Platinum Nanoparticles Decrease Reactive Oxygen Species and Modulate Gene Expression without Alteration of Immune Responses in THP-1 Monocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Platinum Nanoparticles Synthesis and Characterization
2.2. Cell Culture
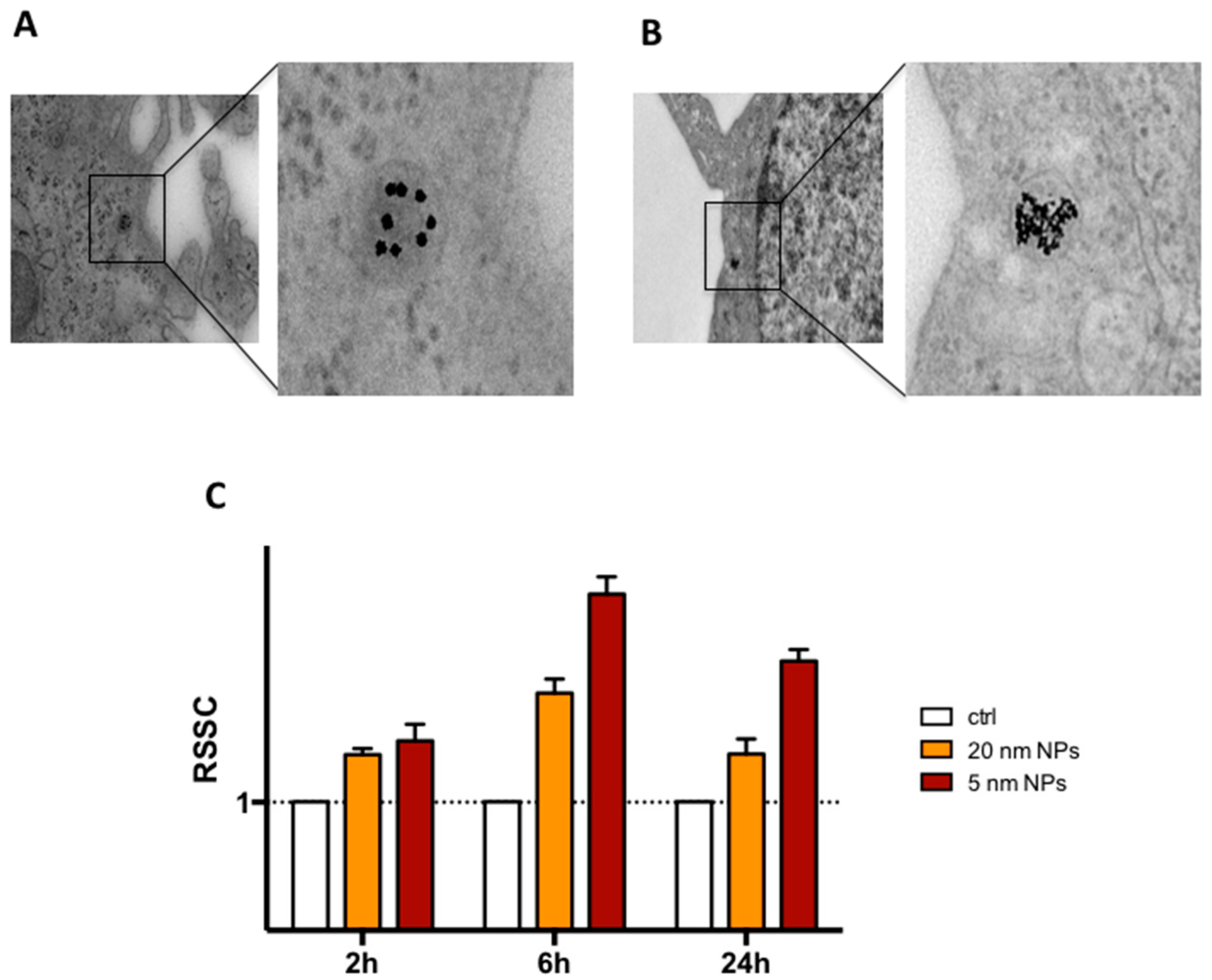
2.3. Transmission Electron Microscopy (TEM) Analysis of Cellular Internalization of PtNPs
2.4. Intracellular Uptake of Pt NPs by Flow Cytometry Analysis
2.5. Annexin-PI Assay
2.6. WST-8 Assay
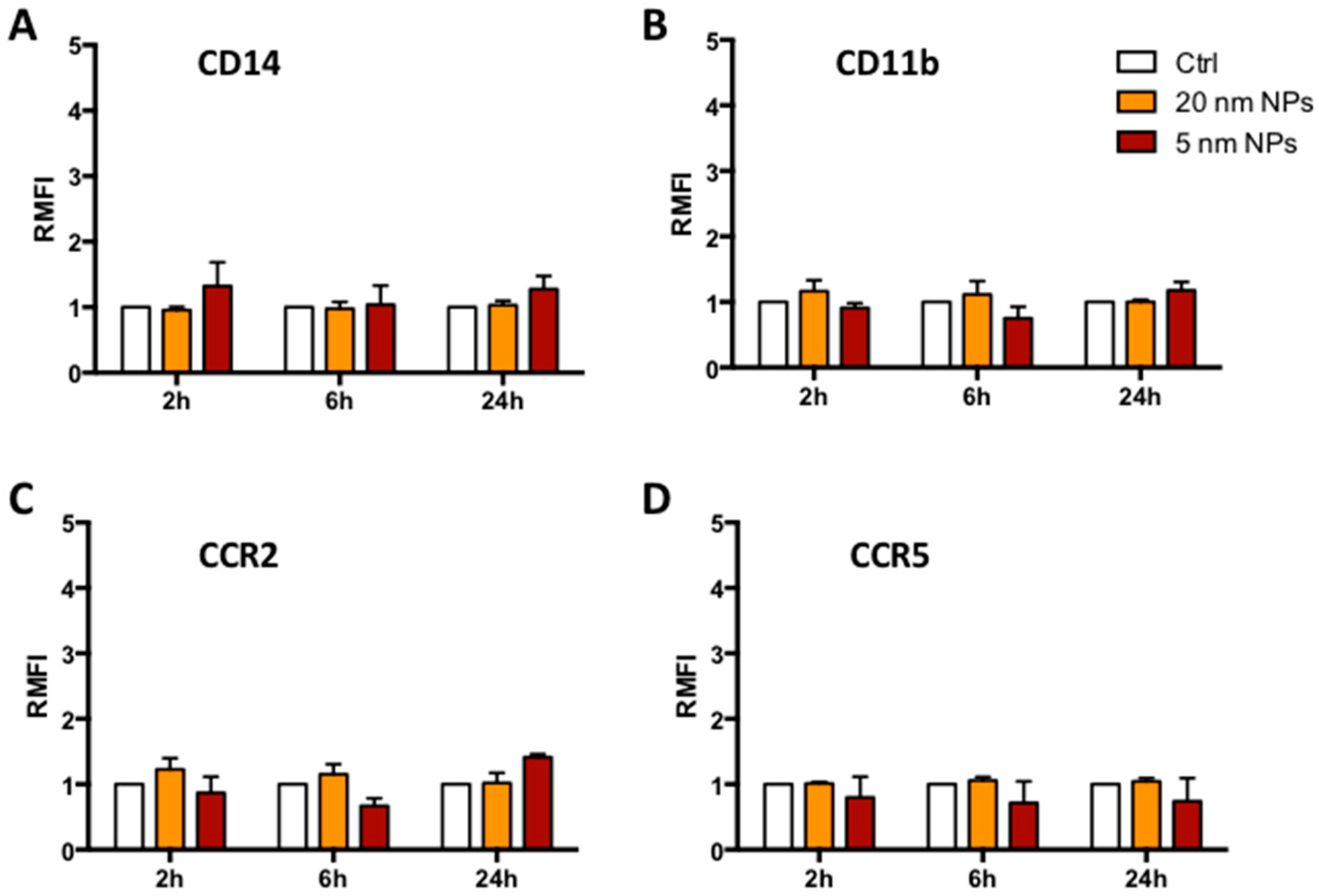
2.7. Receptor Expression
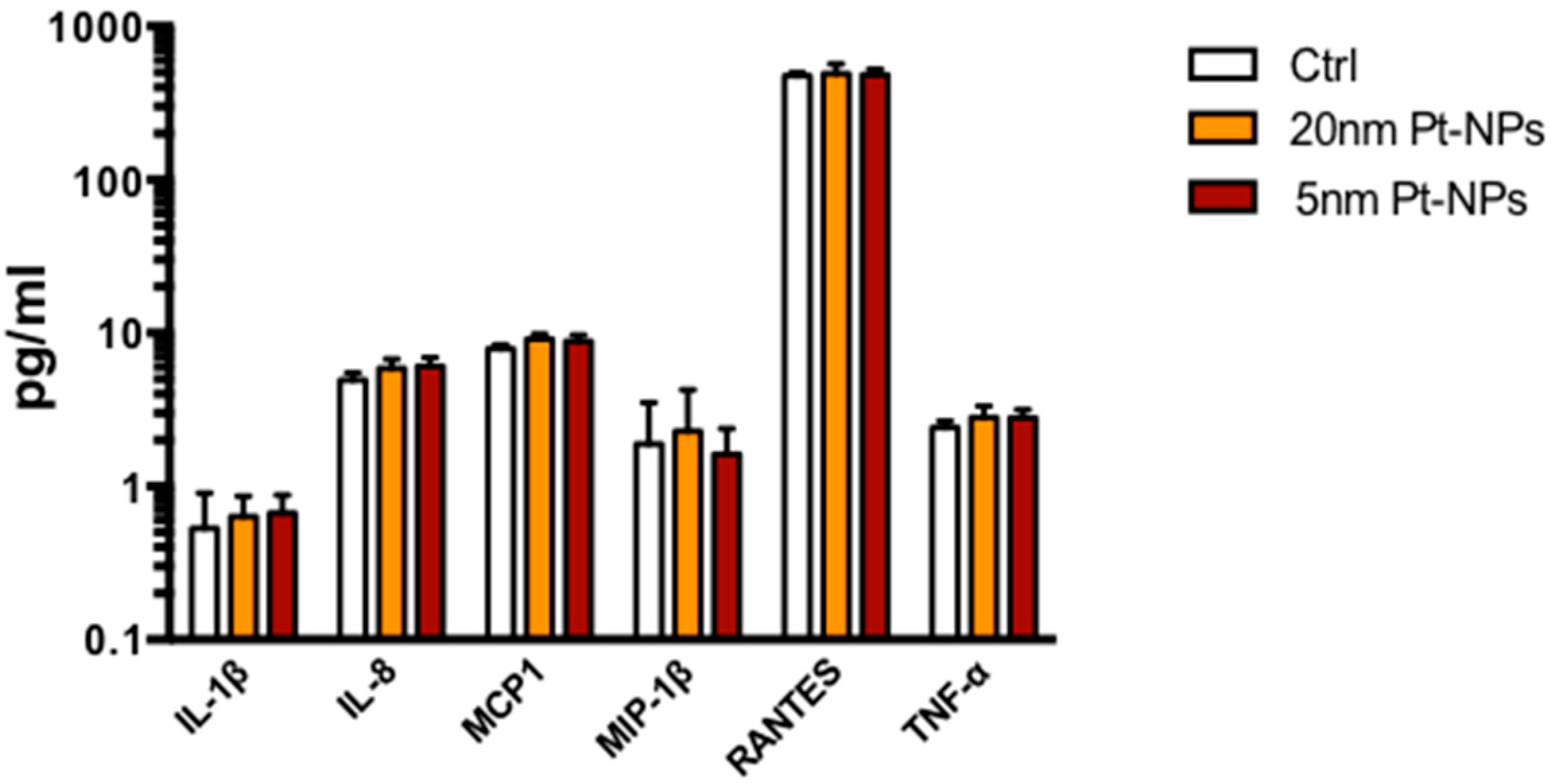
2.8. Inflammatory Cytokine Release
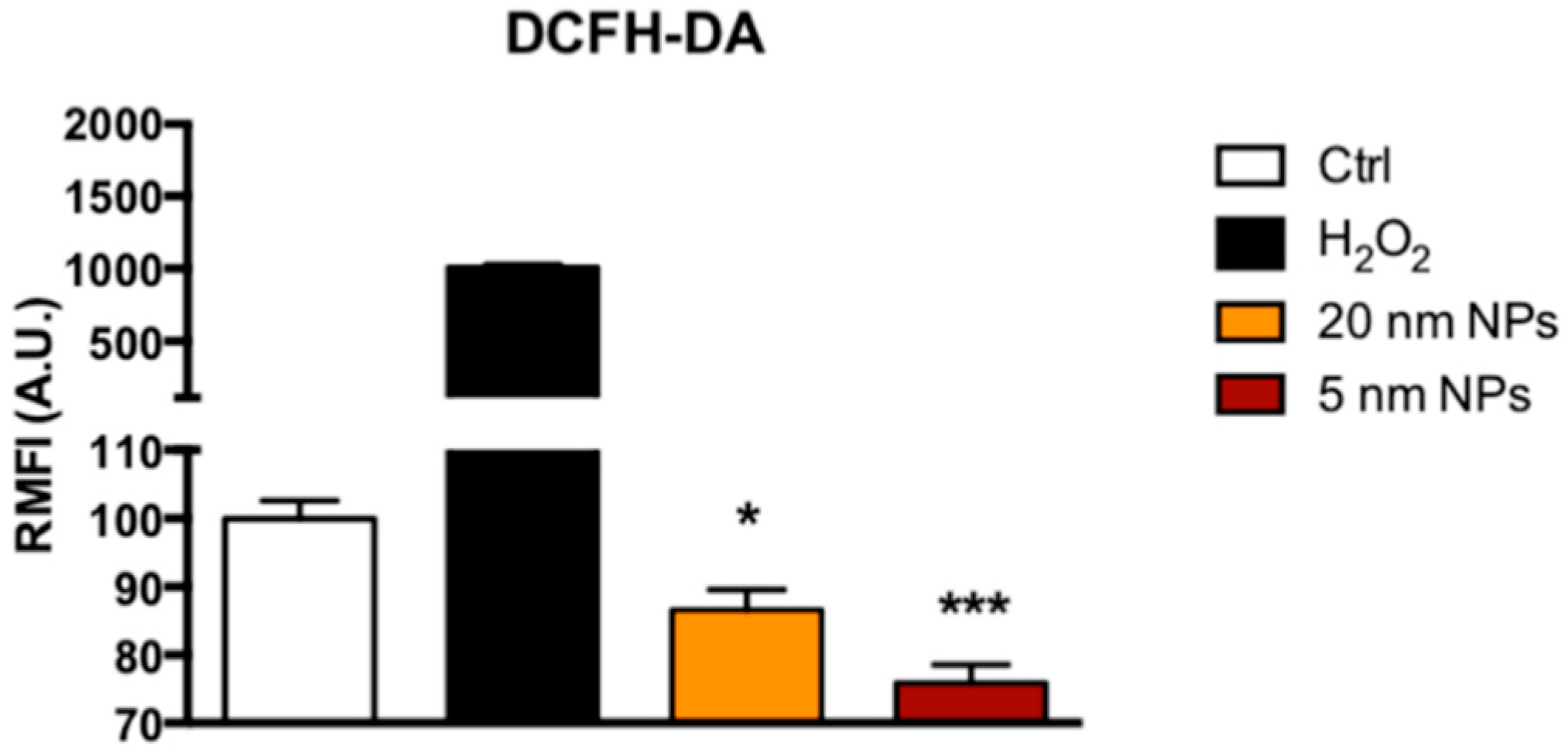
2.9. DCFDH-DA Assay
2.10. RNA Extraction and Microarray Analysis
2.11. Statistical Analysis
3. Results
3.1. Cytocompatibility of PtNPs with THP-1 Monocytes
3.2. THP-1 Monocyte Internalization of PtNPs
3.3. Immune Receptor Expression in the Presence of PtNPs
3.4. Cytokine Expression in the Presence of PTNPs
3.5. Reactive Oxigen Species ( ROS) Scavenging Activity of PtNPs in THP-1 Monocytes
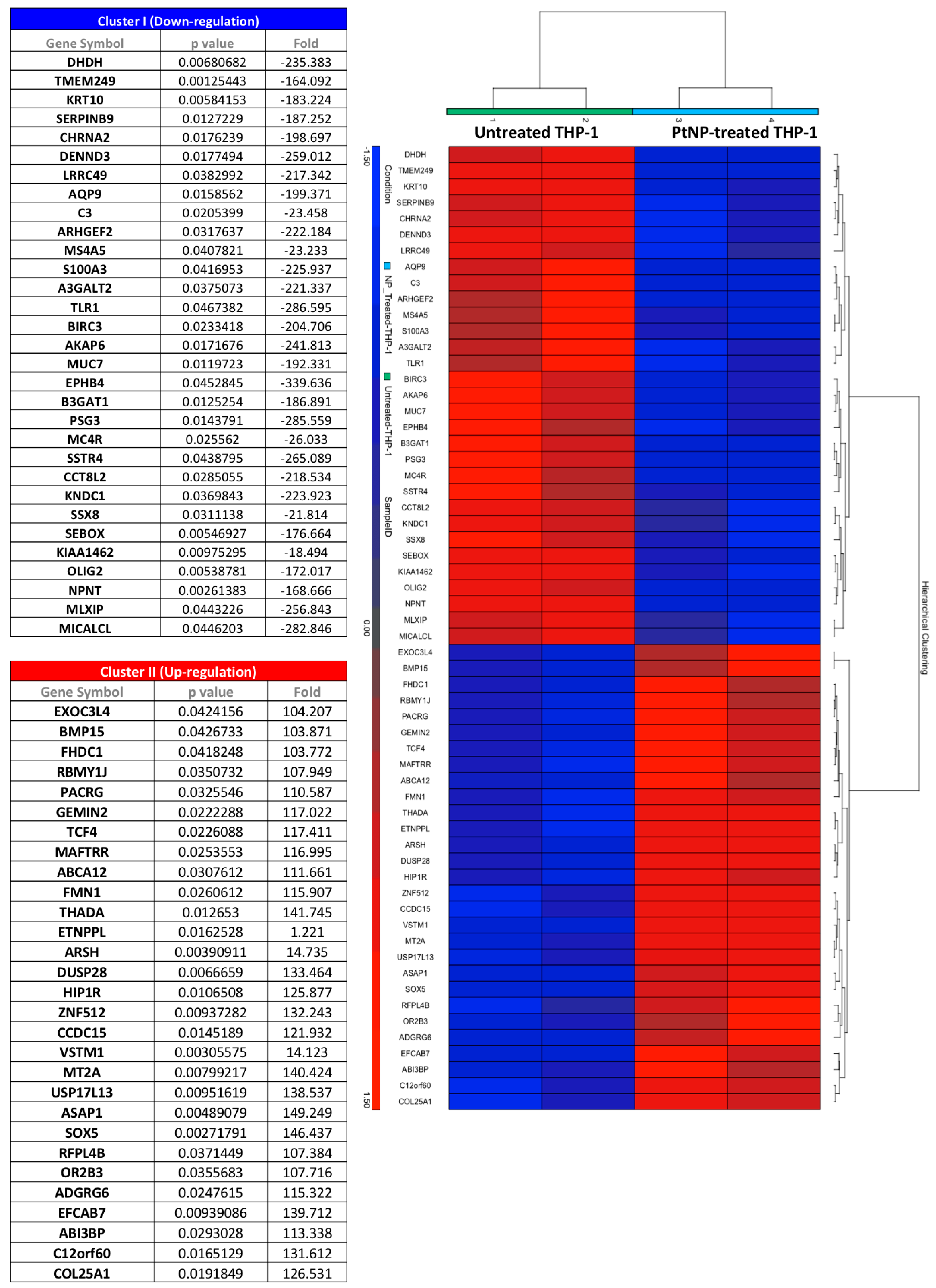
3.6. Gene Transcription in THP-1 Treated with PtNPs
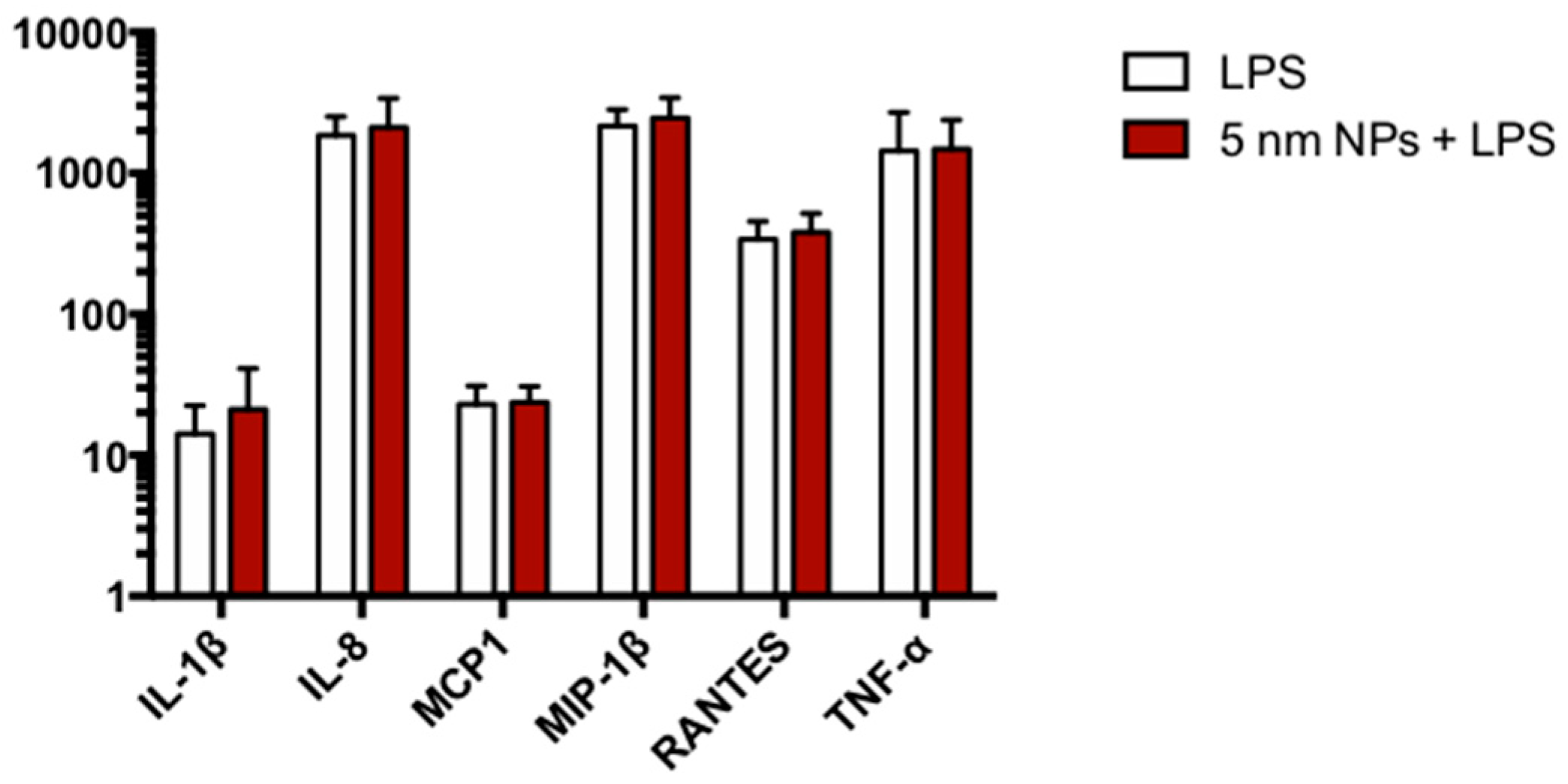
3.7. Cytokine Expression in PtNP-Treated THP-1 after LPS Stimulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Pedone, D.; Moglianetti, M.; De Luca, E.; Bardi, G.; Pompa, P.P. Platinum nanoparticles in nanobiomedicine. Chem. Soc. Rev. 2017, 46, 4951–4975. [Google Scholar] [CrossRef] [PubMed]
- Karakoti, A.; Singh, S.; Dowding, J.M.; Seal, S.; Self, W.T. Redox-active radical scavenging nanomaterials. Chem. Soc. Rev. 2010, 39, 4422. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, K.S.; Husen, A. Green Synthesis, Characterization and Uses of Palladium/Platinum Nanoparticles. Nanoscale Res. Lett. 2016, 11, 482. [Google Scholar] [CrossRef] [PubMed]
- Elder, A.; Yang, H.; Gwiazda, R.; Teng, X.; Thurston, S.; He, H.; Oberdörster, G. Testing Nanomaterials of Unknown Toxicity: An Example Based on Platinum Nanoparticles of Different Shapes. Adv. Mater. 2007, 19, 3124–3129. [Google Scholar] [CrossRef]
- Hamasaki, T.; Kashiwagi, T.; Imada, T.; Nakamichi, N.; Aramaki, S.; Toh, K.; Morisawa, S.; Shimakoshi, H.; Hisaeda, Y.; Shirahata, S. Kinetic Analysis of Superoxide Anion Radical-Scavenging and Hydroxyl Radical-Scavenging Activities of Platinum Nanoparticles. Langmuir 2008, 24, 7354–7364. [Google Scholar] [CrossRef] [PubMed]
- Horie, M.; Kato, H.; Endoh, S.; Fujita, K.; Nishio, K.; Komaba, L.K.; Fukui, H.; Nakamura, A.; Miyauchi, A.; Nakazato, T.; et al. Evaluation of cellular influences of platinum nanoparticles by stable medium dispersion. Metallomics 2011, 3, 1244. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, H.; Haruki, R.; Yamada, K.; Böttcher, C.; Komatsu, T. Hemoglobin–Albumin Cluster Incorporating a Pt Nanoparticle: Artificial O2 Carrier with Antioxidant Activities. PLoS ONE 2014, 9, e110541. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.-K.; Kim, J.-C.; Park, H.-J.; Sul, O.-J.; Lee, M.-H.; Kim, J.-S.; Choi, H.-S. Platinum nanoparticles reduce ovariectomy-induced bone loss by decreasing osteoclastogenesis. Exp. Mol. Med. 2012, 44, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Moglianetti, M.; De Luca, E.; Pedone, D.; Marotta, R.; Catelani, T.; Sartori, B.; Amenitsch, H.; Retta, S.F.; Pompa, P.P. Platinum nanozymes recover cellular ROS homeostasis in an oxidative stress-mediated disease model. Nanoscale 2016, 8, 3739–3752. [Google Scholar] [CrossRef] [PubMed]
- Nomura, M.; Yoshimura, Y.; Kikuiri, T.; Hasegawa, T.; Taniguchi, Y.; Deyama, Y.; Koshiro, K.; Sano, H.; Suzuki, K.; Inoue, N. Platinum nanoparticles suppress osteoclastogenesis through scavenging of reactive oxygen species produced in RAW264.7 cells. J. Pharmacol. Sci. 2011, 117, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, S.; Ozawa, Y.; Watanabe, K.; Izuo, N.; Toda, T.; Yokote, K.; Shimizu, T. Palladium and Platinum Nanoparticles Attenuate Aging-Like Skin Atrophy via Antioxidant Activity in Mice. PLoS ONE 2014, 9, e109288. [Google Scholar] [CrossRef] [PubMed]
- Shiny, P.J.; Mukherjee, A.; Chandrasekaran, N. Haemocompatibility assessment of synthesised platinum nanoparticles and its implication in biology. Bioprocess Biosyst. Eng. 2014, 37, 991–997. [Google Scholar] [CrossRef] [PubMed]
- Yoshihisa, Y.; Zhao, Q.-L.; Hassan, M.A.; Wei, Z.-L.; Furuichi, M.; Miyamoto, Y.; Kondo, T.; Shimizu, T. SOD/catalase mimetic platinum nanoparticles inhibit heat-induced apoptosis in human lymphoma U937 and HH cells. Free Radic. Res. 2011, 45, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Nejdl, L.; Kudr, J.; Moulick, A.; Hegerova, D.; Ruttkay-Nedecky, B.; Gumulec, J.; Cihalova, K.; Smerkova, K.; Dostalova, S.; Krizkova, S.; et al. Platinum nanoparticles induce damage to DNA and inhibit DNA replication. PLoS ONE 2017, 12, e0180798. [Google Scholar] [CrossRef] [PubMed]
- Albanese, A.; Tang, P.S.; Chan, W.C.W. The Effect of Nanoparticle Size, Shape, and Surface Chemistry on Biological Systems. Annu. Rev. Biomed. Eng. 2012, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Crist, R.M.; Grossman, J.H.; Patri, A.K.; Stern, S.T.; Dobrovolskaia, M.A.; Adiseshaiah, P.P.; Clogston, J.D.; McNeil, S.E. Common pitfalls in nanotechnology: lessons learned from NCI’s Nanotechnology Characterization Laboratory. Integr. Biol. (Camb). 2013, 5, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Rehman, M.U.; Yoshihisa, Y.; Miyamoto, Y.; Shimizu, T. The anti-inflammatory effects of platinum nanoparticles on the lipopolysaccharide-induced inflammatory response in RAW 264.7 macrophages. Inflamm. Res. 2012, 61, 1177–1185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbas, A.K.; Lichtman, A.H.; Pillai, S. Cellular and Molecular Immunology; Elsevier: New York, NY, USA, 2015; ISBN 9780323316149. [Google Scholar]
- Ingersoll, M.A.; Platt, A.M.; Potteaux, S.; Randolph, G.J. Monocyte trafficking in acute and chronic inflammation. Trends Immunol. 2011, 32, 470–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fröhlich, E. Cellular elimination of nanoparticles. Environ. Toxicol. Pharmacol. 2016, 46, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Chanput, W.; Mes, J.J.; Wichers, H.J. THP-1 cell line: An in vitro cell model for immune modulation approach. Int. Immunopharmacol. 2014, 23, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Gorczyca, W.; Sun, Z.-Y.; Cronin, W.; Li, X.; Mau, S.; Tugulea, S. Immunophenotypic Pattern of Myeloid Populations by Flow Cytometry Analysis. Methods Cell Biol. 2011, 103, 221–266. [Google Scholar] [PubMed]
- White, G.E.; Iqbal, A.J.; Greaves, D.R. CC Chemokine Receptors and Chronic Inflammation—Therapeutic Opportunities and Pharmacological Challenges. Pharmacol. Rev. 2013, 65, 47–89. [Google Scholar] [CrossRef] [PubMed]
- Guarnieri, D.; Melone, P.; Moglianetti, M.; Marotta, R.; Netti, P.A.; Pompa, P.P. Particle size affects the cytosolic delivery of membranotropic peptide-functionalized platinum nanozymes. Nanoscale 2017, 9, 11288–11296. [Google Scholar] [CrossRef] [PubMed]
- Garrafa, E.; Imberti, L.; Tiberio, G.; Prandini, A.; Giulini, S.M.; Caimi, L. Heterogeneous expression of toll-like receptors in lymphatic endothelial cells derived from different tissues. Immunol. Cell Biol. 2011, 89, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Labbé, K.; McIntire, C.R.; Doiron, K.; Leblanc, P.M.; Saleh, M. Cellular Inhibitors of Apoptosis Proteins cIAP1 and cIAP2 Are Required for Efficient Caspase-1 Activation by the Inflammasome. Immunity 2011, 35, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Van der Burgh, R.; Meeldijk, J.; Jongeneel, L.; Frenkel, J.; Bovenschen, N.; van Gijn, M.; Boes, M. Reduced serpinB9-mediated caspase-1 inhibition can contribute to autoinflammatory disease. Oncotarget 2016, 7, 19265–19271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varecza, Z.; Elekes, K.; László, T.; Perkecz, A.; Pintér, E.; Sándor, Z.; Szolcsányi, J.; Keszthelyi, D.; Szabó, Á.; Sándor, K.; Molnár, T.F.; Szántó, Z.; Pongrácz, J.E.; Helyes, Z. Expression of the Somatostatin Receptor Subtype 4 in Intact and Inflamed Pulmonary Tissues. J. Histochem. Cytochem. 2009, 57, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Lubbers, R.; van Essen, M.F.; van Kooten, C.; Trouw, L.A. Production of complement components by cells of the immune system. Clin. Exp. Immunol. 2017, 188, 183–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsushima, A.; Ogura, H.; Koh, T.; Shimazu, T.; Sugimoto, H. Enhanced Expression of Aquaporin 9 in Activated Polymorphonuclear Leukocytes in Patients With Systemic Inflammatory Response Syndrome. Shock 2014, 42, 322–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Xiaoli, L.; Xu, N.; Li, L.; Lu, Q.; Zhang, J.; Huang, B.; Du, Q. EphrinB2/EphB4 Interaction Promotes Myeloid Leukemia Cell Invasion through RhoA-Mediated Mechanism. Blood 2014, 124, 1018. [Google Scholar]
- Xie, M.; Li, T.; Li, N.; Li, J.; Yao, Q.; Han, W.; Ruan, G. VSTM-v1, a potential myeloid differentiation antigen that is downregulated in bone marrow cells from myeloid leukemia patients. J. Hematol. Oncol. 2015, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Curtis, J.; Luo, Y.; Zenner, H.L.; Cuchet-Lourenço, D.; Wu, C.; Lo, K.; Maes, M.; Alisaac, A.; Stebbings, E.; Liu, J.Z.; et al. Susceptibility to tuberculosis is associated with variants in the ASAP1 gene encoding a regulator of dendritic cell migration. Nat. Genet. 2015, 47, 523–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, K.-I.; Takano, H.; Shimada, A.; Wada, E.; Yanagisawa, R.; Sakurai, M.; Satoh, M.; Yoshikawa, T. Role of metallothionein in coagulatory disturbance and systemic inflammation induced by lipopolysaccharide in mice. FASEB J. 2006, 20, 533–535. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, V. The SoxD transcription factors—Sox5, Sox6, and Sox13—are key cell fate modulators. Int. J. Biochem. Cell Biol. 2010, 42, 429–432. [Google Scholar] [CrossRef] [PubMed]
- Schmidtmann, E.; Anton, T.; Rombaut, P.; Herzog, F.; Leonhardt, H. Determination of local chromatin composition by CasID. Nucleus 2016, 7, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Pusapati, G.V.; Hughes, C.E.; Dorn, K.V.; Zhang, D.; Sugianto, P.; Aravind, L.; Rohatgi, R. EFCAB7 and IQCE Regulate Hedgehog Signaling by Tethering the EVC-EVC2 Complex to the Base of Primary Cilia. Dev. Cell 2014, 28, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Xu, Y.; Yang, K.; Liu, M.; Wei, D.; Zhang, Y.; Shi, X.; Yang, F.; Wang, X.; Liang, S.; et al. THADA gene polymorphism and prostate cancer risk: a meta-analysis. Oncol. Res. Treat. 2014, 37, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhen, Y.-Z.; Lin, Y.-J.; Liu, J.; Wei, J.; Xu, R.; Hu, G. KNDC1 knockdown protects human umbilical vein endothelial cells from senescence. Mol. Med. Rep. 2014, 10, 82–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gatto, F.; Cagliani, R.; Catelani, T.; Guarnieri, D.; Moglianetti, M.; Pompa, P.; Bardi, G. PMA-Induced THP-1 Macrophage Differentiation is Not Impaired by Citrate-Coated Platinum Nanoparticles. Nanomaterials 2017, 7, 332. [Google Scholar] [CrossRef] [PubMed]
- Singha, S.; Kim, D.; Seo, H.; Cho, S.W.; Ahn, K.H. Fluorescence sensing systems for gold and silver species. Chem. Soc. Rev. Chem. Soc. Rev 2015, 44, 4367–4399. [Google Scholar] [CrossRef]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gatto, F.; Moglianetti, M.; Pompa, P.P.; Bardi, G. Platinum Nanoparticles Decrease Reactive Oxygen Species and Modulate Gene Expression without Alteration of Immune Responses in THP-1 Monocytes. Nanomaterials 2018, 8, 392. https://doi.org/10.3390/nano8060392
Gatto F, Moglianetti M, Pompa PP, Bardi G. Platinum Nanoparticles Decrease Reactive Oxygen Species and Modulate Gene Expression without Alteration of Immune Responses in THP-1 Monocytes. Nanomaterials. 2018; 8(6):392. https://doi.org/10.3390/nano8060392
Chicago/Turabian StyleGatto, Francesca, Mauro Moglianetti, Pier Paolo Pompa, and Giuseppe Bardi. 2018. "Platinum Nanoparticles Decrease Reactive Oxygen Species and Modulate Gene Expression without Alteration of Immune Responses in THP-1 Monocytes" Nanomaterials 8, no. 6: 392. https://doi.org/10.3390/nano8060392





