Optical Biosensing System for the Detection of Survivin mRNA in Colorectal Cancer Cells Using a Graphene Oxide Carrier-Bound Oligonucleotide Molecular Beacon
Abstract
:1. Introduction
2. Results and Discussion
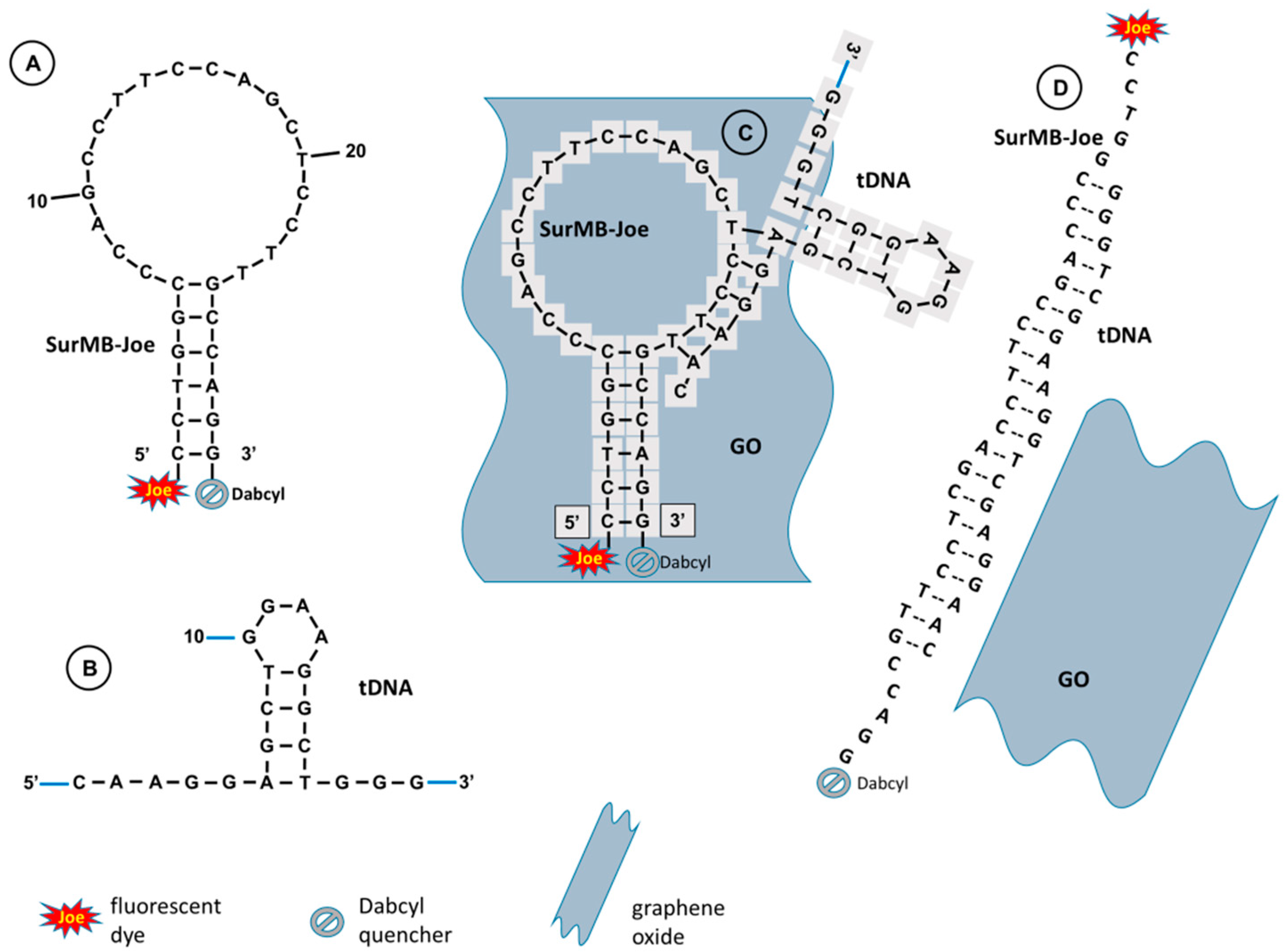
2.1. Hairpin Structures of SurMB-Joe, tDNA Target, and the Hybridization Product
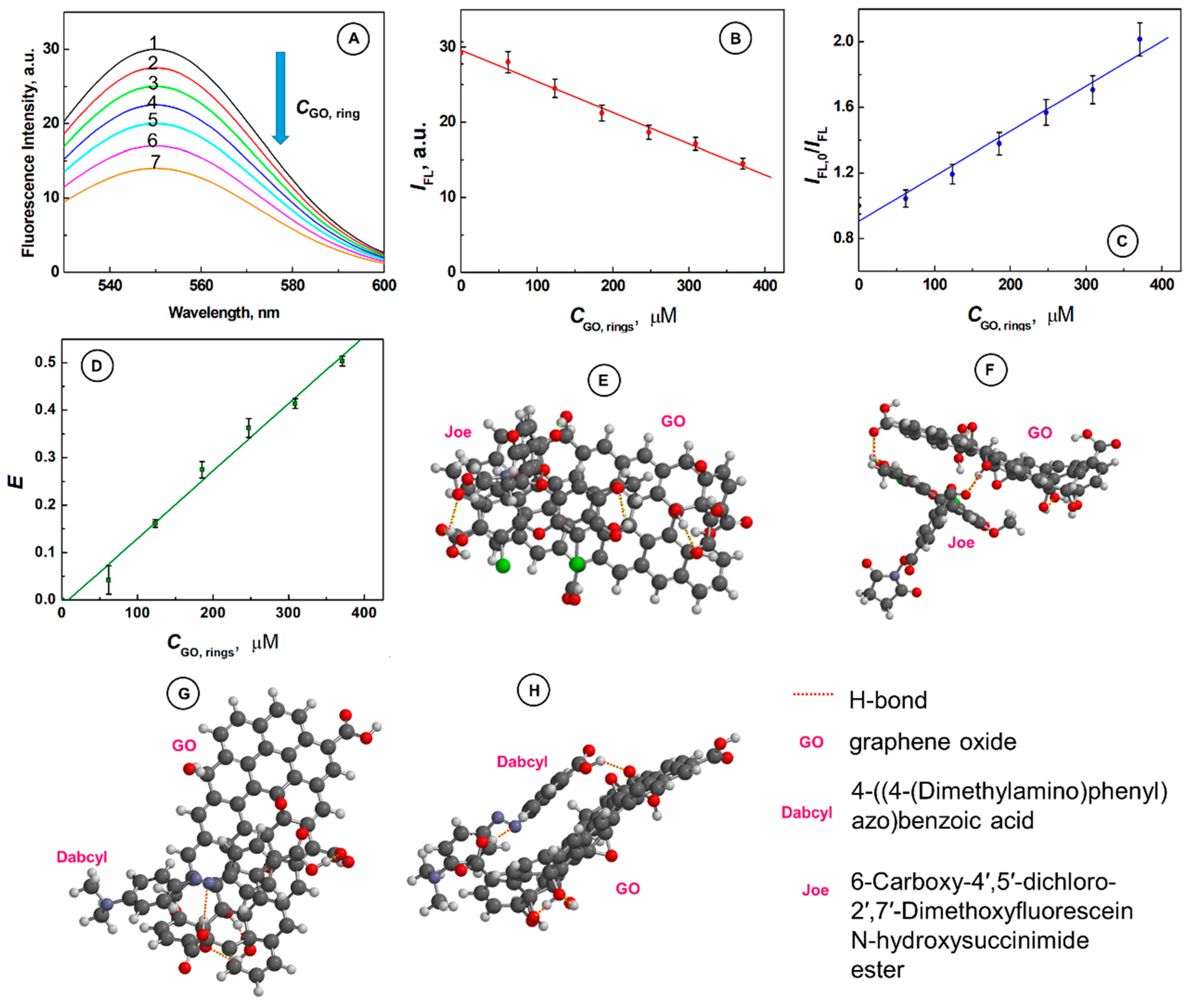
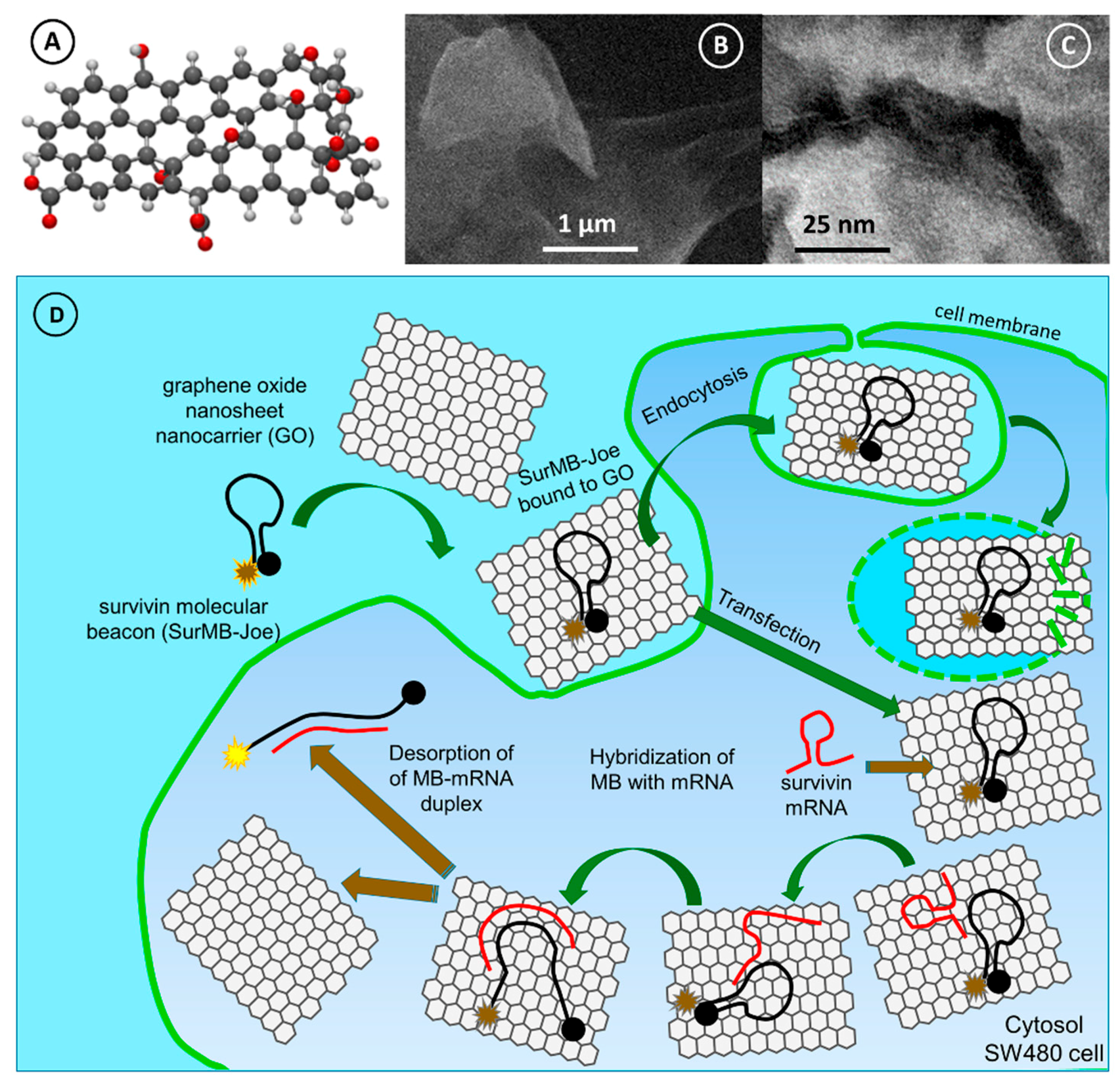
2.2. Supramolecular Interactions of SurMB-Joe Components with GO Nanocarriers
2.3. Desorptive Hybridization of GO-Bound SurMB-Joe with Complementary tDNA Target and Mutants
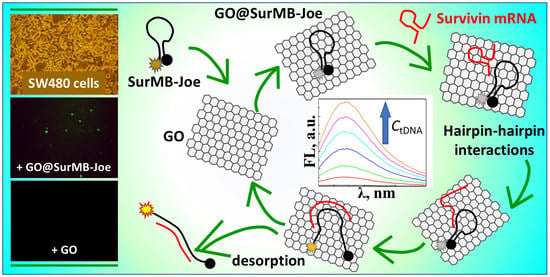
2.4. Detection of Survivin mRNA in Colorectal Cancer Cells SW480 Using GO@SurMB-Joe Nanoprobes
2.5. Mechanism of Hairpin–Hairpin Interactions in Desorptive Hybridization of GO@SurMB-Joe and tDNA
3. Materials and Methods
3.1. Chemicals
3.2. Apparatus
3.3. Cell Culture
3.4. Cell Transfection
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, B.; Ma, L.; Huang, Z.; Hu, H.; Wu, P.; Liu, J. Janus DNA orthogonal adsorption of graphene oxide and metal oxide nanoparticles enabling stable sensing in serum. Mater. Horiz. 2018, 5, 65–69. [Google Scholar] [CrossRef]
- Stobiecka, M.; Dworakowska, B.; Jakiela, S.; Lukasiak, A.; Chalupa, A.; Zembrzycki, K. Sensing of survivin mRNA in malignant astrocytes using graphene oxide nanocarrier-supported oligonucleotide molecular beacons. Sens. Actuators B 2016, 235, 136–145. [Google Scholar] [CrossRef]
- Park, J.S.; Goo, N.-I.; Kim, D.-E. Mechanism of DNA adsorption and desorption on graphene oxide. Langmuir 2014, 30, 12587–12595. [Google Scholar] [CrossRef] [PubMed]
- Stobiecka, M.; Hepel, M. Double-shell gold nanoparticle-based DNA-carriers with poly-L-lysine binding surface. Biomaterials 2011, 32, 3312–3321. [Google Scholar] [CrossRef] [PubMed]
- Hepel, M.; Stobiecka, M. Detection of oxidative stress biomarkers using functional gold nanoparticles. In Fine Particles in Medicine and Pharmacy; Matijević, E., Ed.; Springer: Boston, MA, USA, 2012; pp. 241–281. [Google Scholar]
- Ding, Y.; Jiang, Z.; Saha, K.; Kim, C.S.; Kim, S.T.; Landis, R.F.; Rotello, V.M. Gold nanoparticles for nucleic acid delivery. Mol. Ther. 2014, 22, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.H.; Yoo, H.J.; Kwon, Y.H.; Yoon, H.Y.; Lee, S.G.; Kim, S.R.; Yeom, D.W.; Kang, M.J.; Choi, Y.W. Design of multifunctional liposomal nanocarriers for folate receptor-specific intracellular drug delivery. Mol. Pharm. 2015, 12, 4200–4213. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, K.; Krazinski, B.E.; Kowalczyk, A.E.; Dworakowska, B.; Jakiela, S.; Stobiecka, M. Hairpin–hairpin molecular beacon interactions for detection of survivin mRNA in malignant sw480 cells. ACS Appl. Mater. Interfaces 2018, 10, 17028–17039. [Google Scholar] [CrossRef] [PubMed]
- Pattni, B.S.; Chupin, V.V.; Torchilin, V.P. New developments in liposomal drug delivery. Chem. Rev. 2015, 115, 10938–10966. [Google Scholar] [CrossRef] [PubMed]
- Skalickova, S.; Nejdl, L.; Kudr, J.; Ruttkay-Nedecky, B.; Jimenez, A.M.J.; Kopel, P.; Kremplova, M.; Masarik, M.; Stiborova, M.; Eckschlager, T.; et al. Fluorescence characterization of gold modified liposomes with antisense N-myc DNA bound to the magnetisable particles with encapsulated anticancer drugs (doxorubicin, ellipticine and etoposide). Sensors (Basel) 2016, 16, 290. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Zhang, L.; Yang, T.H.; Wu, H. Stimuli-responsive polymeric micelles for drug delivery and cancer therapy. Int. J. Nanomed. 2018, 13, 2921–2942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stobiecka, M. Exosomes and microvesicles: Extracellular frontiers of intercellular communication. In Encyclopedia of Surface and Colloid Science, 3rd ed.; Somasundaran, P., Ed.; CRC Press, Taylor & Francis: New York, NY, USA, 2016. [Google Scholar]
- Ratajczak, K.; Stobiecka, M. Ternary interactions and energy transfer between fluorescein isothiocyanate, adenosine triphosphate, and graphene oxide nanocarriers. J. Phys. Chem. B 2017, 121, 6822–6830. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Chen, Y.; Li, S.; Lin, S.; Jiang, Y. A graphene-based biosensing platform based on regulated release of an aptameric DNA biosensor. Sensors 2015, 15, 28244–28256. [Google Scholar] [CrossRef] [PubMed]
- Rana, M.; Balcioglu, M.; Robertson, N.; Yigit, M.V. Nano-graphene oxide as a novel platform for monitoring the effect of LNA modification on nucleic acid interactions. Analyst 2014, 139, 714–720. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Song, Y.; Ke, G.; Guan, Z.; Zhang, H.; Lin, Y.; Huang, Y.; Zhu, Z.; Yang, C.J. Graphene oxide protected nucleic acid probes for bioanalysis and biomedicine. Chem. Eur. J. 2013, 19, 10442–10451. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.; Hu, D.; Lin, C.-T.; Li, J.; Lin, Y. Aptamer/graphene oxide nanocomplex for in situ molecular probing in living cells. J. Am. Chem. Soc. 2010, 132, 9274–9276. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Ding, R.; Zhao, X.; Li, Y.; Qu, L.; Pei, H.; Yildirimer, L.; Wu, Z.; Zhang, W. Graphene-based nanomaterials for drug and/or gene delivery, bioimaging, and tissue engineering. Drug Discov. Today 2017, 22, 1302–1317. [Google Scholar] [CrossRef] [PubMed]
- Fong, Y.T.; Chen, C.-H.; Chen, J.-P. Intratumoral delivery of doxorubicin on folate-conjugated graphene oxide by in-situ forming thermo-sensitive hydrogel for breast cancer therapy. Nanomaterials 2017, 7, 388. [Google Scholar] [CrossRef] [PubMed]
- Shabani, R.; Ashjari, M.; Ashtari, K.; Izadyar, F.; Behnam, B.; Khoei, S.; Asghari-Jafarabadi, M.; Koruji, M. Elimination of mouse tumor cells from neonate spermatogonial cells utilizing cisplatin-entrapped folic acid-conjugated poly (lactic-co-glycolic acid) nanoparticles in vitro. Int. J. Nanomed. 2018, 13, 2943–2954. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Xu, X.-L.; Zhang, J.-Z.; Mao, X.-H.; Xie, M.-W.; Cheng, Z.-L.; Lu, L.-J.; Duan, X.-H.; Zhang, L.-M.; Shen, J. Magnetic cationic amylose nanoparticles used to deliver survivin-small interfering RNA for gene therapy of hepatocellular carcinoma in vitro. Nanomaterials 2017, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Kramer, F.R. Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol. 1996, 14, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Kempaiah, R.; Huang, P.-J.J.; Maheshwari, V.; Liu, J. Adsorption and desorption of DNA on graphene oxide studied by fluorescently labeled oligonucleotides. Langmuir 2011, 27, 2731–2738. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, S.; Liu, B.; Wu, J.; Zhou, Y.; He, L.; Ding, J.; Liu, J. Intracellular detection of ATP using an aptamer beacon covalently linked to graphene oxide resisting nonspecific probe displacement. Anal. Chem. 2014, 86, 12229–12235. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.-J.J.; Liu, J. Molecular beacon lighting up on graphene oxide. Anal. Chem. 2012, 84, 4192–4198. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.H.; Yang, H.H.; Zhu, C.L.; Chen, X.; Chen, G.N. A graphene platform for sensing biomolecules. Angew. Chem. Int. Ed. 2009, 48, 4785–4787. [Google Scholar] [CrossRef] [PubMed]
- Caldorera-Moore, M.E.; Liechty, W.B.; Peppas, N.A. Responsive theranostic systems: Integration of diagnostic imaging agents and responsive controlled release drug delivery carriers. Acc. Chem. Res. 2011, 44, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Carpi, S.; Fogli, S.; Giannetti, A.; Adinolfi, B.; Tombelli, S.; Pozzo, E.D.; Vanni, A.; Martinotti, E.; Martini, C.; Breschi, M.C.; et al. Theranostic properties of a survivin-directed molecular beacon in human melanoma cells. PLoS ONE 2014, 9, e114588. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.K.W.; Chow, M.Y.T.; Zhang, Y.; Leung, S.W.S. siRNA versus miRNA as therapeutics for gene silencing. Mol. Ther. Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zangemeister-Wittke, U.; Simon, H.U. An IAP in action: The multiple roles of survivin in differentiation, immunity and malignancy. Cell Cycle 2004, 3, 1121–1123. [Google Scholar] [CrossRef] [PubMed]
- Mobahat, M.; Narendran, A.; Riabowol, K. Survivin as a preferential target for cancer therapy. Int. J. Mol. Sci. 2014, 15, 2494–2516. [Google Scholar] [CrossRef] [PubMed]
- Sarela, A.I.; Macadam, R.C.A.; Farmery, S.M.; Markham, A.F.; Guilloua, P.J. Expression of the antiapoptosis gene, survivin, predicts death from recurrent colorectal carcinoma. Gut 2000, 46. [Google Scholar] [CrossRef]
- Chu, X.-Y.; Chen, L.-B.; Wang, J.-H.; Su, Q.-S.; Yang, J.-R.; Lin, Y.; Xue, L.-J.; Liu, X.-B.; Mo, X.-B. Overexpression of survivin is correlated with increased invasion and metastasis of colorectal cancer. J. Surg. Oncol. 2012, 105, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Jakubowska, K.; Pryczynicz, A.; Dymicka-Piekarska, V.; Famulski, W.; Guzińska-Ustymowicz, K. Immunohistochemical expression and serum level of survivin protein in colorectal cancer patients. Oncol. Lett. 2016, 12, 3591–3597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawasaki, H.; Altieri, D.C.; Lu, C.-D.; Toyoda, M.; Tenjo, T.; Tanigawa, N. Inhibition of apoptosis by survivin predicts shorter survival rates in colorectal cancer. Cancer Res. 1998, 58, 5071–5074. [Google Scholar] [PubMed]
- Piao, Y.; Liu, F.; Seo, T.S. A novel molecular beacon bearing a graphite nanoparticle as a nanoquencher for in situ mRNA detection in cancer cells. ACS Appl. Mater. Interfaces 2012, 4, 6785–6789. [Google Scholar] [CrossRef] [PubMed]
- Stobiecka, M.; Chalupa, A. DNA strand replacement mechanism in molecular beacons encoded for the detection of cancer biomarkers. J. Phys. Chem. B 2016, 120, 4782–4790. [Google Scholar] [CrossRef] [PubMed]
- Olie, R.; Simões-Wüst, A.; Baumann, B.; Leech, S.; Fabbro, D.; Stahel, R.; Zangemeister-Wittke, U. A novel antisense oligonucleotide targeting survivin expression induces apoptosis and sensitizes lung cancer cells to chemotherapy. Cancer Res. 2000, 60, 2805–2809. [Google Scholar] [PubMed]
- Liu, B.; Sun, Z.; Zhang, X.; Liu, J. Mechanisms of DNA sensing on graphene oxide. Anal. Chem. 2013, 85, 7987–7993. [Google Scholar] [CrossRef] [PubMed]
- Stobiecka, M.; Hepel, M. Multimodal coupling of optical transitions and plasmonic oscillations in rhodamine B modified gold nanoparticles. Phys. Chem. Chem. Phys. 2011, 13, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Duan, G.; Zhang, Y.; Luan, B.; Weber, J.K.; Zhou, R.W.; Yang, Z.; Zhao, L.; Xu, J.; Luo, J.; Zhou, R. Graphene-induced pore formation on cell membranes. Sci. Rep. 2017, 7, 42767. [Google Scholar] [CrossRef] [PubMed]
- Nitin, N.; Santangelo, P.J.; Kim, G.; Nie, S.; Bao, G. Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells. Nucleic Acids Res. 2004, 32, e58. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Graber, Z.T.; Baumgart, T.; Stone, H.A.; Cohen, A.E. Lipid-gel model of biological membranes. Biophys. J. 2018, 114, 115a. [Google Scholar] [CrossRef]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ratajczak, K.; Krazinski, B.E.; Kowalczyk, A.E.; Dworakowska, B.; Jakiela, S.; Stobiecka, M. Optical Biosensing System for the Detection of Survivin mRNA in Colorectal Cancer Cells Using a Graphene Oxide Carrier-Bound Oligonucleotide Molecular Beacon. Nanomaterials 2018, 8, 510. https://doi.org/10.3390/nano8070510
Ratajczak K, Krazinski BE, Kowalczyk AE, Dworakowska B, Jakiela S, Stobiecka M. Optical Biosensing System for the Detection of Survivin mRNA in Colorectal Cancer Cells Using a Graphene Oxide Carrier-Bound Oligonucleotide Molecular Beacon. Nanomaterials. 2018; 8(7):510. https://doi.org/10.3390/nano8070510
Chicago/Turabian StyleRatajczak, Katarzyna, Bartlomiej E. Krazinski, Anna E. Kowalczyk, Beata Dworakowska, Slawomir Jakiela, and Magdalena Stobiecka. 2018. "Optical Biosensing System for the Detection of Survivin mRNA in Colorectal Cancer Cells Using a Graphene Oxide Carrier-Bound Oligonucleotide Molecular Beacon" Nanomaterials 8, no. 7: 510. https://doi.org/10.3390/nano8070510