Development of a Pseudomonas aeruginosa Agmatine Biosensor
Abstract
:1. Introduction

2. Experimental Section
2.1. Bacterial Strains and Plasmids
| Parent strain | Phenotype | Genotype | Source or reference |
|---|---|---|---|
| PA14 | Wild-type | Wild-type | [18] |
| WT Agmatine Reporter | aguRB-CTX-Lux | This work | |
| WT Reporter Vector Control | CTX-Lux | This work | |
| AgDi Knockout | aguA:gm, ∆agu2ABCA’ | [12] | |
| AgDi Knockout, Agmatine Reporter | aguA:gm, ∆agu2ABCA’, AguRB-CTX-Lux | This work | |
| AgDi Knockout, Reporter Vector Control | aguA:gm, ∆agu2ABCA’, CTX-Lux | This work | |
| Arginine Decarboxylase Knockout | ∆speA | This work | |
| Arginine Decarboxylase Knockout, Agmatine Reporter | ∆speA, AguRB-CTX-Lux | This work | |
| Arginine Decarboxylase Knockout, Reporter Vector Control | ∆speA, CTX-Lux | This work | |
| AgDi Knockout, Arginine Decarboxylase Knockout | aguA:gm, ∆agu2ABCA’, ∆speA | This work | |
| Agmatine Biosensor-AgDi Knockout, Arginine Decarboxylase Knockout, Agmatine Reporter | aguA:gm, ∆agu2ABCA’, ∆speA, AguR-CTX-Lux | This work | |
| AgDi Knockout, Arginine Decarboxylase Knockout, Reporter Vector Control | aguA:gm, ∆agu2ABCA’, ∆speA, CTX-Lux | This work | |
| E. coli Top10 | Competent cells for cloning purposes | F- mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ- | Invitrogen |
| E. coli SM10 | Vehicle for conjugative mating to P. aeruginosa | KmR, thi-1, thr, leu, tonA, lacY, supE, recA::RP4-2-Tc::Mu, pir. | [19] |
| Plasmid | Description | Features | Source or reference |
|---|---|---|---|
| Mini-ctx-lux | luxCDABE based reporter vector with site specific integration at attB site in P. aeruginosa chromosome | See Reference | [20] |
| pEX18-Ap | Gene replacement vector | See Reference | [17] |
| CTXnoT7 | Contains Lux operon, T7 promoter removed to reduce background expression | No T7 | This work |
| AguRB-lux | Contains Lux operon induced by agmatine via the inserted promotion system of the primary agmatine deiminase, T7 promoter removed to reduce background expression | No T7, AguRB promoter fusion to luxCDABE | This work |
| pBAD202 | Arabinose induced expression vector | See manufacturer | Invitrogen |
| SpeBAD | speA from PA14 cloned into MCS of pBAD202 | Arabinose induced expression vector of SpeA | This work |
| speA KO | pEX18-Ap based cloning vector with speA knockout construct | Designed for disruption of chromosomal speA in P. aeruginosa | This work |
2.2. Development of the Agmatine Biosensor
2.3. Mass Spectrometry Measurement of Agmatine
2.4. Bioassay Technique
2.5. Spinal Cord Infection
3. Results and Discussion
3.1. Construction of a Bioluminescent Agmatine Reporter Construct for Use in P. aeruginosa
3.2. Behavior of Biosensor Construct in Mutants of the Arginine Decarboxylase and Agmatine Deiminase Pathways
3.3. Validation of the ΔspeA, aguA:gm, Δagu2ABCA’, aguRB:lux Mutant as an Agmatine Biosensor
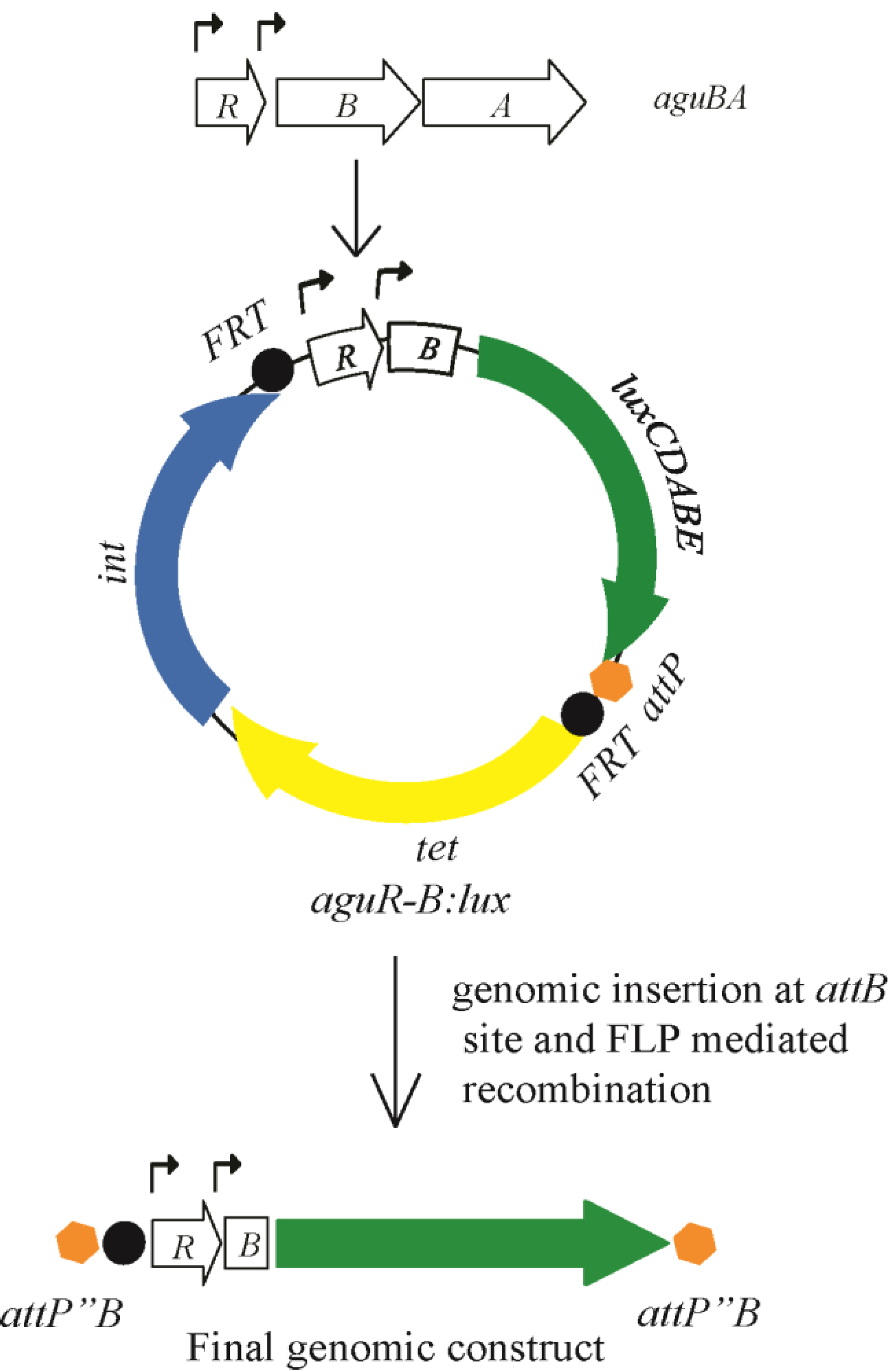


3.4. Use of the Agmatine Biosensor in Biologic Assays
3.4.1. Rapid Detection of Agmatine Secretion by Clinical P. aeruginosa Isolates

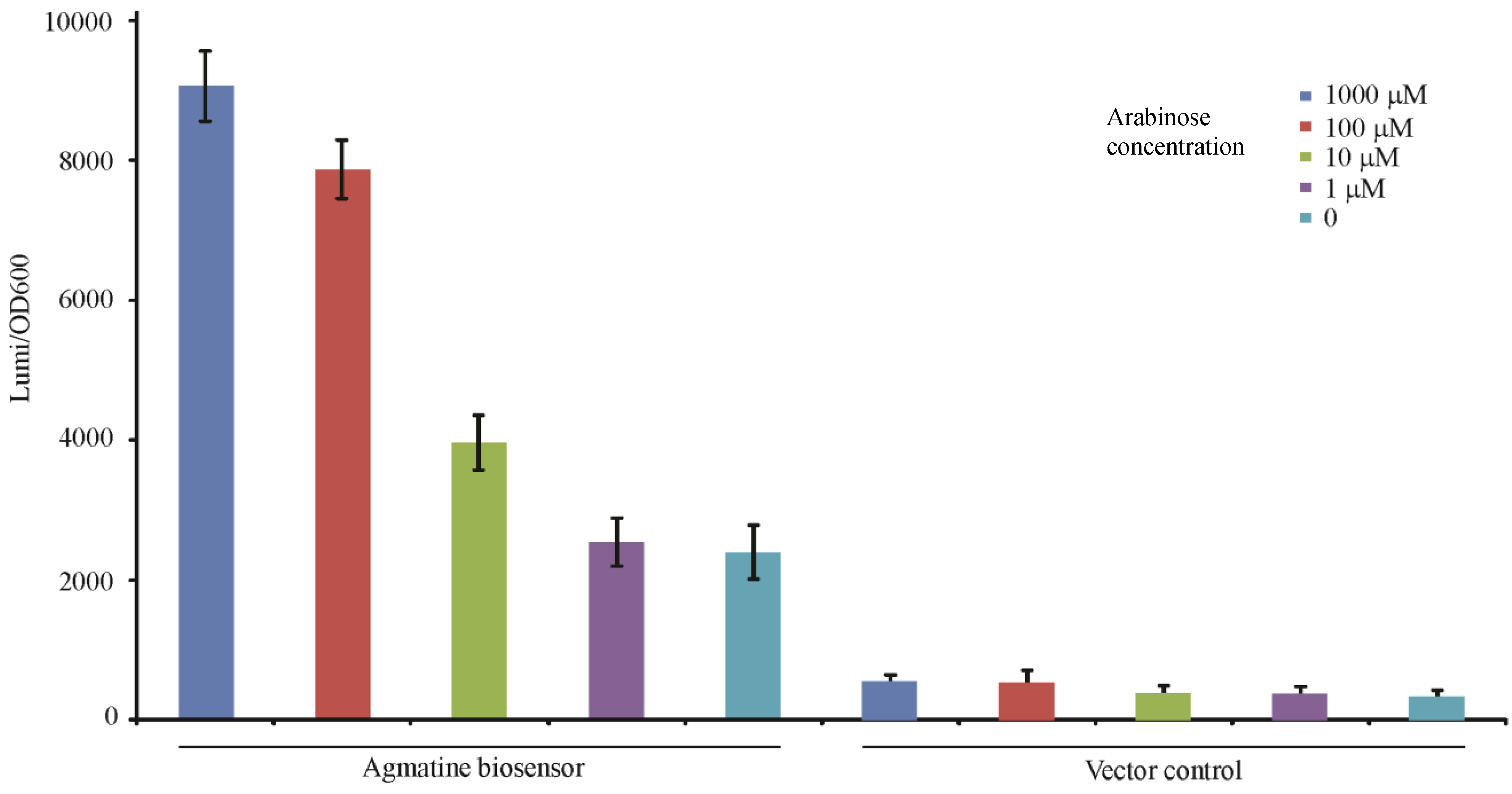
3.4.2. Detection of Arginine Decarboxylase Enzyme Activity
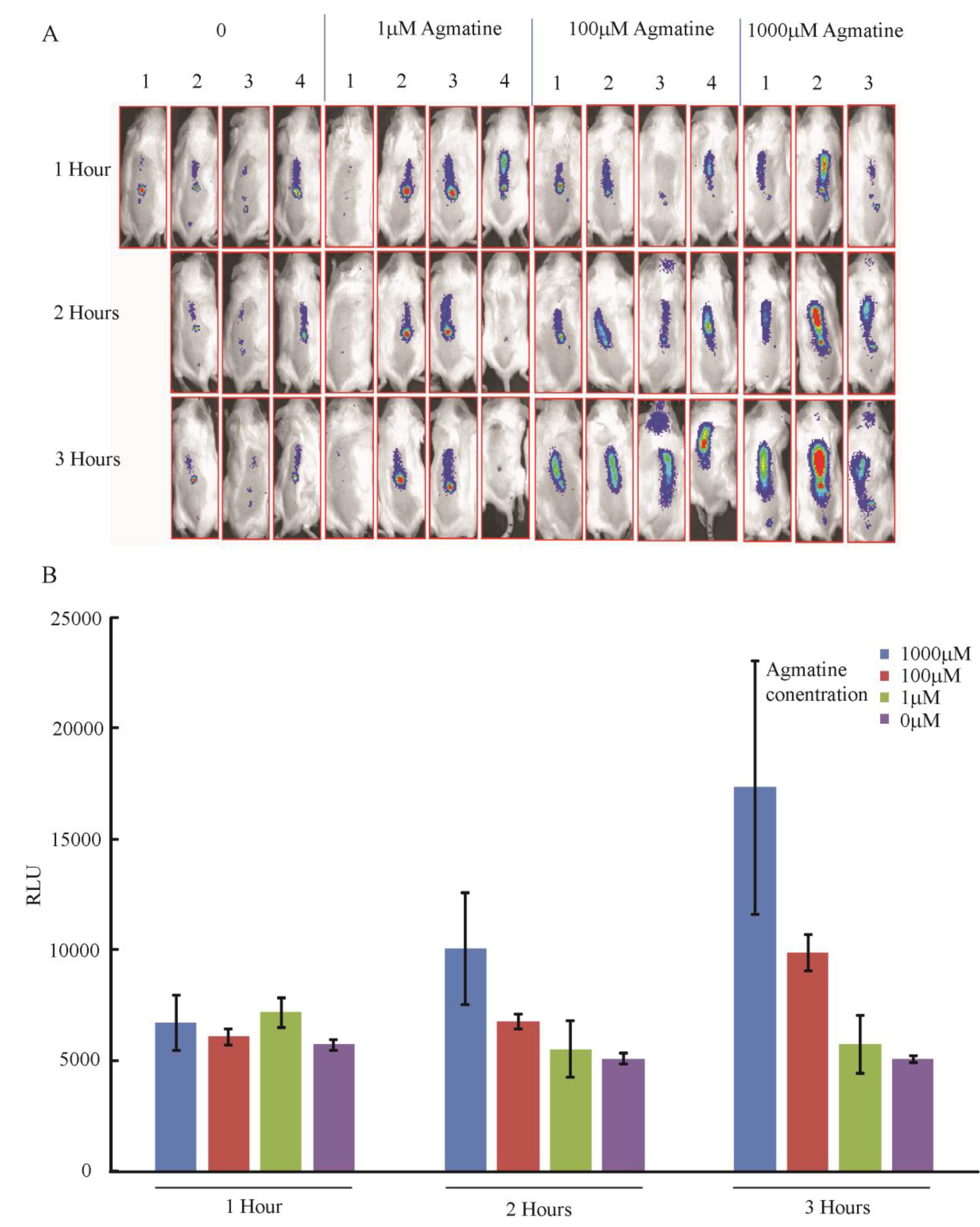
3.4.3. In Vivo Detection of Agmatine during Infection in Mouse Spinal Cords
4. Conclusions
Acknowledgments
Ethics Statement
Author Contributions
Conflicts of Interest
References
- Wortham, B.W.; Patel, C.N.; Oliveira, M.A. Polyamines in bacteria: Pleiotropic effects yet specific mechanisms. Adv. Exp. Med. Biol. 2007, 603, 106–115. [Google Scholar] [PubMed]
- Gugliucci, A. Polyamines as clinical laboratory tools. Clin. Chim. Acta 2004, 344, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Piletz, J.E.; Aricioglu, F.; Cheng, J.T.; Fairbanks, C.A.; Gilad, V.H.; Haenisch, B.; Halaris, A.; Hong, S.; Lee, J.E.; Li, J.; et al. Agmatine: Clinical applications after 100 years in translation. Drug Discov. Today 2013, 18, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Wade, C.L.; Eskridge, L.L.; Nguyen, H.O.; Kitto, K.F.; Stone, L.S.; Wilcox, G.; Fairbanks, C.A. Immunoneutralization of agmatine sensitizes mice to micro-opioid receptor tolerance. J. Pharmacol. Exp. Ther. 2009, 331, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Reis, D.J.; Regunathan, S. Agmatine: An endogenous ligand at imidazoline receptors is a novel neurotransmitter. Ann. N. Y. Acad. Sci. 1999, 881, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Regunathan, S.; Piletz, J.E. Regulation of inducible nitric oxide synthase and agmatine synthesis in macrophages and astrocytes. Ann. N. Y. Acad. Sci. 2003, 1009, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Mun, C.H.; Lee, W.T.; Park, K.A.; Lee, J.E. Regulation of endothelial nitric oxide synthase by agmatine after transient global cerebral ischemia in rat brain. Anat. Cell Biol. 2010, 43, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Janowitz, T.; Kneifel, H.; Piotrowski, M. Identification and characterization of plant agmatine iminohydrolase, the last missing link in polyamine biosynthesis of plants. FEBS Lett. 2003, 544, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Jubault, M.; Hamon, C.; Gravot, A.; Lariagon, C.; Delourme, R.; Bouchereau, A.; Manzanares-Dauleux, M.J. Differential regulation of root arginine catabolism and polyamine metabolism in clubroot-susceptible and partially resistant Arabidopsis genotypes. Plant Physiol. 2008, 146, 2008–2019. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.H.; Lu, C.D. Polyamine effects on antibiotic susceptibility in bacteria. Antimicrob. Agents Chemother. 2007, 51, 2070–2077. [Google Scholar] [CrossRef] [PubMed]
- Patel, C.N.; Wortham, B.W.; Lines, J.L.; Fetherston, J.D.; Perry, R.D.; Oliveira, M.A. Polyamines are essential for the formation of plague biofilm. J. Bacteriol. 2006, 188, 2355–2363. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.J.; Du, R.H.; Calcutt, M.W.; Abdolrasulnia, R.; Christman, B.W.; Blackwell, T.S. Discovery of an operon that participates in agmatine metabolism and regulates biofilm formation in Pseudomonas aeruginosa. Mol. Microbiol. 2010, 76, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Chou, H.T.; Kwon, D.H.; Hegazy, M.; Lu, C.D. Transcriptome analysis of agmatine and putrescine catabolism in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2008, 190, 1966–1975. [Google Scholar] [CrossRef] [PubMed]
- Nakada, Y.; Jiang, Y.; Nishijyo, T.; Itoh, Y.; Lu, C.D. Molecular characterization and regulation of the aguBA operon, responsible for agmatine utilization in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2001, 183, 6517–6524. [Google Scholar] [CrossRef] [PubMed]
- Nakada, Y.; Itoh, Y. Identification of the putrescine biosynthetic genes in Pseudomonas aeruginosa and characterization of agmatine deiminase and N-carbamoylputrescine amidohydrolase of the arginine decarboxylase pathway. Microbiology 2003, 149, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Rahme, L.G.; Stevens, E.J.; Wolfort, S.F.; Shao, J.; Tompkins, R.G.; Ausubel, F.M. Common virulence factors for bacterial pathogenicity in plants and animals. Science 1995, 268, 1899–1902. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.T.; Karkhoff-Schweizer, R.R.; Kutchma, A.J.; Schweizer, H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 1998, 212, 77–86. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Baldini, R.L.; Deziel, E.; Saucier, M.; Zhang, Q.; Liberati, N.T.; Lee, D.; Urbach, J.; Goodman, H.M.; Rahme, L.G. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc. Natl. Acad. Sci. USA 2004, 101, 2530–2535. [Google Scholar] [CrossRef] [PubMed]
- De Lorenzo, V.; Timmis, K.N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994, 235, 386–405. [Google Scholar] [PubMed]
- Becher, A.; Schweizer, H.P. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 2000, 29, 948–950, 952. [Google Scholar] [PubMed]
- Hylden, J.L.; Wilcox, G.L. Intrathecal morphine in mice: A new technique. Eur. J. Pharmacol. 1980, 67, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Fairbanks, C.A. Spinal delivery of analgesics in experimental models of pain and analgesia. Adv. Drug Deliv. Rev. 2003, 55, 1007–1041. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.W.; Mahajan-Miklos, S.; Ausubel, F.M. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 1999, 96, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Yull, F.E.; Han, W.; Jansen, E.D.; Everhart, M.B.; Sadikot, R.T.; Christman, J.W.; Blackwell, T.S. Bioluminescent detection of endotoxin effects on HIV-1 LTR-driven transcription in vivo. J. Histochem. Cytochem. 2003, 51, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Bocan, T.M.; Panchal, R.G.; Bavari, S. Applications of in vivo imaging in the evaluation of the pathophysiology of viral and bacterial infections and in development of countermeasures to BSL3/4 pathogens. Mol. Imaging Biol. 2014. [Google Scholar] [CrossRef]
- Van Oosten, M.; Schafer, T.; Gazendam, J.A.; Ohlsen, K.; Tsompanidou, E.; de Goffau, M.C.; Harmsen, H.J.; Crane, L.M.; Lim, E.; Francis, K.P.; et al. Real-time in vivo imaging of invasive- and biomaterial-associated bacterial infections using fluorescently labelled vancomycin. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef]
- Williams, B.J.; Dehnbostel, J.; Blackwell, T.S. Pseudomonas aeruginosa: Host defence in lung diseases. Respirology 2010, 15, 1037–1056. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gilbertsen, A.; Williams, B. Development of a Pseudomonas aeruginosa Agmatine Biosensor. Biosensors 2014, 4, 387-402. https://doi.org/10.3390/bios4040387
Gilbertsen A, Williams B. Development of a Pseudomonas aeruginosa Agmatine Biosensor. Biosensors. 2014; 4(4):387-402. https://doi.org/10.3390/bios4040387
Chicago/Turabian StyleGilbertsen, Adam, and Bryan Williams. 2014. "Development of a Pseudomonas aeruginosa Agmatine Biosensor" Biosensors 4, no. 4: 387-402. https://doi.org/10.3390/bios4040387




