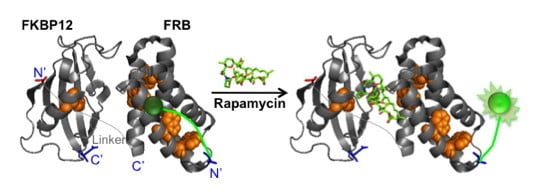
A Signal-On Fluorosensor Based on Quench-Release Principle for Sensitive Detection of Antibiotic Rapamycin
Abstract
:1. Introduction
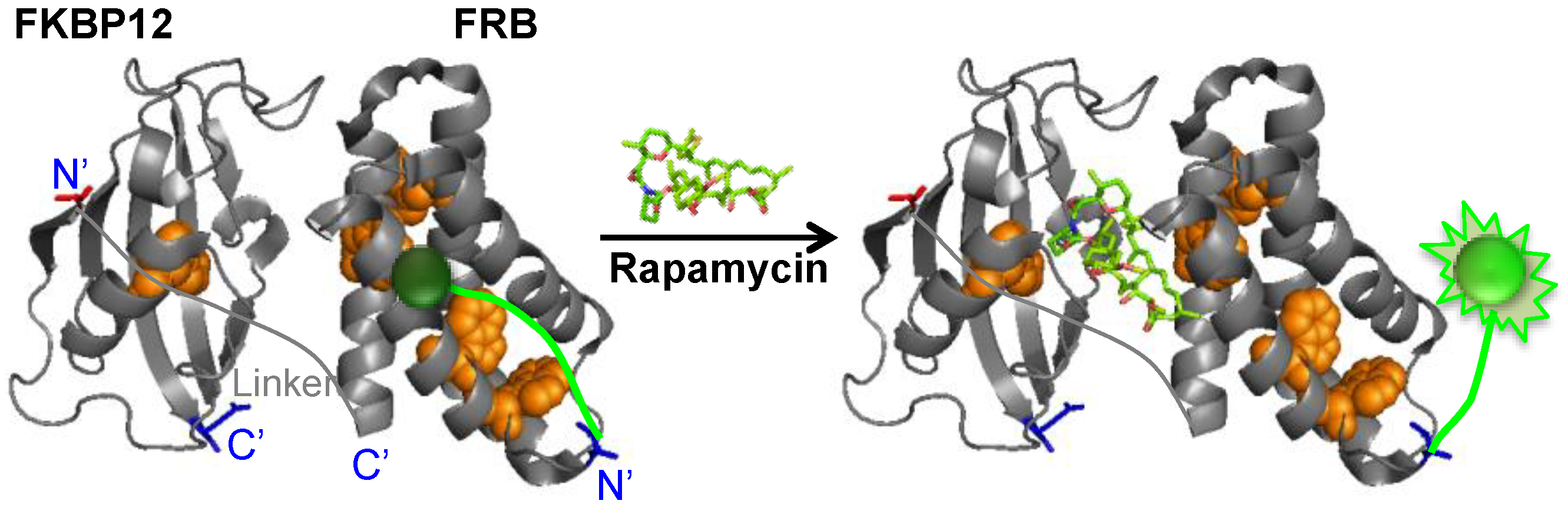
2. Experimental Section
2.1. Construction of Q’-Body Genes
| Primer Name | Nucleotide Sequence (5'-3') |
|---|---|
| FKBP-G3S2-Back | GGTGGCGGTTCAGGTGGCGGTTCAATGGGCGTCCAGGTCGAG |
| FKBP-linkFor | GCTGCCACCTCCGCCTGAACCGCCTCCACCCTCCAGTTTCAGCAGCTCCAC |
| FRB-linkBack | GGCGGAGGTGGCAGCGGCGGTGGCGGGTCGACGCTCATTAGAGTGGCAATC |
| FRB-For | ATGAGAACCCCCCCCGAGCTGTTTTGAGATTCGTCT |
| G3S2-amb-Back | TAGTCTAATGAGACCGGTGGCGGTTCAGGTGGC |
| Sal-link-amb-For | CCACTCTAATGAGCGTCGACCCGCCACCGCCGCTGCCCTATCCGCCTGAACCGCC |
| FRB-G3S2-Back | GGTGGCGGTTCAGGTGGCGGTTCACTCATTAGAGTGGCAATC |
| FRB-linkFor | TCCGCCTGSSCCGCCTCCACCGAGCTGTTTTGAGATTCGTC |
| FKBP-link-Back | GGTGGCAGCGGCGGAGGTGGCAGCGGCGGTGGCGGGTCGACGATGGGCGTCCAGGTCGGAG |
| FKBP-For | ATGAGAACCCCCCCCCTCCAGTTTCAGCAGCTCCAC |
| FKBP-link-amb-Back | GGTGGCAGCGGCGGTTAGGGCAGCGGCGGTGGCGGGTCGACGATGGGCGTCCAGGTCGAG |
2.2. Synthesis and Purification of Q’-Bodies
2.3. Fluorescence Measurements
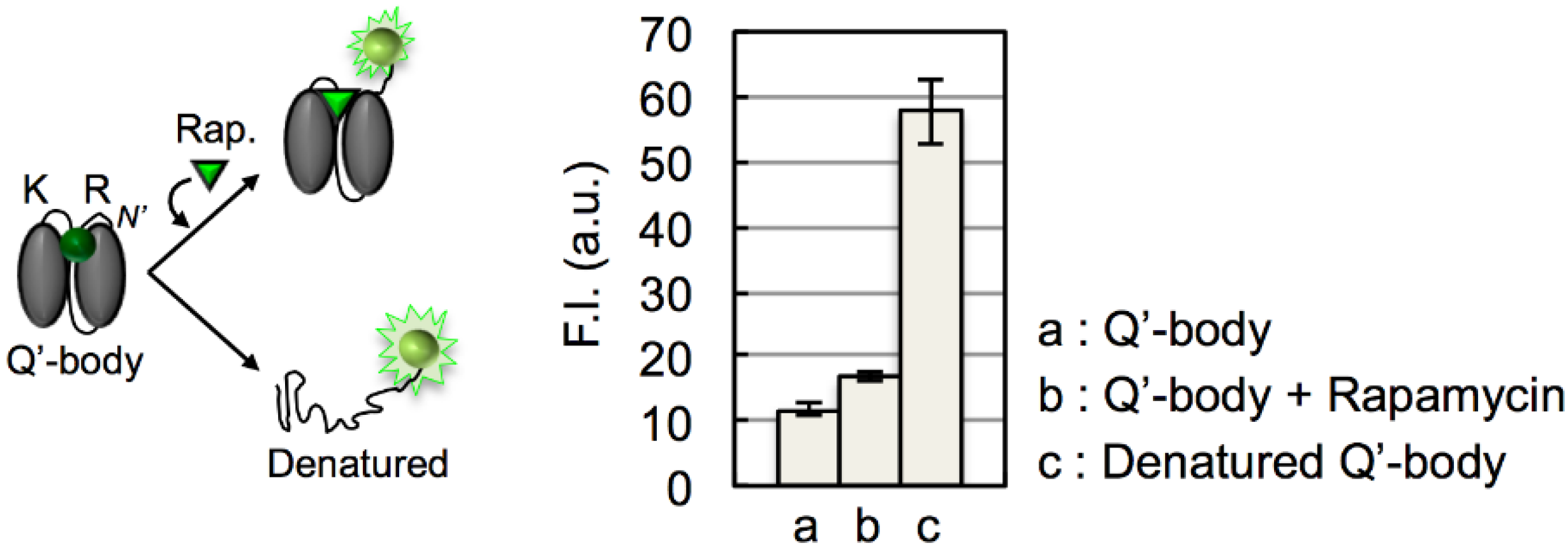
3. Results and Discussion
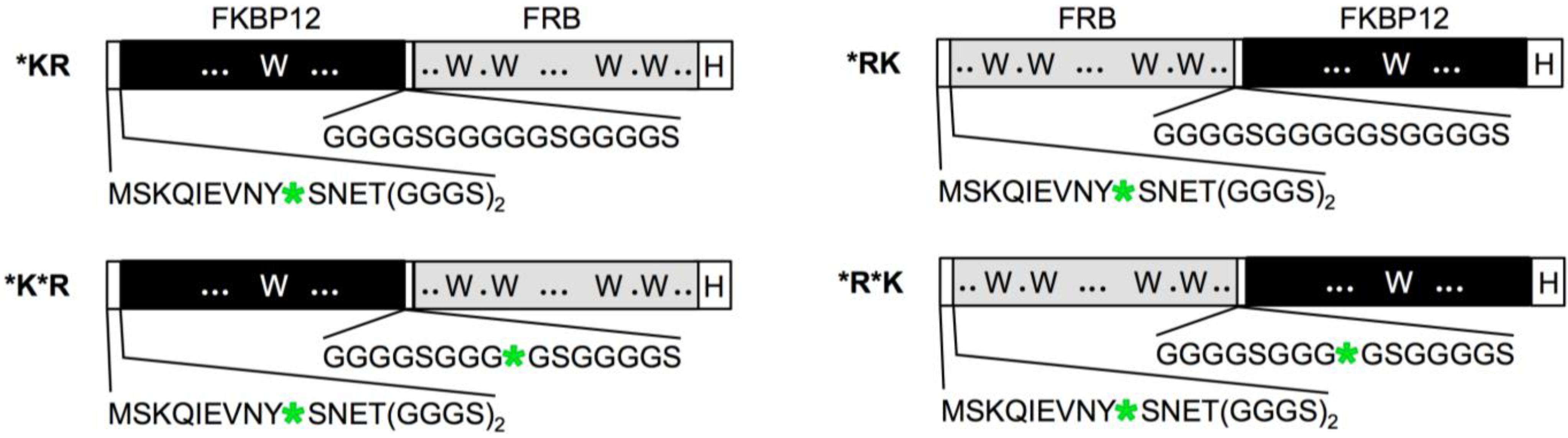

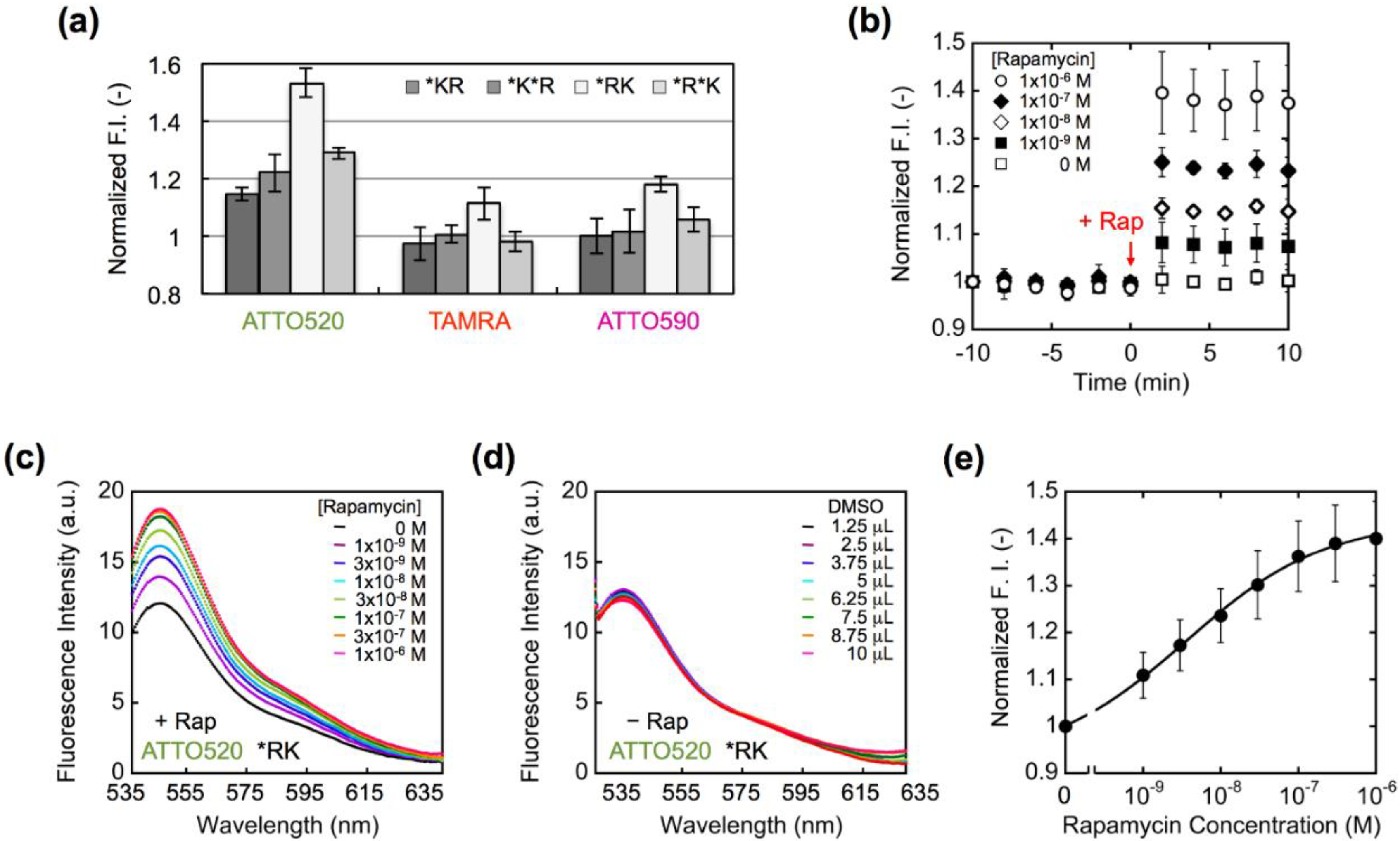
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Vezina, C.; Kudelski, A.; Sehgal, S.N. Rapamycin (ay-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. 1975, 28, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, S.N. Sirolimus: Its discovery, biological properties, and mechanism of action. Transplant. Proc. 2003, 35, 7S–14S. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar]
- Banaszynski, L.A.; Liu, C.W.; Wandless, T.J. Characterization of the FKBP.Rapamycin.FRB ternary complex. J. Am. Chem. Soc. 2005, 127, 4715–4721. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zheng, X.F.; Brown, E.J.; Schreiber, S.L. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc. Natl. Acad. Sci. USA 1995, 92, 4947–4951. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Chen, J.; Schreiber, S.L.; Clardy, J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human frap. Science 1996, 273, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Fingar, D.C.; Richardson, C.J.; Tee, A.R.; Cheatham, L.; Tsou, C.; Blenis, J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 2004, 24, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Alocilja, E.C.; Radke, S.M. Market analysis of biosensors for food safety. Biosens. Bioelectron. 2003, 18, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Rasooly, A.; Jacobson, J. Development of biosensors for cancer clinical testing. Biosens. Bioelectron. 2006, 21, 1851–1858. [Google Scholar] [CrossRef] [PubMed]
- Ibraheem, A.; Campbell, R.E. Designs and applications of fluorescent protein-based biosensors. Curr. Opin. Chem. Biol. 2010, 14, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Ohba, Y.; Fujioka, Y.; Nakada, S.; Tsuda, M. Fluorescent protein-based biosensors and their clinical applications. Prog. Mol. Biol. Transl. Sci. 2013, 113, 313–348. [Google Scholar] [PubMed]
- Tainaka, K.; Sakaguchi, R.; Hayashi, H.; Nakano, S.; Liew, F.F.; Morii, T. Design strategies of fluorescent biosensors based on biological macromolecular receptors. Sensors 2010, 10, 1355–1376. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Puhl, H.L., 3rd; Ikeda, S.R. Estimating protein-protein interaction affinity in living cells using quantitative forster resonance energy transfer measurements. J. Biomed. Opt. 2007, 12. [Google Scholar] [CrossRef]
- Cui, B.; Wang, Y.; Song, Y.; Wang, T.; Li, C.; Wei, Y.; Luo, Z.Q.; Shen, X. Bioluminescence resonance energy transfer system for measuring dynamic protein-protein interactions in bacteria. mBio 2014, 5, e01050–e01014. [Google Scholar] [CrossRef] [PubMed]
- Galarneau, A.; Primeau, M.; Trudeau, L.E.; Michnick, S.W. Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nat. Biotechnol. 2002, 20, 619–622. [Google Scholar] [CrossRef] [PubMed]
- Cabantous, S.; Nguyen, H.B.; Pedelacq, J.D.; Koraichi, F.; Chaudhary, A.; Ganguly, K.; Lockard, M.A.; Favre, G.; Terwilliger, T.C.; Waldo, G.S. A new protein-protein interaction sensor based on tripartite split-gfp association. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Luker, K.E.; Smith, M.C.; Luker, G.D.; Gammon, S.T.; Piwnica-Worms, H.; Piwnica-Worms, D. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc. Natl. Acad. Sci. USA 2004, 101, 12288–12293. [Google Scholar] [CrossRef] [PubMed]
- Ohmuro-Matsuyama, Y.; Chung, C.I.; Ueda, H. Demonstration of protein-fragment complementation assay using purified firefly luciferase fragments. BMC Biotechnol. 2013, 13. [Google Scholar] [CrossRef] [PubMed]
- Abe, R.; Jeong, H.J.; Arakawa, D.; Dong, J.; Ohashi, H.; Kaigome, R.; Saiki, F.; Yamane, K.; Takagi, H.; Ueda, H. Ultra q-bodies: Quench-based antibody probes that utilize dye-dye interactions with enhanced antigen-dependent fluorescence. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Abe, R.; Ohashi, H.; Iijima, I.; Ihara, M.; Takagi, H.; Hohsaka, T.; Ueda, H. “Quenchbodies”: Quench-based antibody probes that show antigen-dependent fluorescence. J. Am. Chem. Soc. 2011, 133, 17386–17394. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.J.; Ohmuro-Matsuyama, Y.; Ohashi, H.; Ohsawa, F.; Tatsu, Y.; Inagaki, M.; Ueda, H. Detection of vimentin serine phosphorylation by multicolor quenchbodies. Biosens. Bioelectron. 2013, 40, 17–23. [Google Scholar] [CrossRef]
- Jeong, H.J.; Ueda, H. Strategy for making a superior quenchbody to proteins: Effect of the fluorophore position. Sensors 2014, 14, 13285–13297. [Google Scholar] [CrossRef] [PubMed]
- Ueda, H.; Dong, J. From fluorescence polarization to quenchbody: Recent progress in fluorescent reagentless biosensors based on antibody and other binding proteins. Biochim. Biophys. Acta 2014, 1844, 1951–1959. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, D.A.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. Aust. Assoc. Clin. Biochem. 2008, 29, S49–S52. [Google Scholar]
- Hohsaka, T.; Sisido, M. Incorporation of non-natural amino acids into proteins. Curr. Opin. Chem. Biol. 2002, 6, 809–815. [Google Scholar] [CrossRef]
- Mansoor, S.E.; Dewitt, M.A.; Farrens, D.L. Distance mapping in proteins using fluorescence spectroscopy: The tryptophan-induced quenching (TRIQ) method. Biochemistry 2010, 49, 9722–9731. [Google Scholar] [CrossRef] [PubMed]
- DiJoseph, J.F.; Fluhler, E.; Armstrong, J.; Sharr, M.; Sehgal, S.N. Therapeutic blood levels of sirolimus (rapamycin) in the allografted rat. Transplantation 1996, 62, 1109–1112. [Google Scholar] [CrossRef] [PubMed]
- Cloughesy, T.F.; Yoshimoto, K.; Nghiemphu, P.; Brown, K.; Dang, J.; Zhu, S.; Hsueh, T.; Chen, Y.; Wang, W.; Youngkin, D.; et al. Antitumor activity of rapamycin in a phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008, 5, e8. [Google Scholar] [CrossRef]
- Rodriguez, A.E.; Rodriguez Alemparte, M.; Vigo, C.F.; Fernandez Pereira, C.; Llaurado, C.; Vetcher, D.; Pocovi, A.; Ambrose, J. Role of oral rapamycin to prevent restenosis in patients with de novo lesions undergoing coronary stenting: Results of the argentina single centre study (orar trial). Heart 2005, 91, 1433–1437. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, F.; Heit, A.; Dreher, S.; Eisenacher, K.; Mages, J.; Haas, T.; Krug, A.; Janssen, K.P.; Kirschning, C.J.; Wagner, H. Mammalian target of rapamycin (mTOR) orchestrates the defense program of innate immune cells. Eur. J. Immunol. 2008, 38, 2981–2992. [Google Scholar] [CrossRef] [PubMed]
Appendix
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, H.-J.; Itayama, S.; Ueda, H. A Signal-On Fluorosensor Based on Quench-Release Principle for Sensitive Detection of Antibiotic Rapamycin. Biosensors 2015, 5, 131-140. https://doi.org/10.3390/bios5020131
Jeong H-J, Itayama S, Ueda H. A Signal-On Fluorosensor Based on Quench-Release Principle for Sensitive Detection of Antibiotic Rapamycin. Biosensors. 2015; 5(2):131-140. https://doi.org/10.3390/bios5020131
Chicago/Turabian StyleJeong, Hee-Jin, Shuya Itayama, and Hiroshi Ueda. 2015. "A Signal-On Fluorosensor Based on Quench-Release Principle for Sensitive Detection of Antibiotic Rapamycin" Biosensors 5, no. 2: 131-140. https://doi.org/10.3390/bios5020131