.L.; Jin, Y.J.; Burakoff, S.J. Cytosolic tyrosine dephosphorylation of STAT5. Potential role of SHP-2 in STAT5 regulation. J. Biol. Chem. 2000, 275, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Patterson, K.I.; Brummer, T.; O’brien, P.M.; Daly, R.J. Dual-specificity phosphatases: Critical regulators with diverse cellular targets. Biochem. J. 2009, 418, 475–489. [Google Scholar] [CrossRef] [PubMed]
- Barajas-Espinosa, A.; Basye, A.; Angelos, M.G.; Chen, C.A. Modulation of p38 kinase by DUSP4 is important in regulating cardiovascular function under oxidative stress. Free Radic. Biol. Med. 2015, 89, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.Y.; Lin, Y.C.; Hsiao, W.Y.; Liao, F.H.; Huang, P.Y.; Tan, T.H. DUSP4 deficiency enhances CD25 expression and CD4 + T-cell proliferation without impeding T-cell development. Eur. J. Immunol. 2012, 42, 476–488. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, W.Y.; Lin, Y.C.; Liao, F.H.; Chan, Y.C.; Huang, C.Y. Dual-specificity phosphatase 4 regulates STAT5 protein stability and helper T cell polarization. PLoS ONE 2015, 10, e0145880. [Google Scholar] [CrossRef] [PubMed]
- Schmid, C.A.; Robinson, M.D.; Scheifinger, N.A.; Müller, S.; Cogliatti, S.; Tzankov, A.; Müller, A. DUSP4 deficiency caused by promoter hypermethylation drives JNK signaling and tumor cell survival in diffuse large B cell lymphoma. J. Exp. Med. 2015, 212, 775–792. [Google Scholar] [CrossRef] [PubMed]
- Guan, K.L.; Butch, E. Isolation and characterization of a novel dual specific phosphatase, HVH2, which selectively dephosphorylates the mitogen-activated protein kinase. J. Biol. Chem. 1995, 270, 7197–7203. [Google Scholar] [PubMed]
- Chu, Y.; Solski, P.A.; Khosravi-Far, R.; Der, C.J.; Kelly, K. The mitogen-activated protein kinase phosphatases PAC1, MKP-1, and MKP-2 have unique substrate specificities and reduced activity in vivo toward the ERK2 sevenmaker mutation. J. Biol. Chem. 1996, 271, 6497–6501. [Google Scholar] [PubMed]
- Marshall, C.J. Specificity of receptor tyrosine kinase signaling: Transient versus sustained extracellular signal-regulated kinase activation. Cell 1995, 80, 179–185. [Google Scholar] [CrossRef]
- Chiarugi, P.; Taddei, M.L.; Schiavone, N.; Papucci, L.; Giannoni, E.; Fiaschi, T.; Capaccioli, S.; Raugei, G.; Ramponi, G. LMW-PTP is a positive regulator of tumor onset and growth. Oncogene 2004, 23, 3905–3914. [Google Scholar] [CrossRef] [PubMed]
- Rigacci, S.; Guidotti, V.; Parri, M.; Berti, A. Modulation of STAT5 interaction with LMW-PTP during early megakaryocyte differentiation. Biochemistry 2008, 47, 1482–1489. [Google Scholar] [CrossRef] [PubMed]
- Rigacci, S.; Talini, D.; Berti, A. LMW-PTP associates and dephosphorylates STAT5 interacting with its C-terminal domain. Biochem. Biophys. Res. Commun. 2003, 312, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Sutherland, K.D.; Sum, E.Y.M.; Olayioye, M.; Wittlin, S.; Tang, T.K.; Lindeman, G.J.; Visvader, J.E. CPAP is a novel stat5-interacting cofactor that augments stat5-mediated transcriptional activity. Mol. Endocrinol. Balt. Md 2002, 16, 2019–2033. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Feng, Y.; Ye, K. Src-family tyrosine kinase fyn phosphorylates phosphatidylinositol 3-kinase enhancer-activating Akt, preventing its apoptotic cleavage and promoting cell survival. Cell Death Differ. 2007, 14, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.-B.; Liu, X.; Ensslin, M.A.; Dillehay, D.L.; Ormandy, C.J.; Sohn, P.; Serra, R.; Ye, K. PIKE-A is required for prolactin-mediated STAT5a activation in mammary gland development. EMBO J. 2010, 29, 956–968. [Google Scholar] [CrossRef] [PubMed]
- Smit, L.; Van Der Horst, G.; Borst, J. Sos, Vav, and C3G participate in B cell receptor-induced signaling pathways and differentially associate with Shc-Grh2, Crk, and Crk-L Adaptors. J. Biol. Chem. 1996, 271, 8564–8569. [Google Scholar] [CrossRef] [PubMed]
- Quelle, F.W.; Wang, D.; Nosaka, T.; Thierfelder, W.E.; Stravopodis, D.; Weinstein, Y.; Ihle, J.N. Erythropoietin induces activation of Stat5 through association with specific tyrosines on the receptor that are not required for a mitogenic response. Mol. Cell. Biol. 1996, 16, 1622–1631. [Google Scholar] [CrossRef] [PubMed]
- Miyakawa, Y.; Oda, A.; Druker, B.J.; Miyazaki, H.; Handa, M.; Ohashi, H.; Ikeda, Y. Thrombopoietin induces tyrosine phosphorylation of Stat3 and Stat5 in human blood platelets. Blood 1996, 87, 439–446. [Google Scholar] [PubMed]
- Ozaki, K.; Oda, A.; Wakao, H.; Rhodes, J.; Druker, B.J.; Ishida, A.; Wakui, M.; Okamoto, S.; Morita, K.; Handa, M.; et al. Thrombopoietin induces association of Crkl with STAT5 but not STAT3 in human platelets. Blood 1998, 92, 4652–4662. [Google Scholar] [PubMed]
- Rosa Santos, S.C.; Dumon, S.; Mayeux, P.; Gisselbrecht, S.; Gouilleux, F. Cooperation between STAT5 and phosphatidylinositol 3-kinase in the IL-3-dependent survival of a bone marrow derived cell line. Oncogene 2000, 19, 1164–1172. [Google Scholar] [CrossRef] [PubMed]
- Nyga, R.; Pecquet, C.; Harir, N.; Gu, H.; Dhennin-Duthille, I.; Régnier, A.; Gouilleux-Gruart, V.; Lassoued, K.; Gouilleux, F. Activated STAT5 proteins induce activation of the PI 3-kinase/Akt and Ras/MAPK pathways via the Gab2 scaffolding adapter. Biochem. J. 2005, 390, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Hosui, A.; Kimura, A.; Yamaji, D.; Zhu, B.; Na, R.; Hennighausen, L. Loss of STAT5 causes liver fibrosis and cancer development through increased TGF-{beta} and STAT3 activation. J. Exp. Med. 2009, 206, 819–831. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Able, A.A.; Burrell, J.A.; Stephens, J.M. STAT5-Interacting Proteins: A Synopsis of Proteins that Regulate STAT5 Activity. Biology 2017, 6, 20. https://doi.org/10.3390/biology6010020
Able AA, Burrell JA, Stephens JM. STAT5-Interacting Proteins: A Synopsis of Proteins that Regulate STAT5 Activity. Biology. 2017; 6(1):20. https://doi.org/10.3390/biology6010020
Chicago/Turabian StyleAble, Ashley A., Jasmine A. Burrell, and Jacqueline M. Stephens. 2017. "STAT5-Interacting Proteins: A Synopsis of Proteins that Regulate STAT5 Activity" Biology 6, no. 1: 20. https://doi.org/10.3390/biology6010020




