Separation of Bacteria, Protozoa and Carbon Nanotubes by Density Gradient Centrifugation
Abstract
:1. Introduction
2. Results and Discussion
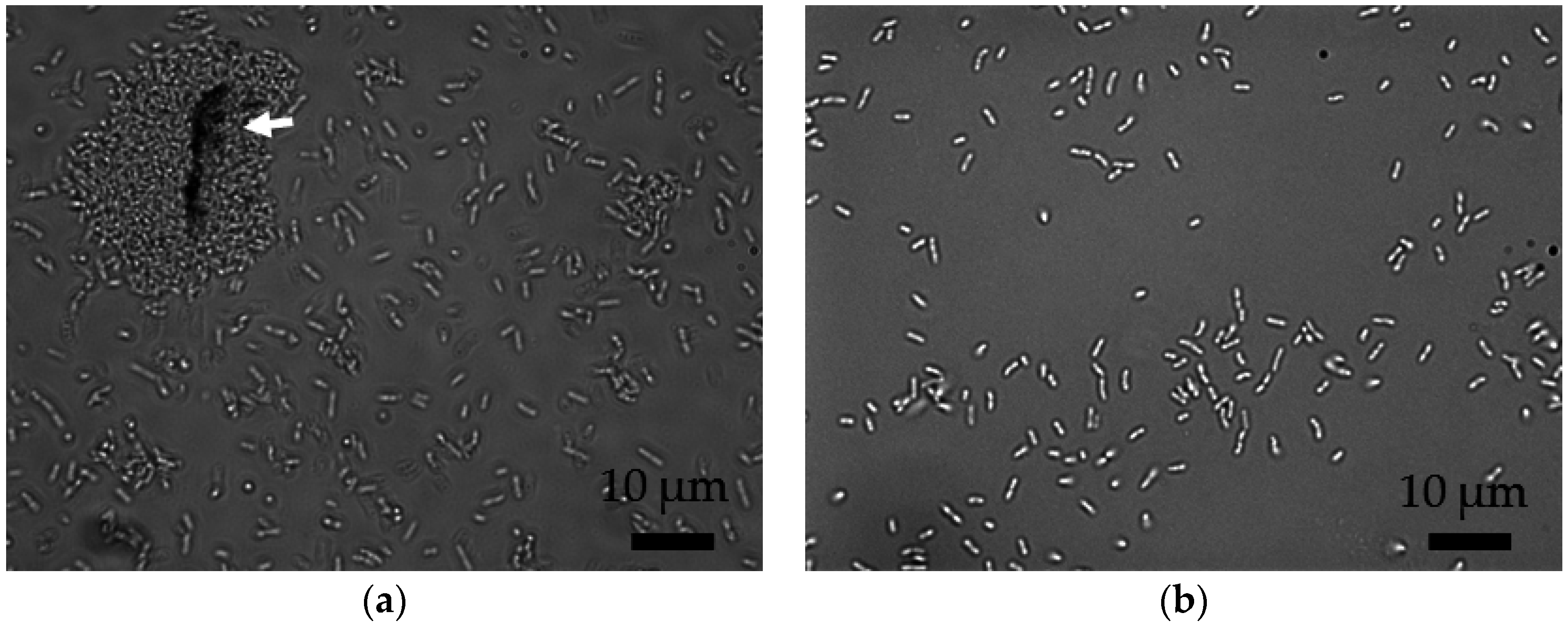
2.1. MWCNT Characteristics and Associations with Bacteria
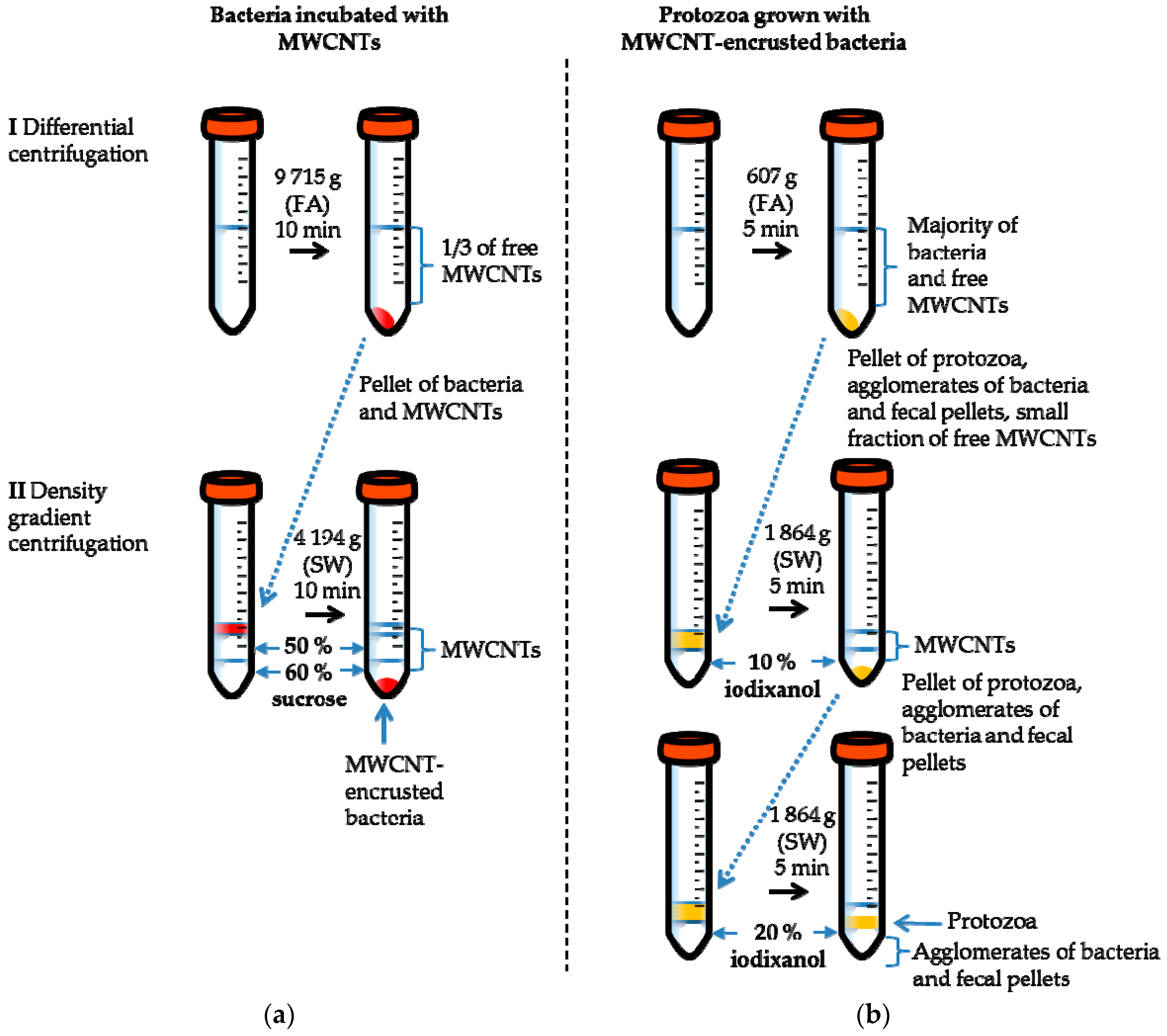
2.2. Separation of MWCNT-Associated Bacteria from Unbound MWCNTs
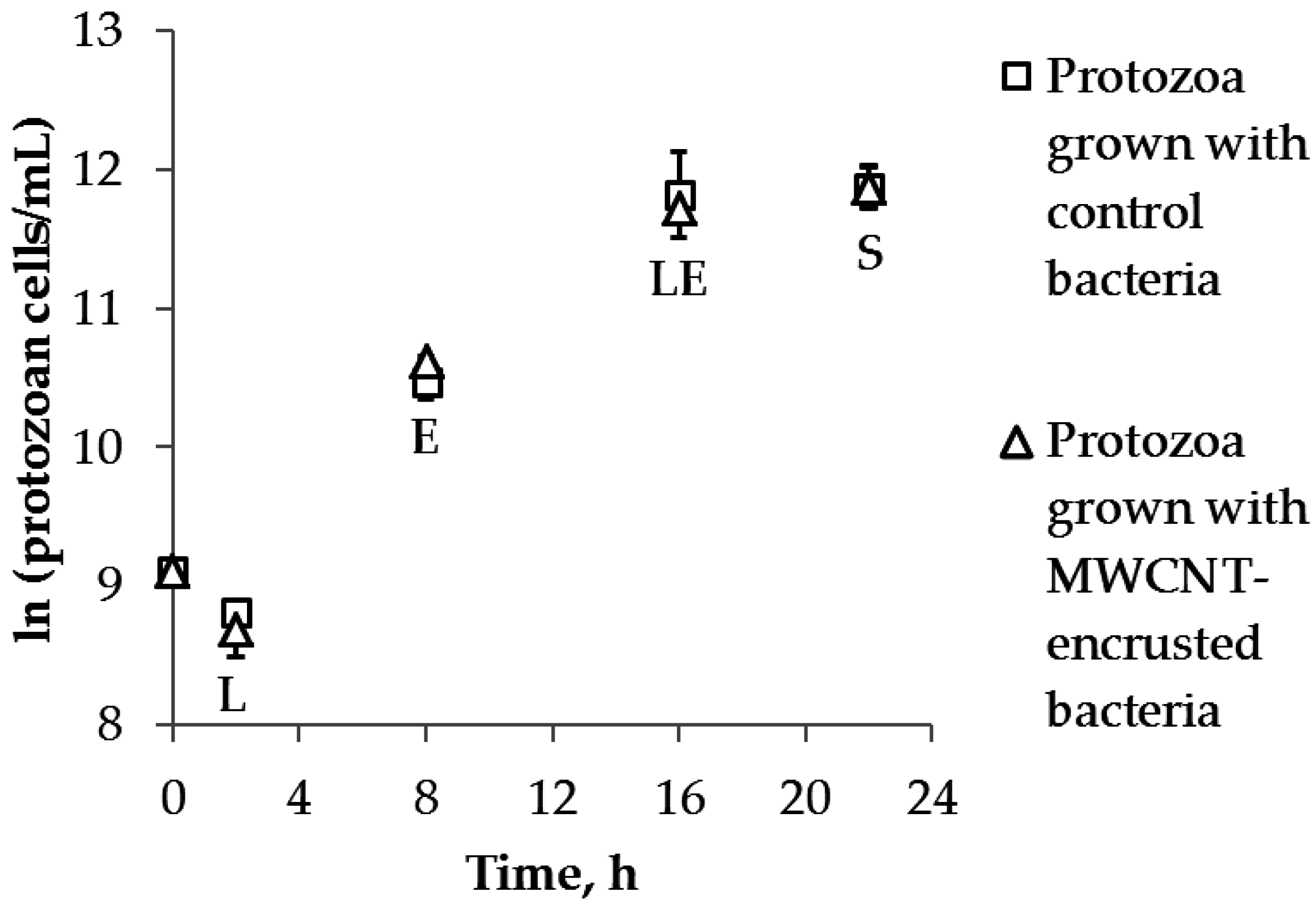
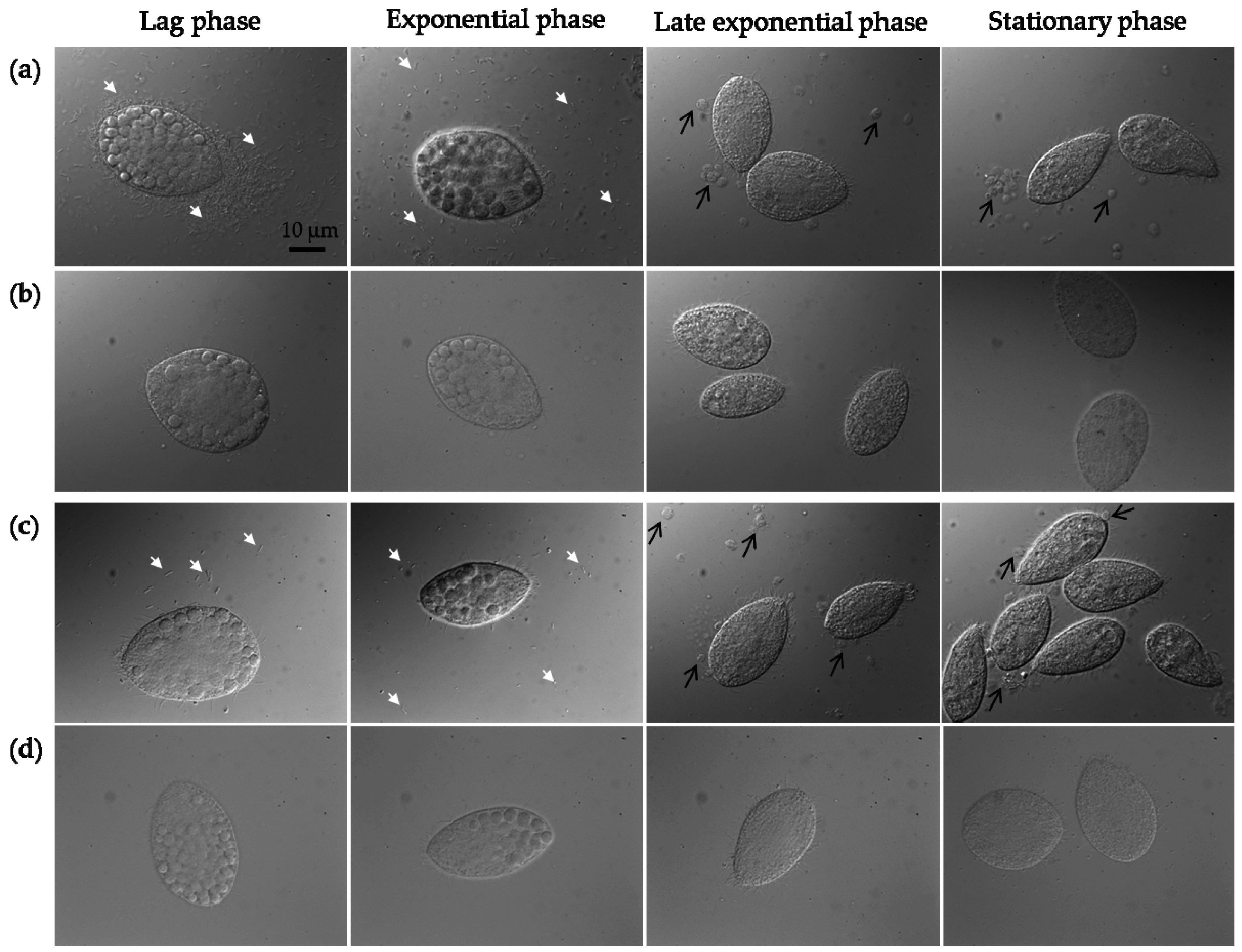
2.3. Protozoan Growth by Predation of Bacteria with and without MWCNTs
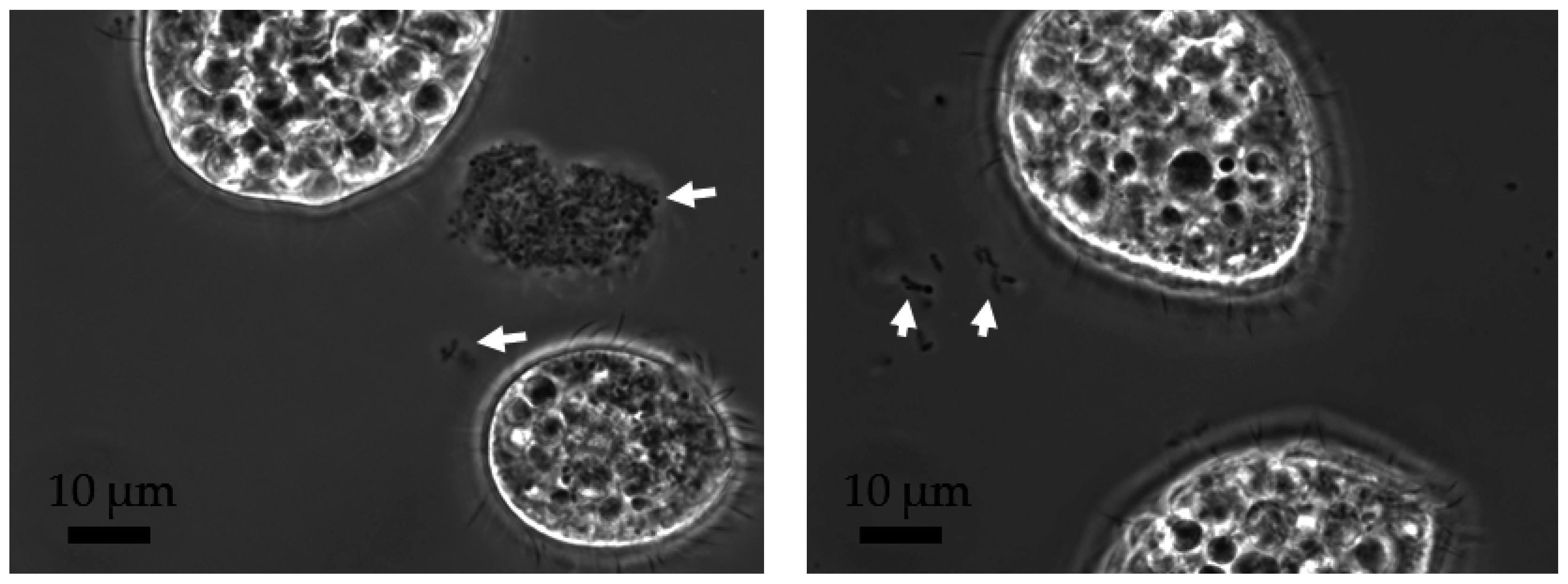
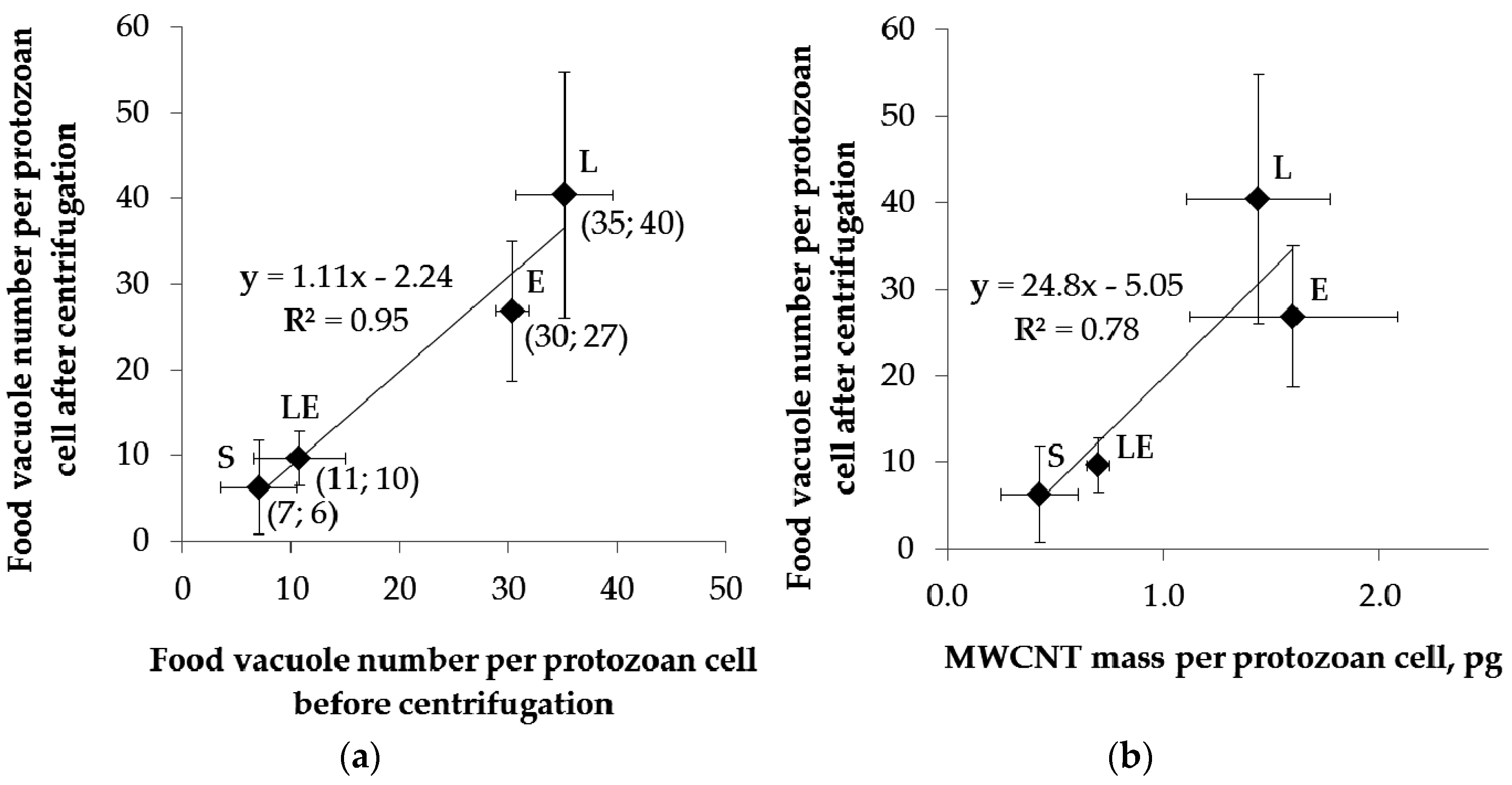
2.4. Separation of Protozoa from Bacteria, Fecal Pellets, and MWCNTs
3. Materials and Methods
3.1. MWCNT Synthesis and Characterization
3.2. MWCNT Suspension Preparation and Characterization
3.3. P. aeruginosa Culturing and Incubation with MWCNTs
3.4. Differential Centrifugation and Sucrose Density Gradient Centrifugation of Bacteria
3.5. Theoretical Estimations of Centrifugal Separation Parameters
3.6. Culturing of T. thermophila and Exposure to MWCNTs
3.7. Differential Centrifugation of Protozoan Exposures and Density Gradient Centrifugation in Iodixanol Solutions
3.8. Determining 14C-MWCNT Localization in the Sucrose Gradient
3.9. Determining 14C-MWCNT Localization in the Iodixanol Media
3.10. Liquid Scintillation Counting (LSC)
3.11. Protozoan and Bacterial Cell Counting, Optical Microscopy, and Food Vacuole Counting
3.12. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 14C-MWCNT | 14C-labeled multiwall carbon nanotubes |
| BET | Brunauer–Emmett–Teller |
| CNT | carbon nanotubes |
| CV | coefficient of variation |
| LSC | liquid scintillation counting |
| MWCNT | multiwall carbon nanotubes |
| NP | nanoparticles |
| SWCNT | single-wall carbon nanotubes |
| TGA | thermogravimetric analysis |
References
- Juganson, K.; Ivask, A.; Blinova, I.; Mortimer, M.; Kahru, A. NanoE-Tox: New and in-depth database concerning ecotoxicity of nanomaterials. Beilstein J. Nanotechnol. 2015, 6, 1788–1804. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L. Nanomaterials in agricultural production: Benefits and possible threats? In Sustainable Nanotechnology and the Environment: Advances and Achievements; Shamim, N., Sharma, V.K., Eds.; American Chemical Society: Washington, DC, USA, 2013; Volume 1124, pp. 73–90. [Google Scholar]
- Holden, P.A.; Schimel, J.P.; Godwin, H.A. Five reasons to use bacteria when assessing manufactured nanomaterial environmental hazards and fates. Curr. Opin. Biotechnol. 2014, 27, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Holden, P.A.; Nisbet, R.M.; Lenihan, H.S.; Miller, R.J.; Cherr, G.N.; Schimel, J.P.; Gardea-Torresdey, J.L. Ecological nanotoxicology: Integrating nanomaterial hazard considerations across the subcellular, population, community, and ecosystems levels. Acc. Chem. Res. 2013, 46, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, M.; Kahru, A.; Slaveykova, V.I. Uptake, localization and clearance of quantum dots in ciliated protozoa Tetrahymena thermophila. Environ. Pollut. 2014, 190, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, M.; Kasemets, K.; Kahru, A. Toxicity of ZnO and CuO nanoparticles to ciliated protozoa Tetrahymena thermophila. Toxicology 2010, 269, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, M.; Kasemets, K.; Vodovnik, M.; Marinsek-Logar, R.; Kahru, A. Exposure to CuO nanoparticles changes the fatty acid composition of protozoa Tetrahymena thermophila. Environ. Sci. Technol. 2011, 45, 6617–6624. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.Y.; Powell, B.A.; Mortimer, M.; Ke, P.C. Adaptive interactions between zinc oxide nanoparticles and Chlorella sp. Environ. Sci. Technol. 2012, 46, 12178–12185. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.G.; Ouyang, S.H.; Mu, L.; An, J.; Zhou, Q. Effects of graphene oxide and oxidized carbon nanotubes on the cellular division, microstructure, uptake, oxidative stress, and metabolic profiles. Environ. Sci. Technol. 2015, 49, 10825–10833. [Google Scholar] [CrossRef] [PubMed]
- Werlin, R.; Priester, J.H.; Mielke, R.E.; Kramer, S.; Jackson, S.; Stoimenov, P.K.; Stucky, G.D.; Cherr, G.N.; Orias, E.; Holden, P.A. Biomagnification of cadmium selenide quantum dots in a simple experimental microbial food chain. Nat. Nanotechnol. 2011, 6, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.; Sequeira, D.; Kolen’ko, Y.V.; Pinto, I.M.; Petrovykh, D.Y. Analytical protocols for separation and electron microscopy of nanoparticles interacting with bacterial cells. Anal. Chem. 2015, 87, 4641–4648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerrillo, C.; Barandika, G.; Igartua, A.; Areitioaurtena, O.; Uranga, N.; Mendoza, G. Colloidal stability and ecotoxicity of multiwalled carbon nanotubes: Influence of select organic matters. Environ. Toxicol. Chem. 2016, 35, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.S.Y.; Nasser, F.; St-Denis, C.H.; Mandal, H.S.; Ghafari, P.; Hadjout-Rabi, N.; Bols, N.C.; Tang, X. Carbon nanotube compared with carbon black: Effects on bacterial survival against grazing by ciliates and antimicrobial treatments. Nanotoxicology 2013, 7, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Garrison, C.E.; Bochdansky, A.B. A simple separation method for downstream biochemical analysis of aquatic microbes. J. Microbiol. Meth. 2015, 111, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Berk, S.G.; Guerry, P.; Colwell, R.R. Separation of small ciliate protozoa from bacteria by sucrose gradient centrifugation. Appl. Environ. Microbiol. 1976, 31, 450–452. [Google Scholar] [PubMed]
- Zhang, Y.C.; Shi, Y.F.; Liou, Y.H.; Sawvel, A.M.; Sun, X.H.; Cai, Y.; Holden, P.A.; Stucky, G.D. High performance separation of aerosol sprayed mesoporous TiO2 sub-microspheres from aggregates via density gradient centrifugation. J. Mater. Chem. 2010, 20, 4162–4167. [Google Scholar] [CrossRef]
- Lee, S.H.; Salunke, B.K.; Kim, B.S. Sucrose density gradient centrifugation separation of gold and silver nanoparticles synthesized using Magnolia kobus plant leaf extracts. Biotechnol. Bioprocess E 2014, 19, 169–174. [Google Scholar] [CrossRef]
- Deng, X.Y.; Xiong, D.; Wang, H.F.; Chen, D.D.; Jiao, Z.; Zhang, H.J.; Wu, M.H. Bulk enrichment and separation of multi-walled carbon nanotubes by density gradient centrifugation. Carbon 2009, 47, 1608–1610. [Google Scholar] [CrossRef]
- Pinheiro, M.D.O.; Power, M.E.; Butler, B.J.; Dayeh, V.R.; Slawson, R.; Lee, L.E.J.; Lynn, D.H.; Bols, N.C. Use of Tetrahymena thermophila to study the role of protozoa in inactivation of viruses in water. Appl. Environ. Microbiol. 2007, 73, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Rhiem, S.; Riding, M.J.; Baumgartner, W.; Martin, F.L.; Semple, K.T.; Jones, K.C.; Schaffer, A.; Maes, H.M. Interactions of multiwalled carbon nanotubes with algal cells: Quantification of association, visualization of uptake, and measurement of alterations in the composition of cells. Environ. Pollut. 2015, 196, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Kryuchkova, M.; Danilushkina, A.; Lvov, Y.; Fakhrullin, R. Evaluation of toxicity of nanoclays and graphene oxide in vivo: A Paramecium caudatum study. Environ. Sci. Nano 2016, 3, 442–452. [Google Scholar] [CrossRef]
- Aruoja, V.; Pokhrel, S.; Sihtmae, M.; Mortimer, M.; Madler, L.; Kahru, A. Toxicity of 12 metal-based nanoparticles to algae, bacteria and protozoa. Environ. Sci. Nano 2015, 2, 630–644. [Google Scholar] [CrossRef]
- Eroglu, E.; Melis, A. “Density equilibrium” method for the quantitative and rapid in situ determination of lipid, hydrocarbon, or biopolymer content in microorganisms. Biotechnol. Bioeng. 2009, 102, 1406–1415. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, M.; Petersen, E.J.; Buchholz, B.A.; Orias, E.; Holden, P.A. Bioaccumulation of multiwall carbon nanotubes in Tetrahymena thermophila by direct feeding or trophic transfer. Environ. Sci. Technol. 2016, 50, 8876–8885. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Petersen, E.J.; Habteselassie, M.Y.; Mao, L.; Huang, Q. Degradation of multiwall carbon nanotubes by bacteria. Environ. Pollut. 2013, 181, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Petersen, E.J.; Huang, Q. Phase distribution of 14C-labeled multiwalled carbon nanotubes in aqueous systems containing model solids: Peat. Environ. Sci. Technol. 2011, 45, 1356–1362. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, C.A.; Craps, M.; Czerw, R.; Carroll, D.L. The effects of high energy probe sonication on the thermoelectric power of large diameter multiwalled carbon nanotubes synthesized by chemical vapor deposition. Synth. Met. 2013, 184, 68–72. [Google Scholar] [CrossRef]
- Livshts, M.A.; Khomyakova, E.; Evtushenko, E.G.; Lazarev, V.N.; Kulemin, N.A.; Semina, S.E.; Generozov, E.V.; Govorun, V.M. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.C.A.; Bavishi, A.; Ogbonna, N.; Maddux, S.; Choudhary, M. Use of the sucrose gradient method for bacterial cell cycle synchronization. J. Microbiol. Biol. Educ. 2012, 13, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Priester, J.H.; van de Werfhorst, L.C.; Ge, Y.; Adeleye, A.S.; Tomar, S.; Tom, L.M.; Piceno, Y.M.; Andersen, G.L.; Holden, P.A. Effects of TiO2 and Ag nanoparticles on polyhydroxybutyrate biosynthesis by activated sludge bacteria. Environ. Sci. Technol. 2014, 48, 14712–14720. [Google Scholar] [CrossRef] [PubMed]
- Pilizota, T.; Shaevitz, J.W. Plasmolysis and cell shape depend on solute outer-membrane permeability during hyperosmotic shock in E. coli. Biophys. J. 2013, 104, 2733–2742. [Google Scholar] [CrossRef] [PubMed]
- Lemes, A.C.L.; Santos, A.; Durán, N. Bacterial remediation from effluent containing multi-walled carbon nanotubes. J. Phys. Conf. Ser. 2011, 304, 012023. [Google Scholar] [CrossRef]
- Moon, H.M.; Kim, J.W. Carbon nanotube clusters as universal bacterial adsorbents and magnetic separation agents. Biotechnol. Prog. 2010, 26, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Application Sheet C51. Purification of Toxoplasma Gondii from Cell Cultures; Axis-Shield: Dundee, Scotland, UK, 2013.
- Ford, T.; Graham, J.; Rickwood, D. Iodixanol: A nonionic iso-osmotic centrifugation medium for the formation of self-generated gradients. Anal. Biochem. 1994, 220, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Petersen, E.J.; Huang, Q.G.; Weber, W.J. Bioaccumulation of radio-labeled carbon nanotubes by Eisenia foetida. Environ. Sci. Technol. 2008, 42, 3090–3095. [Google Scholar] [CrossRef] [PubMed]
- Petersen, E.J.; Huang, Q.G.; Weber, W.J. Relevance of octanol-water distribution measurements to the potential ecological uptake of multi-walled carbon nanotubes. Environ. Toxicol. Chem. 2010, 29, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- ISO/TS 11308:2011(E). Nanotechnologies—Characterization of Single-Wall Carbon Nanotubes Using Thermogravimetric Analysis. Available online: https://www.iso.org/obp/ui/#iso:std:iso:ts:11308:ed-1:v1:en.
- Taurozzi, J.S.; Hackley, V.A.; Wiesner, M.R. Ultrasonic dispersion of nanoparticles for environmental, health and safety assessment—Issues and recommendations. Nanotoxicology 2011, 5, 711–729. [Google Scholar] [CrossRef] [PubMed]
- Priester, J.H.; Stoimenov, P.K.; Mielke, R.E.; Webb, S.M.; Ehrhardt, C.; Zhang, J.P.; Stucky, G.D.; Holden, P.A. Effects of soluble cadmium salts versus CdSe quantum dots on the growth of planktonic Pseudomonas aeruginosa. Environ. Sci. Technol. 2009, 43, 2589–2594. [Google Scholar] [CrossRef] [PubMed]
- Horst, A.M.; Neal, A.C.; Mielke, R.E.; Sislian, P.R.; Suh, W.H.; Madler, L.; Stucky, G.D.; Holden, P.A. Dispersion of TiO2 nanoparticle agglomerates by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2010, 76, 7292–7298. [Google Scholar] [CrossRef] [PubMed]
- Mielke, R.E.; Priester, J.H.; Werlin, R.A.; Gelb, J.; Horst, A.M.; Orias, E.; Holden, P.A. Differential growth of and nanoscale TiO2 accumulation in Tetrahymena thermophila by direct feeding versus trophic transfer from Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2013, 79, 5616–5624. [Google Scholar] [CrossRef] [PubMed]
- Holden, P.A.; LaMontagne, M.G.; Bruce, A.K.; Miller, W.G.; Lindow, S.E. Assessing the role of Pseudomonas aeruginosa surface-active gene expression in hexadecane biodegradation in sand. Appl. Environ. Microbiol. 2002, 68, 2509–2518. [Google Scholar] [CrossRef] [PubMed]
- Atlas, R.M. Handbook of Media for Environmental Microbiology, 2nd ed.; Taylor & Francis Group: Boca Raton, FL, USA, 2005. [Google Scholar]
- Carney, R.P.; Kim, J.Y.; Qian, H.F.; Jin, R.C.; Mehenni, H.; Stellacci, F.; Bakr, O.M. Determination of nanoparticle size distribution together with density or molecular weight by 2D analytical ultracentrifugation. Nat. Commun. 2011, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priester, J.H.; Olson, S.G.; Webb, S.M.; Neu, M.P.; Hersman, L.E.; Holden, P.A. Enhanced exopolymer production and chromium stabilization in Pseudomonas putida unsaturated biofilms. Appl. Environ. Microbiol. 2006, 72, 1988–1996. [Google Scholar] [CrossRef] [PubMed]
- ImageJ. Image Processing and Analysis in Java. National Institutes of Health (NIH): Bethesda, MD, USA. Available online: http://imagej.nih.gov/ij/ (accessed on 23 September 2015).
- Petersen, E.J.; Flores-Cervantes, D.X.; Bucheli, T.D.; Elliott, L.C.C.; Fagan, J.A.; Gogos, A.; Hanna, S.; Kagi, R.; Mansfield, E.; Bustos, A.R.M.; et al. Quantification of carbon nanotubes in environmental matrices: Current capabilities, case studies, and future prospects. Environ. Sci. Technol. 2016, 50, 4587–4605. [Google Scholar] [CrossRef] [PubMed]
- MEIAF: Micro-Environmental Imaging and Analysis Facility. Available online: http://www.bren.ucsb.edu/facilities/MEIAF/ (accessed on 28 September 2016).

| Component of the System | Diameter, m |
|---|---|
| MWCNTs in Dryl’s medium | 1.7 × 10−7 ± 2 × 10−9 (0.21) a |
| MWCNTs in bacterial growth medium (half-strength 21C) | 3.2 × 10−7 ± 1 × 10−8 (0.24) a |
| Bacteria (P. aeruginosa) | 8.9 × 10−7 b |
| Protozoa (T. thermophila) | 2.4 × 10−5 b |
| Fecal pellets of protozoa | 4.0 × 10−6 c |
| Bacterial agglomerates | <1 × 10−5 and >8.9 × 10−7 d |
| 1. Centrifugation Step | 2. Medium | 3. Medium Volume, mL | 4. Path Length of the Particles in the Medium, m | 5. Centrifugation Time, s | 6. Rotor Maximum Radius, m | 7. Rotor Minimum Radius, m a | 8. Relative Centrifugal Force (RCF) b | 9. Calculated Particle Diameter (m, Equation (2)) with Expected Sedimentation (Y/N) | |
|---|---|---|---|---|---|---|---|---|---|
| MWCNTs | Bacteria | ||||||||
| Differential centrifugation | Half-strength 21C | 10 | 0.08 | 600 | 0.1359 | 0.0559 | 9715 | 2.8 × 10−7 (Y) c | 7.3 × 10−7 (Y) d |
| Density gradient centrifugation | Dryl’s | 0.5 | 0.003 | 2.7 × 10−7 (N) e | 7.1 × 10−7 (Y) d | ||||
| 50% sucrose | 2 | 0.014 | 8.4 × 10−7 (N) e | 1.6 × 10−6 (N) f | |||||
| 60% sucrose | 2 | 0.027 | 1.4 × 10−6 (N) e | 4.3 × 10−6 (N) f | |||||
| 4.5 | 0.044 | 600 | 0.1854 | 0.1414 | 4194 | ||||
| 1. Centrifugation Step | 2. Medium | 3. Medium Volume, mL | 4. Path Length of the Particles in the Medium, m | 5. Centrifugation Time, s | 6. Rotor Maximum Radius, m | 7. Rotor Minimum Radius, m a | 8. Relative Centrifugal Force (RCF) b | 9. Calculated Particle Diameter (m, Equation (2)) with Expected Sedimentation (Y/N) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MWCNTs c | Bacteria d | Bacterial Agglomerates e | Protozoa d | Fecal Pellets of Protozoa d | ||||||||
| Differential centrifugation | Dryl’s | 10 | 0.08 | 300 | 0.1359 | 0.0559 | 607 | 1.6 × 10−6 (N) f | 4.1 × 10−6 (N) h | 4.1 × 10−6 (Y) h | 4.7 × 10−6 (Y) j | 4.1 × 10−6 (Y) h |
| Density gradient centrifugation I | Dryl’s | 1 | 0.006 | 4.9 × 10−7 (N) f | 1.3 × 10−6 (N) h | 1.3 × 10−6 (Y) h | 1.5 × 10−6 (Y) j | 1.3 × 10−6 (Y) h | ||||
| 10% iodixanol | 2 | 0.027 | 7.0 × 10−7 (N) f | 1.7 × 10−6 (N) i | 1.6 × 10−6 (Y) i | 2.0 × 10−6 (Y) k | 1.6 × 10−6 (Y) i | |||||
| 3 | 0.033 | 300 | 0.1854 | 0.1524 | 1864 | |||||||
| Density gradient centrifugation II | Dryl’s | 0.5 | 0.003 | NA | NA | 1.2 × 10−6 (Y) h | 1.4 × 10−6 (Y) j | 1.2 × 10−6 (Y) h | ||||
| 20% iodixanol | 2 | 0.027 | NA | NA | 3.1 × 10−6 (Y) i | NC | 3.1 × 10−6 (Y) i | |||||
| 2.5 | 0.03 | 300 | 0.1854 | 0.1554 | 1864 | |||||||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mortimer, M.; Petersen, E.J.; Buchholz, B.A.; Holden, P.A. Separation of Bacteria, Protozoa and Carbon Nanotubes by Density Gradient Centrifugation. Nanomaterials 2016, 6, 181. https://doi.org/10.3390/nano6100181
Mortimer M, Petersen EJ, Buchholz BA, Holden PA. Separation of Bacteria, Protozoa and Carbon Nanotubes by Density Gradient Centrifugation. Nanomaterials. 2016; 6(10):181. https://doi.org/10.3390/nano6100181
Chicago/Turabian StyleMortimer, Monika, Elijah J. Petersen, Bruce A. Buchholz, and Patricia A. Holden. 2016. "Separation of Bacteria, Protozoa and Carbon Nanotubes by Density Gradient Centrifugation" Nanomaterials 6, no. 10: 181. https://doi.org/10.3390/nano6100181






