Mechanisms of Cellular Internalization of Quantum Dot® Conjugated Bone Formation Mimetic Peptide CK2.3
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. CK2.3 Peptide
2.1.2. Carboxyl Quantum dot®s (Qdot®s)
2.2. Methods
2.2.1. Conjugation of CK2.3 to Qdot®s
2.2.2. Size Exclusion Chromatography (SEC)
2.2.3. UV/VIS Spectroscopy
2.2.4. FTIR Spectroscopy
2.2.5. Cell Culture
2.2.6. Von Kossa Assay
2.2.7. Immunofluorescence Labelling of C2C12 Cells and Analysis
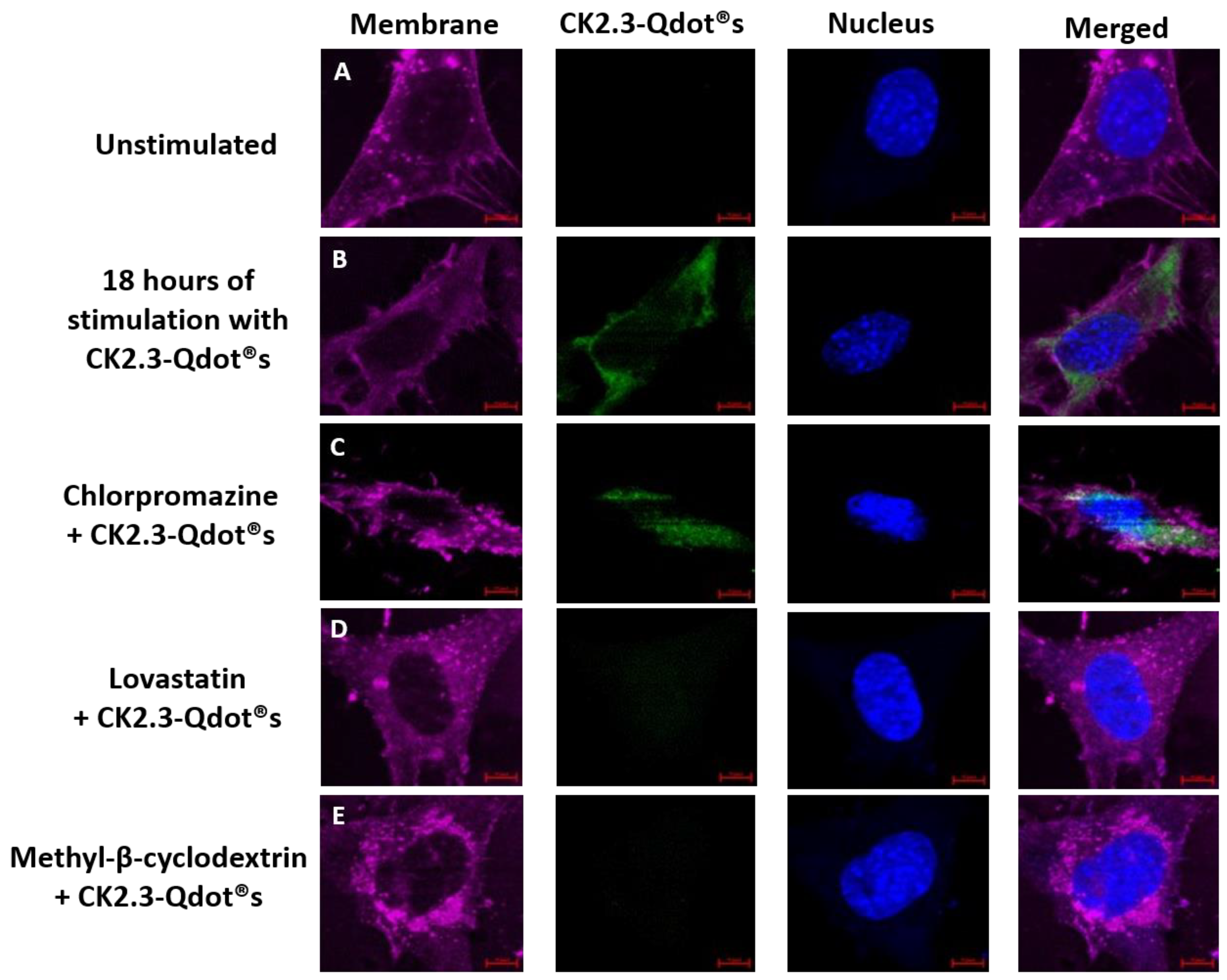
2.2.8. Inhibition of CCPs- and Caveolae-Mediated Endocytosis
3. Results
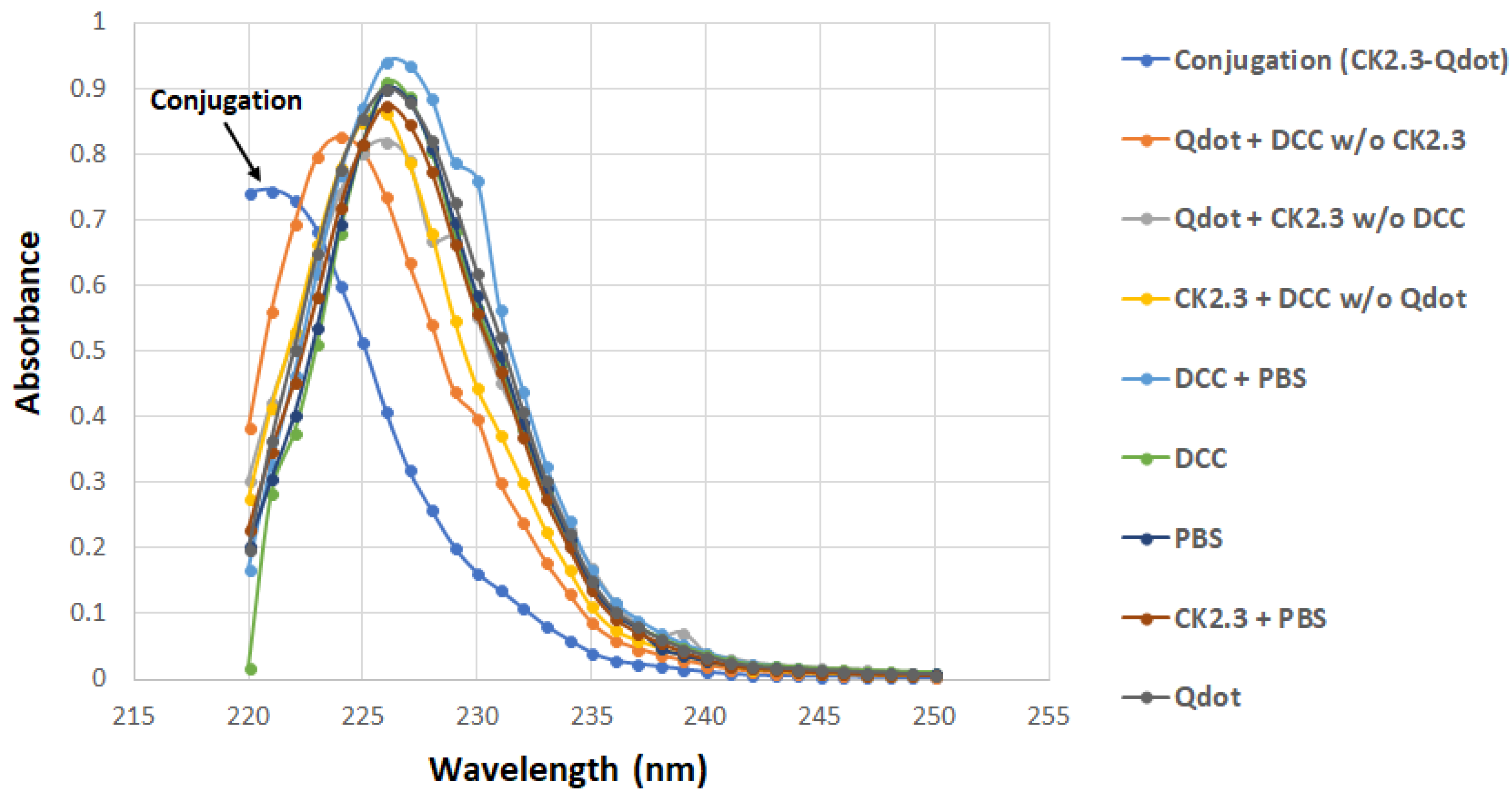
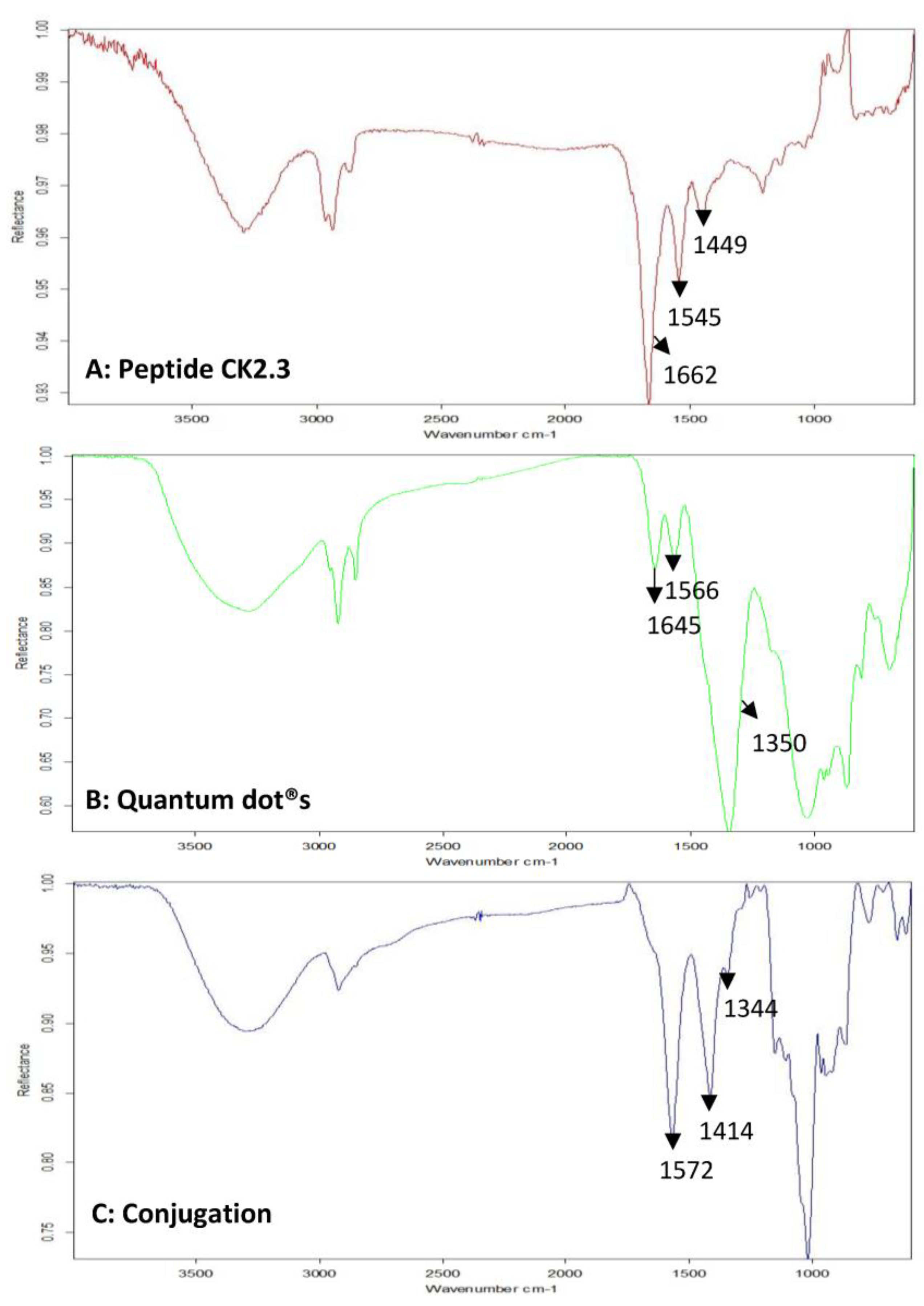
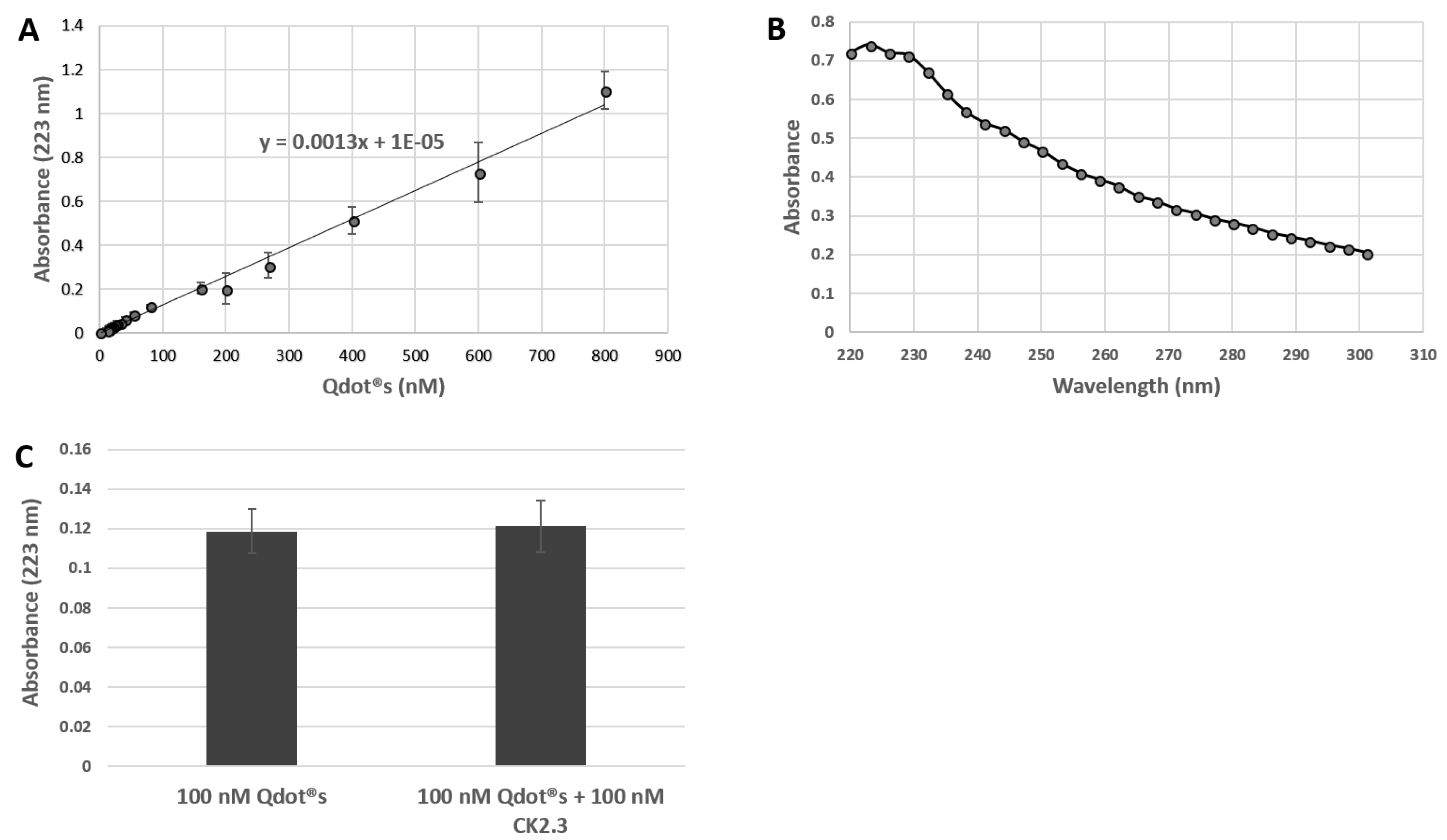
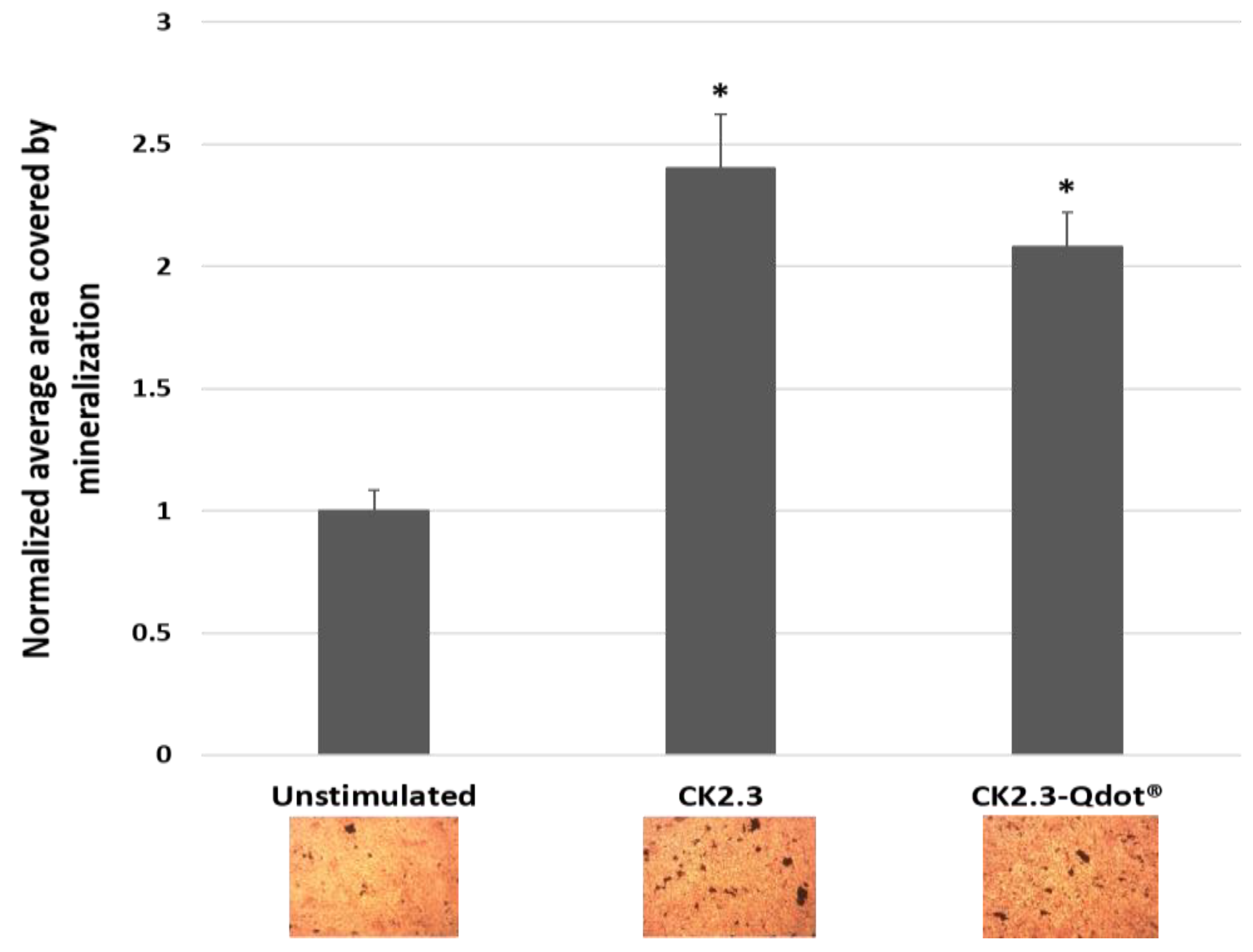
3.1. Development of a Biologically Active and Fluorescently Tagged CK2.3-Qdot®s Probe
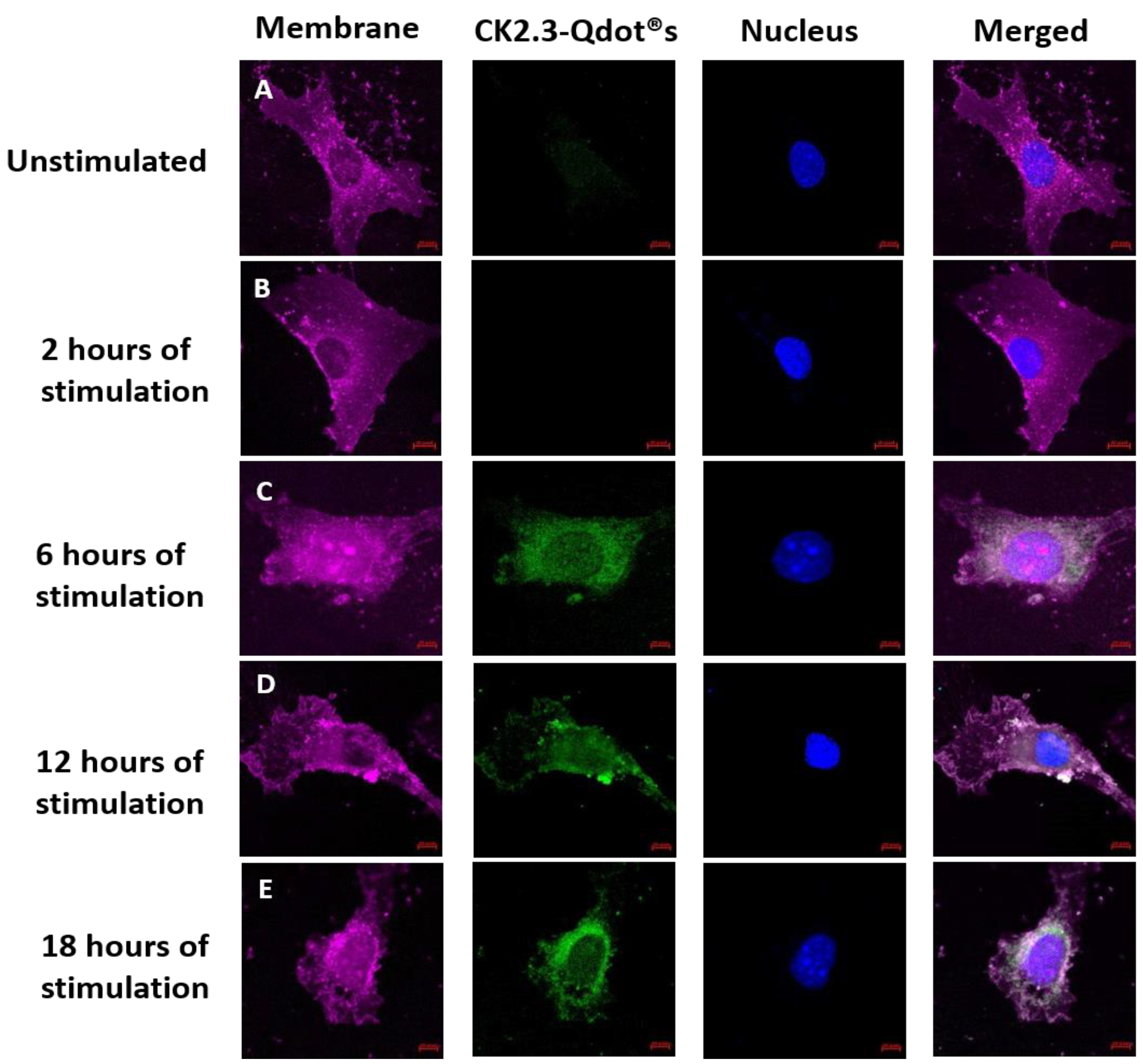
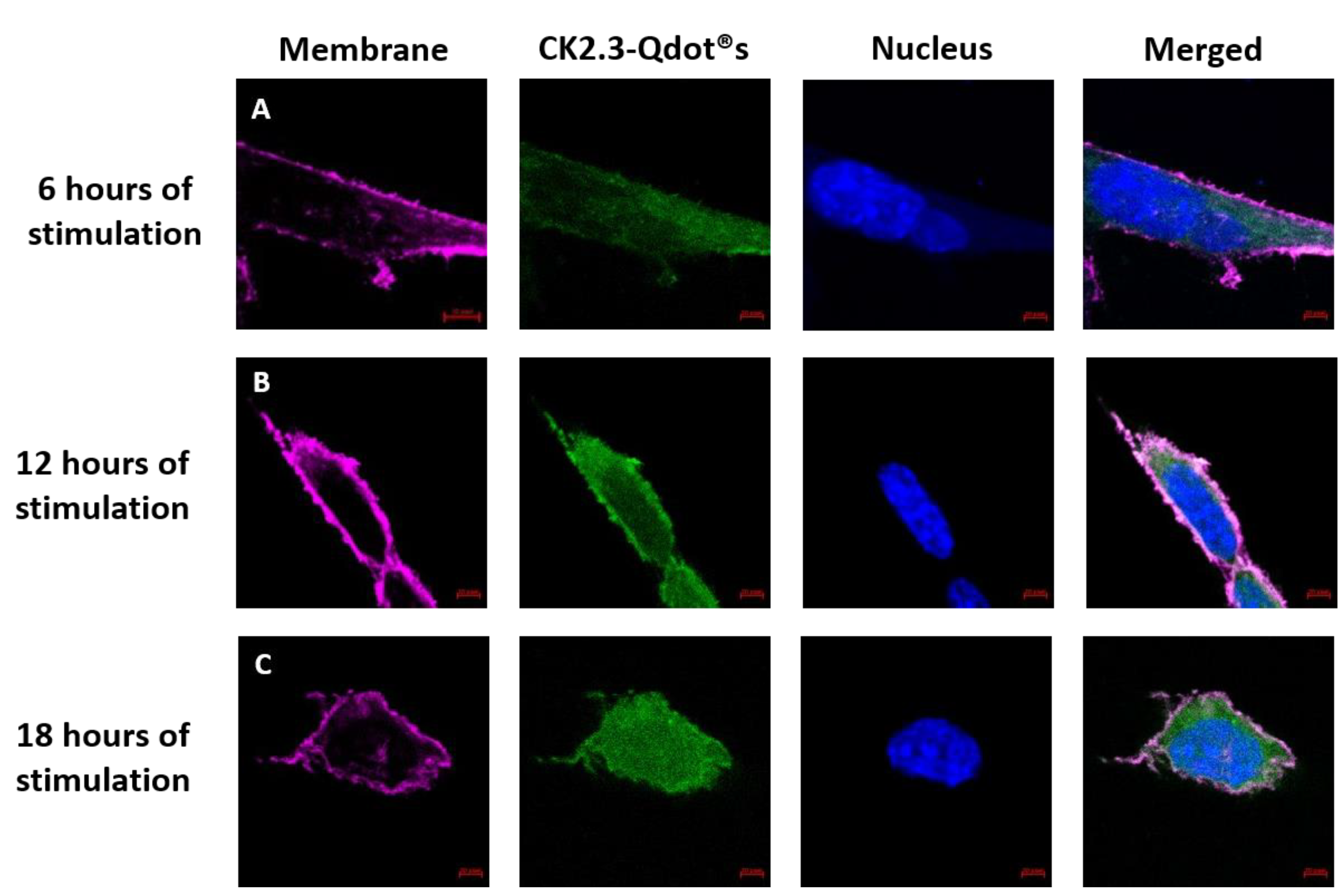
3.2. Time-Dependent Uptake of CK2.3-Qdot®s by C2C12 Cells
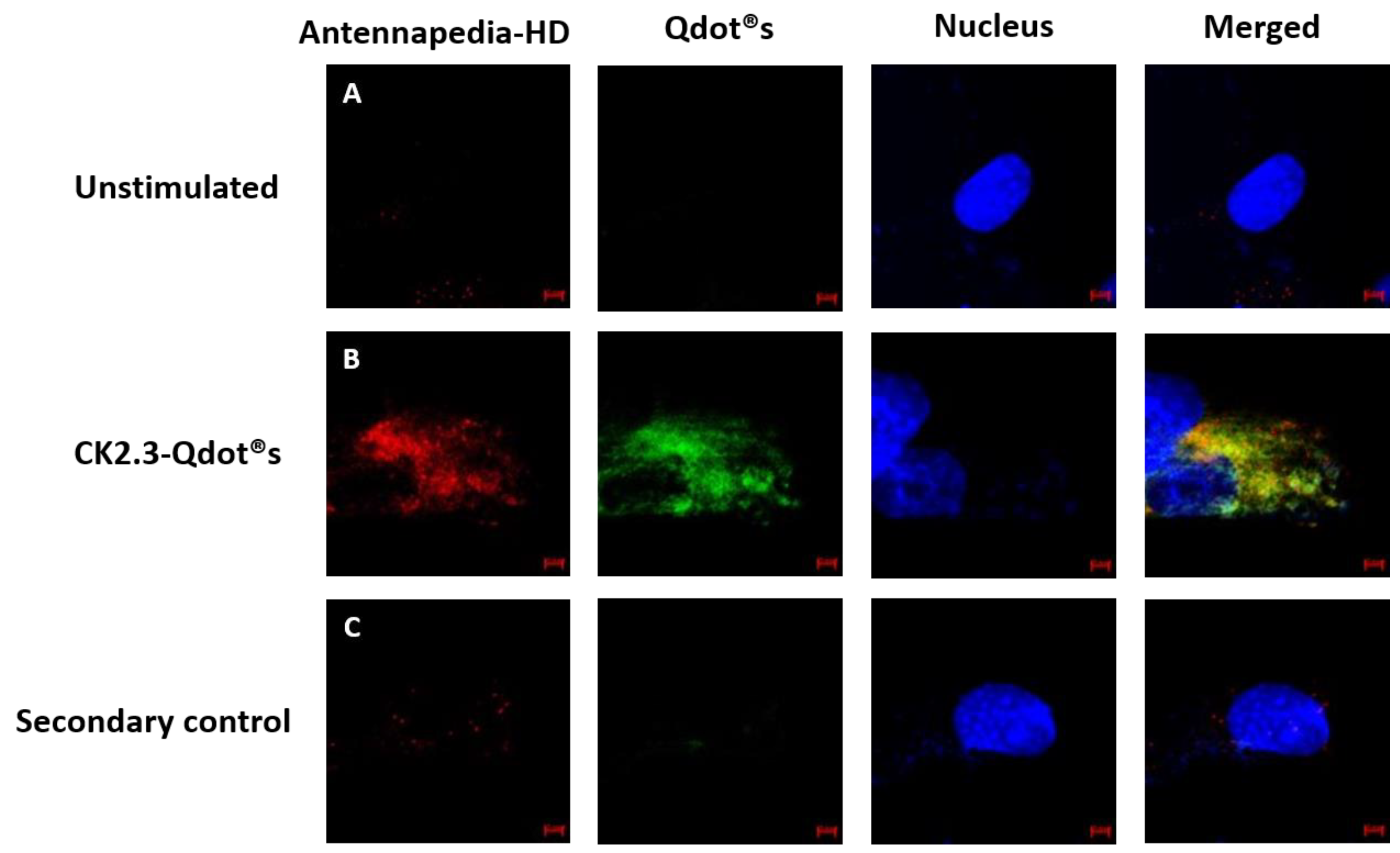
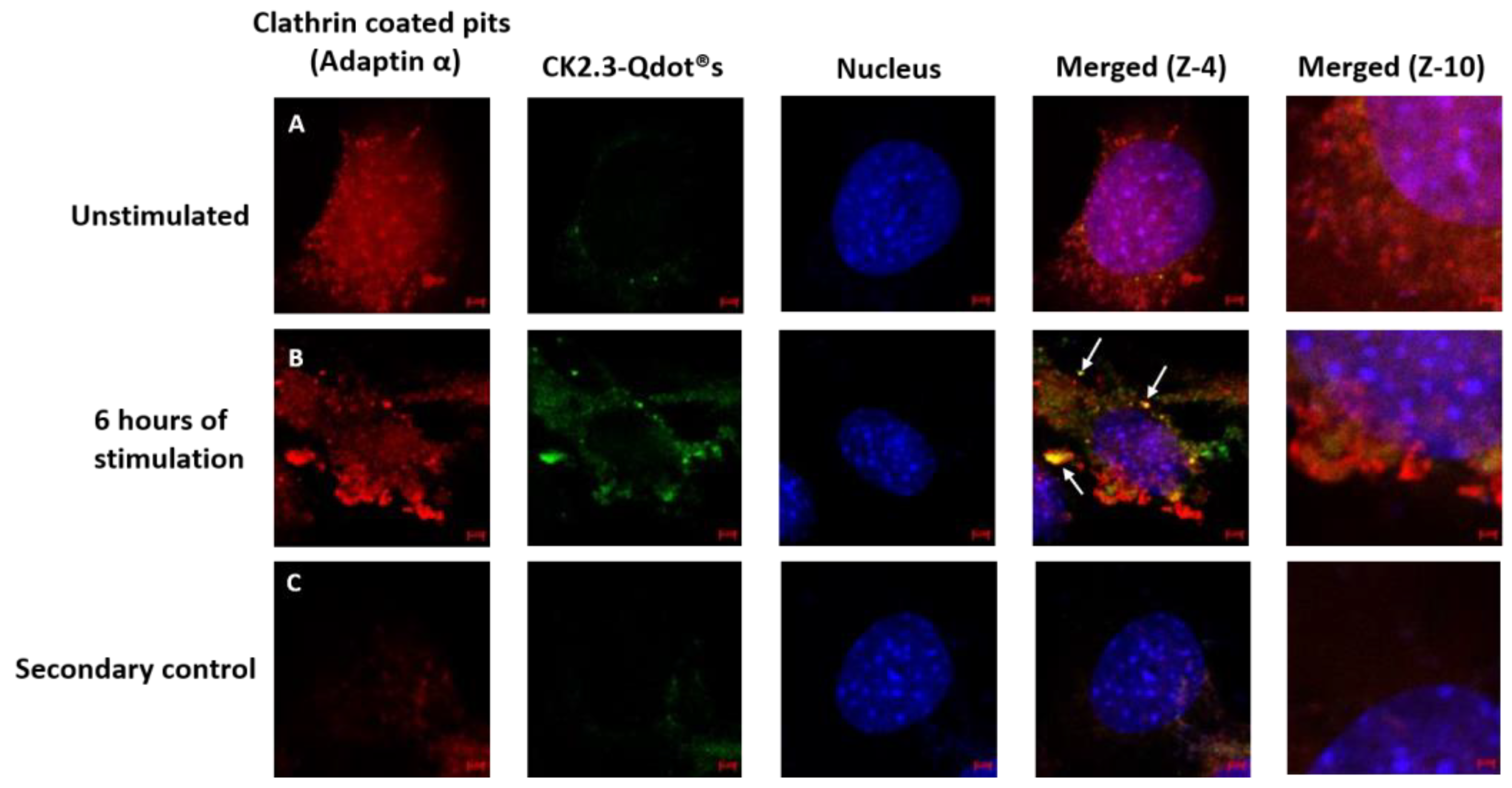
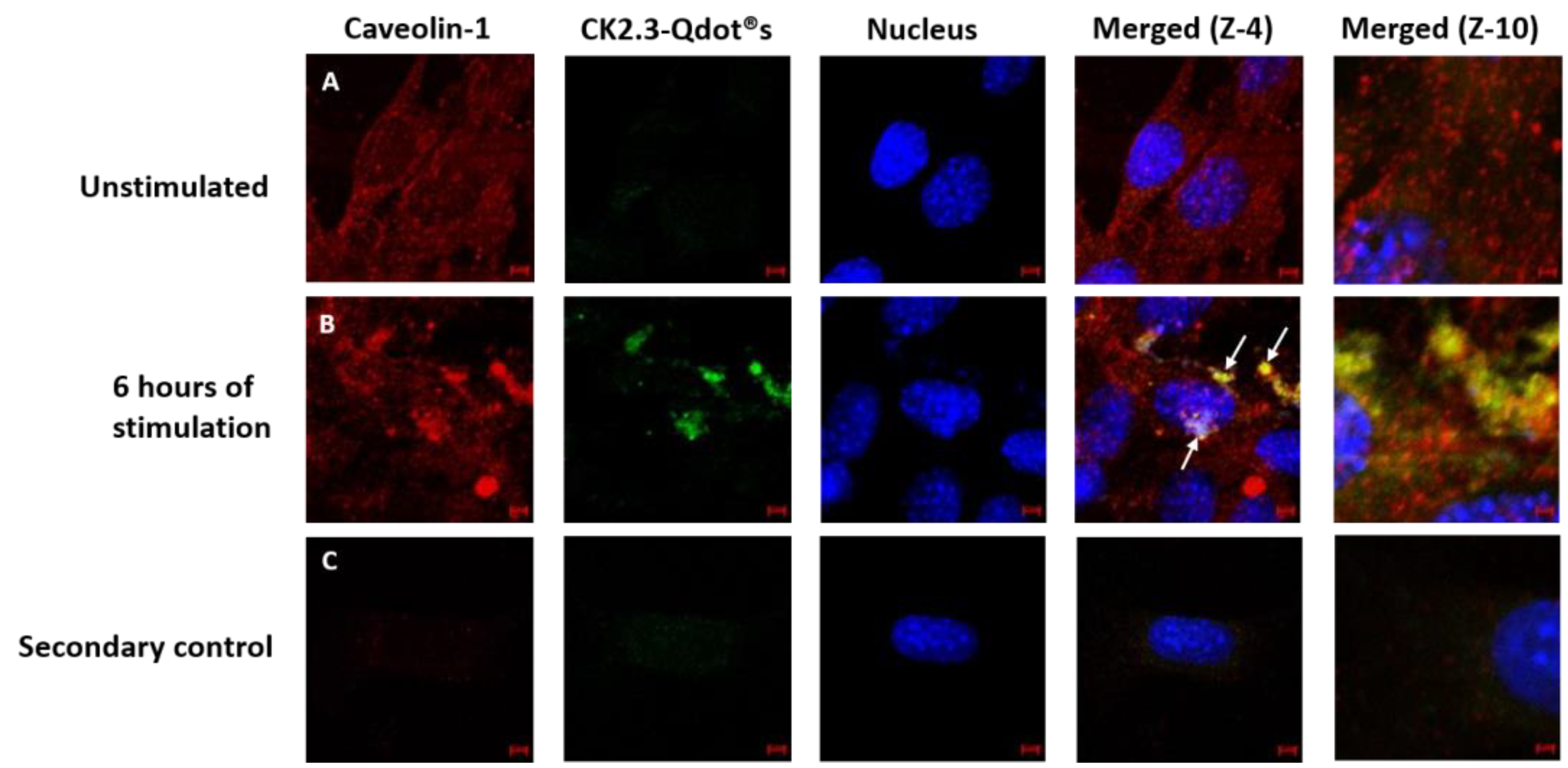
3.3. Caveolae Mediated Endocytosis Is Utilized by CK2.3-Qdot®s for Its Internalization
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cosman, F.; de Beur, S.J.; LeBoff, M.S.; Lewiecki, E.M.; Tanner, B.; Randall, S.; Lindsay, R. Clinician’s Guide to Prevention and Treatment of Osteoporosis. Osteoporos. Int. 2014, 25, 2359–2381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christodoulou, C.; Cooper, C. What is osteoporosis? Postgrad. Med. J. 2003, 79, 133–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cauley, J.A. Public Health Impact of Osteoporosis. J. Gerontol. Ser. A 2013, 68, 1243–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosman, F.; Lindsay, R. Therapeutic potential of parathyroid hormone. Curr. Osteoporos. Rep. 2004, 2, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Drake, M.T.; Clarke, B.L.; Khosla, S. Bisphosphonates: Mechanism of action and role in clinical practice, Mayo Clinic Proceedings. Mayo Clin. Proceed. 2008, 83, 1032–1045. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, M.; Smith, C.L. Molecular mechanisms of selective estrogen receptor modulator (SERM) action. J. Pharmacol. Exp. Ther. 2000, 295, 431–437. [Google Scholar] [PubMed]
- Beck, G.R.J.; Sullivan, E.C.; Moran, E.; Zerler, B. Relationship between alkaline phosphatase levels, osteopontin expression, and mineralization in differentiating MC3T3-E1 osteoblasts. J. Cell. Biochem. 1998, 68, 269–280. [Google Scholar] [CrossRef]
- Jeremiah, M.P.; Unwin, B.K.; Greenawald, M.H.; Casiano, V.E. Diagnosis and management of osteoporosis. Am. Fam. Physician 2015, 92, 261–268. [Google Scholar] [PubMed]
- Meggio, F.; Pinna, L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003, 17, 349–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litchfield, D.W. Protein kinase CK2: structure, Regulation and role in cellular decisions of life and death. Biochem. J. 2003, 369, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Turowec, J.P.; Duncan, J.S.; French, A.C.; Gyenis, L.; St. Denis, N.A.; Vilk, G.; Litchfield, D.W. Protein kinase CK2 is a constitutively active enzyme that promotes cell survival: strategies to identify CK2 substrates and manipulate its activity in mammalian cells. Methods Enzymol. 2010, 484, 471–493. [Google Scholar] [PubMed]
- Gotz, C.; Montenarh, M. Protein kinase CK2 in development and differentiation. Biomed Rep. 2017, 6, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Guerra, B.; Issinger, O.G. Protein kinase CK2 and its role in cellular proliferation, development and pathology. Electrophoresis 1999, 20, 391–408. [Google Scholar] [CrossRef]
- Bragdon, B.; Thinakaran, S.; Moseychuk, O.; King, D.; Young, K.; Litchfield, D.W.; Petersen, N.O.; Nohe, A. Casein kinase 2 beta-subunit is a regulator of bone morphogenetic protein 2 signaling. Biophys. J. 2010, 99, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Moseychuk, O.; Akkiraju, H.; Dutta, J.; D’Angelo, A.; Bragdon, B.; Duncan, R.L.; Nohe, A. Inhibition of CK2 binding to BMPRIa induces C2C12 differentiation into osteoblasts and adipocytes. J. Cell. Commun. Signal 2013, 7, 265–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bragdon, B.; Thinakaran, S.; Moseychuk, O.; Gurski, L.; Bonor, J.; Price, C.; Wang, L.; Beamer, W.G.; Nohe, A. Casein kinase 2 regulates in vivo bone formation through its interaction with bone morphogenetic protein receptor type Ia. Bone 2011, 49, 944–954. [Google Scholar] [CrossRef] [PubMed]
- Lisberg, A.; Ellis, R.; Nicholson, K.; Moku, P.; Swarup, A.; Dhurjati, P.; Nohe, A. Mathematical modeling of the effects of CK2. 3 on mineralization in osteoporotic bone. CPT: Pharmacomet. Syst. Pharmacol. 2017, 6, 208–215. [Google Scholar]
- Akkiraju, H.; Bonor, J.; Olli, K.; Bowen, C.; Bragdon, B.; Coombs, H.; Donahue, L.R.; Duncan, R.; Nohe, A. Systemic injection of CK2.3, a novel peptide acting downstream of bone morphogenetic protein receptor BMPRIa, leads to increased trabecular bone mass. J. Orthop. Res. 2015, 33, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Johnson, I. Molecular Probes Handbook, 11th ed.; Life Technologies Corporation: Carlsbad, CA, USA, 2010. [Google Scholar]
- Mukherjee, S.; Menegazzo, N.; Booksh, K.; Dhurjati, P.; Smorodin, V.; Nohe, A. Synthesis of L-Cysteine stabilized silver nanoparticles and their effects on cell Vviability. Adv. Sci. Lett. 2012, 6, 26–33. [Google Scholar] [CrossRef]
- Bonor, J.; Reddy, V.; Akkiraju, H.; Dhurjati, P.; Nohe, A. Synthesis and characterization of L-Lysine conjugated silver nanoparticles smaller than 10 nM. Adv. Sci. Eng. Med. 2014, 6, 942–947. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, T.; Bakhshi, R.; Petrova, D.; Pocock, R.; Imani, M.; Seifalian, A.M. Biological applications of quantum dots. Biomaterials 2007, 28, 4717–4732. [Google Scholar] [CrossRef] [PubMed]
- Medintz, I.L.; Mattoussi, H.; Clapp, A.R. Potential clinical applications of quantum dots. Int. J. Nanomed. 2008, 3, 151–167. [Google Scholar]
- Resch-Genger, U.; Grabolle, M.; Cavaliere-Jaricot, S.; Nitschke, R.; Nann, T. Quantum dots versus organic dyes as fluorescent labels. Nat. Meth. 2008, 5, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.; Dent, A. Bioconjugation, 1st ed.; Macmillan: London, UK, 1998. [Google Scholar]
- Ruosi, C.; Querques, F.; Granata, F.; Colella, G.; Liccardo, S.; Lombardo, B.; Pastore, L. Cellular and animal models for the identification of osteoporosis determinants increasing vertebral compression fractures risk. J. Osteopor. Phys. Act. 2015, 3, 2. [Google Scholar]
- Yang, Q.; Jian, J.; Abramson, S.B.; Huang, X. Inhibitory effects of iron on bone morphogenetic protein 2–induced osteoblastogenesis. J. Bone Miner. Res. 2011, 26, 1188–1196. [Google Scholar] [CrossRef] [PubMed]
- Kartsogiannis, V.; Ng, K.W. Cell lines and primary cell cultures in the study of bone cell biology. Mol. Cell. Endocrinol. 2004, 228, 79–102. [Google Scholar] [CrossRef] [PubMed]
- Bilezikian, J.P. Principles of Bone Biology, 2nd ed.; Acad. Press: San Diego, CA, USA, 2002. [Google Scholar]
- Namiki, M.; Akiyama, S.; Katagiri, T.; Suzuki, A.; Ueno, N.; Yamaji, N.; Rosen, V.; Wozney, J.M.; Suda, T. A kinase domain-truncated type I receptor blocks bone morphogenetic protein-2-induced signal transduction in C2C12 myoblasts. J. Biol. Chem. 1997, 272, 22046–22052. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, J.K.; Mattoussi, H.; Mauro, J.M.; Simon, S.M. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat. Biotechnol. 2003, 21, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Harush-Frenkel, O.; Rozentur, E.; Benita, S.; Altschuler, Y. Surface charge of nanoparticles determines their endocytic and transcytotic pathway in polarized MDCK cells. Biomacromolecules 2008, 9, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.C.; Nie, S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science 1998, 281, 2016–2018. [Google Scholar] [CrossRef] [PubMed]
- Lidke, D.S.; Nagy, P.; Heintzmann, R.; Arndt-Jovin, D.J.; Post, J.N.; Grecco, H.E.; Jares-Erijman, E.A.; Jovin, T.M. Quantum dot ligands provide new insights into erbB/HER receptor–mediated signal transduction. Nat. Biotechnol. 2004, 22, 198. [Google Scholar] [CrossRef] [PubMed]
- Bharali, D.J.; Lucey, D.W.; Jayakumar, H.; Pudavar, H.E.; Prasad, P.N. Folate-receptor-mediated delivery of InP quantum dots for bioimaging using confocal and two-photon microscopy. J. Am. Chem. Soc. 2005, 127, 11364–11371. [Google Scholar] [CrossRef] [PubMed]
- Derossi, D.; Calvet, S.; Trembleau, A.; Brunissen, A.; Chassaing, G.; Prochiantz, A. Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J. Biol. Chem. 1996, 271, 18188–18193. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.W.; Monteiro-Riviere, N.A. Mechanisms of quantum dot nanoparticle cellular uptake. Toxicol. Sci. 2009, 110, 138–155. [Google Scholar] [CrossRef] [PubMed]
- Nohe, A.; Petersen, N.O. Image Correlation Spectroscopy. Sci. STKE 2007, 417, pl7. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Zhu, Y.L.; Zhou, Y.Y.; Liang, G.F.; Wang, Y.Y.; Hu, F.H.; Xiao, Z.D. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J. Biol. Chem. 2014, 289, 22258–22267. [Google Scholar] [CrossRef] [PubMed]
- Vercauteren, D.; Vandenbroucke, R.E.; Jones, A.T.; Rejman, J.; Demeester, J.; De Smedt, S.C.; Sanders, N.N.; Braeckmans, K. The use of inhibitors to study endocytic pathways of gene carriers: optimization and pitfalls. Mol. Ther. 2010, 18, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Hartung, A.; Bitton-Worms, K.; Rechtman, M.M.; Wenzel, V.; Boergermann, J.H.; Hassel, S.; Henis, Y.I.; Knaus, P. Different routes of bone morphogenic protein (BMP) receptor endocytosis influence BMP signaling. Mol. Cell. Biol. 2006, 26, 7791–7805. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, J.L.; Basu, S.K.; Brown, M.S. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 1983, 98, 241–260. [Google Scholar] [PubMed]
- Petersen, E.N.; Chung, H.; Nayebosadri, A.; Hansen, S.B. Kinetic disruption of lipid rafts is a mechanosensor for phospholipase D. Nat. Commun. 2016, 7, 13873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, P.; Koza, S.; Bouvier, E.S.P. Size-exclusion chromatography for the analysis of protein biotherapeutics and their aggregates. J. Liq. Chromatogr. Relat. Technol. 2012, 35, 2923–2950. [Google Scholar] [PubMed]
- Nielsen, E.B.; Schellman, J.A. The absorption spectra of simple amides and peptides. J. Phys. Chem. 1967, 71, 2297–2304. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Vodnar, D.C.; Paucean, A.; Dulf, F.V.; Socaciu, C. HPLC characterization of lactic acid formation and FTIR fingerprint of probiotic bacteria during fermentation processes. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 2010, 38, 109. [Google Scholar]
- Coates, J. Interpretation of infrared spectra, a practical approach. In Encyclopedia of Analytical Chemistry, 1st ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2000; Volume 12, pp. 10815–10837. [Google Scholar]
- Gershevitz, O.; Sukenik, C.N. In situ FTIR-ATR analysis and titration of carboxylic acid-terminated SAMs. J. Am. Chem. Soc. 2004, 126, 482–483. [Google Scholar] [CrossRef] [PubMed]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [PubMed]
- Haris, P.I.; Robillard, G.T.; van Dijk, A.A.; Chapman, D. Potential of 13C and 15N labeling for studying protein-protein interactions using Fourier transform infrared spectroscopy. Biochemistry 1992, 31, 6279–6284. [Google Scholar] [CrossRef] [PubMed]
- Myshakina, N.S.; Ahmed, Z.; Asher, S.A. Dependence of amide vibrations on hydrogen bonding. J. Phys. Chem. B 2008, 112, 11873–11877. [Google Scholar] [CrossRef] [PubMed]
- Milner-White, E. The partial charge of the nitrogen atom in peptide bonds. Protein Sci. 1997, 6, 2477–2482. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Chen, X. Preparation of peptide-conjugated quantum dots for tumor vasculature-targeted imaging. Nat. Protoc. 2008, 3, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Maye, P.; Rowe, D.W. Examination of Mineralized Nodule Formation in Living Osteoblastic Cultures Using Fluorescent Dyes. Biotechnol. Prog. 2006, 22, 1697–1701. [Google Scholar] [CrossRef] [PubMed]
- Dom, G.; Shaw-Jackson, C.; Matis, C.; Bouffioux, O.; Picard, J.J.; Prochiantz, A.; Mingeot-Leclercq, M.; Brasseur, R.; Rezsohazy, R. Cellular uptake of Antennapedia Penetratin peptides is a two-step process in which phase transfer precedes a tryptophan-dependent translocation. Nucleic Acids Res. 2002, 31, 556–561. [Google Scholar] [CrossRef]
- Joliot, A.; Pernelle, C.; Deagostini-Bazin, H.; Prochiantz, A. Antennapedia homeobox peptide regulates neural morphogenesis. Proc. Natl. Acad. Sci. USA 1991, 88, 1864–1868. [Google Scholar] [CrossRef] [PubMed]
- Perez, F.; Joliot, A.; Bloch-Gallego, E.; Zahraoui, A.; Triller, A.; Prochiantz, A. Antennapedia homeobox as a signal for the cellular internalization and nuclear addressing of a small exogenous peptide. J. Cell Sci. 1992, 102, 717–722. [Google Scholar] [PubMed]
- Derossi, D.; Joliot, A.H.; Chassaing, G.; Prochiantz, A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J. Biol. Chem. 1994, 269, 10444–10450. [Google Scholar] [PubMed]
- Rodal, S.K.; Skretting, G.; Garred, O.; Vilhardt, F.; van Deurs, B.; Sandvig, K. Extraction of cholesterol with methyl-beta-cyclodextrin perturbs formation of clathrin-coated endocytic vesicles. Mol. Biol. Cell 1999, 10, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Bonor, J.; Adams, E.L.; Bragdon, B.; Moseychuk, O.; Czymmek, K.J.; Nohe, A. Initiation of BMP2 signaling in domains on the plasma membrane. J. Cell. Physiol. 2012, 227, 2880–2888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nohe, A.; Keating, E.; Underhill, T.M.; Knaus, P.; Petersen, N.O. Effect of the distribution and clustering of the type IA BMP receptor (ALK3) with the type II BMP receptor on the activation of signalling pathways. J. Cell. Sci. 2003, 116, 3277–3284. [Google Scholar] [CrossRef] [PubMed]
- Bragdon, B.; D’Angelo, A.; Gurski, L.; Bonor, J.; Schultz, K.L.; Beamer, W.G.; Rosen, C.J.; Nohe, A. Altered plasma membrane dynamics of bone morphogenetic protein receptor type Ia in a low bone mass mouse model. Bone 2012, 50, 189–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nohe, A.; Keating, E.; Underhill, T.M.; Knaus, P.; Petersen, N.O. Dynamics and interaction of caveolin-1 isoforms with BMP-receptors. J. Cell Sci. 2005, 118, 643. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, S.; Bragdon, B.; Moseychuk, O.; Bonor, J.; Dhurjati, P.; Nohe, A. Caveolae regulate Smad signaling as verified by novel imaging and system biology approaches. J. Cell. Physiol. 2013, 228, 1060–1069. [Google Scholar] [CrossRef] [PubMed]
- Bonor, J.C.; Schaefer, R.J.; Menegazzo, N.; Booksh, K.; Nohe, A.G. Design of 1,25 dihydroxyvitamin D3 coupled quantum dots, a novel imaging tool. J. Nanosci. Nanotechnol. 2012, 12, 2185. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.S.; Bichelberger, M.A.; Colin, D.Y.; Robitaille, R.; Pelletier, J.N.; Masson, J. Monitoring methotrexate in clinical samples from cancer patients during chemotherapy with a LSPR-based competitive sensor. Analyst 2012, 137, 4742–4750. [Google Scholar] [CrossRef] [PubMed]
- McMahon, H.T.; Boucrot, E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2011, 12, 517. [Google Scholar] [CrossRef] [PubMed]
- Nabi, I.R.; Le, P.U. Caveolae/raft-dependent endocytosis. J. Cell Biol. 2003, 161, 673–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Gehring, W. Cellular uptake of the Antennapedia homeodomain polypeptide by macropinocytosis. Biochem. Biophys. Res. Commun. 2014, 443, 1136–1140. [Google Scholar] [CrossRef] [PubMed]
- Clift, M.J.; Rothen-Rutishauser, B.; Brown, D.M.; Duffin, R.; Donaldson, K.; Proudfoot, L.; Guy, K.; Stone, V. The impact of different nanoparticle surface chemistry and size on uptake and toxicity in a murine macrophage cell line. Toxicol. Appl. Pharmacol. 2008, 232, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Colton, H.M.; Falls, J.G.; Ni, H.; Kwanyuen, P.; Creech, D.; McNeil, E.; Casey, W.M.; Hamilton, G.; Cariello, N.F. Visualization and quantitation of peroxisomes using fluorescent nanocrystals: treatment of rats and monkeys with fibrates and detection in the liver. Toxicol. Sci. 2004, 80, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Ballou, B.; Lagerholm, B.C.; Ernst, L.A.; Bruchez, M.P.; Waggoner, A.S. Noninvasive imaging of quantum dots in mice. Bioconjug. Chem. 2004, 15, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Cui, Y.; Levenson, R.M.; Chung, L.W.; Nie, S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat. Biotechnol. 2004, 22, 969. [Google Scholar] [CrossRef] [PubMed]
- Voura, E.B.; Jaiswal, J.K.; Mattoussi, H.; Simon, S.M. Tracking metastatic tumor cell extravasation with quantum dot nanocrystals and fluorescence emission-scanning microscopy. Nat. Med. 2004, 10, 993. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Hanaki, K.; Suzuki, K.; Yamamoto, K. Applications of T-lymphoma labeled with fluorescent quantum dots to cell tracing markers in mouse body. Biochem. Biophys. Res. Commun. 2004, 314, 46–53. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vrathasha, V.; Booksh, K.; Duncan, R.L.; Nohe, A. Mechanisms of Cellular Internalization of Quantum Dot® Conjugated Bone Formation Mimetic Peptide CK2.3. Nanomaterials 2018, 8, 513. https://doi.org/10.3390/nano8070513
Vrathasha V, Booksh K, Duncan RL, Nohe A. Mechanisms of Cellular Internalization of Quantum Dot® Conjugated Bone Formation Mimetic Peptide CK2.3. Nanomaterials. 2018; 8(7):513. https://doi.org/10.3390/nano8070513
Chicago/Turabian StyleVrathasha, Vrathasha, Karl Booksh, Randall L. Duncan, and Anja Nohe. 2018. "Mechanisms of Cellular Internalization of Quantum Dot® Conjugated Bone Formation Mimetic Peptide CK2.3" Nanomaterials 8, no. 7: 513. https://doi.org/10.3390/nano8070513




