Assembling Deep Neural Networks for Medical Compound Figure Detection
Abstract
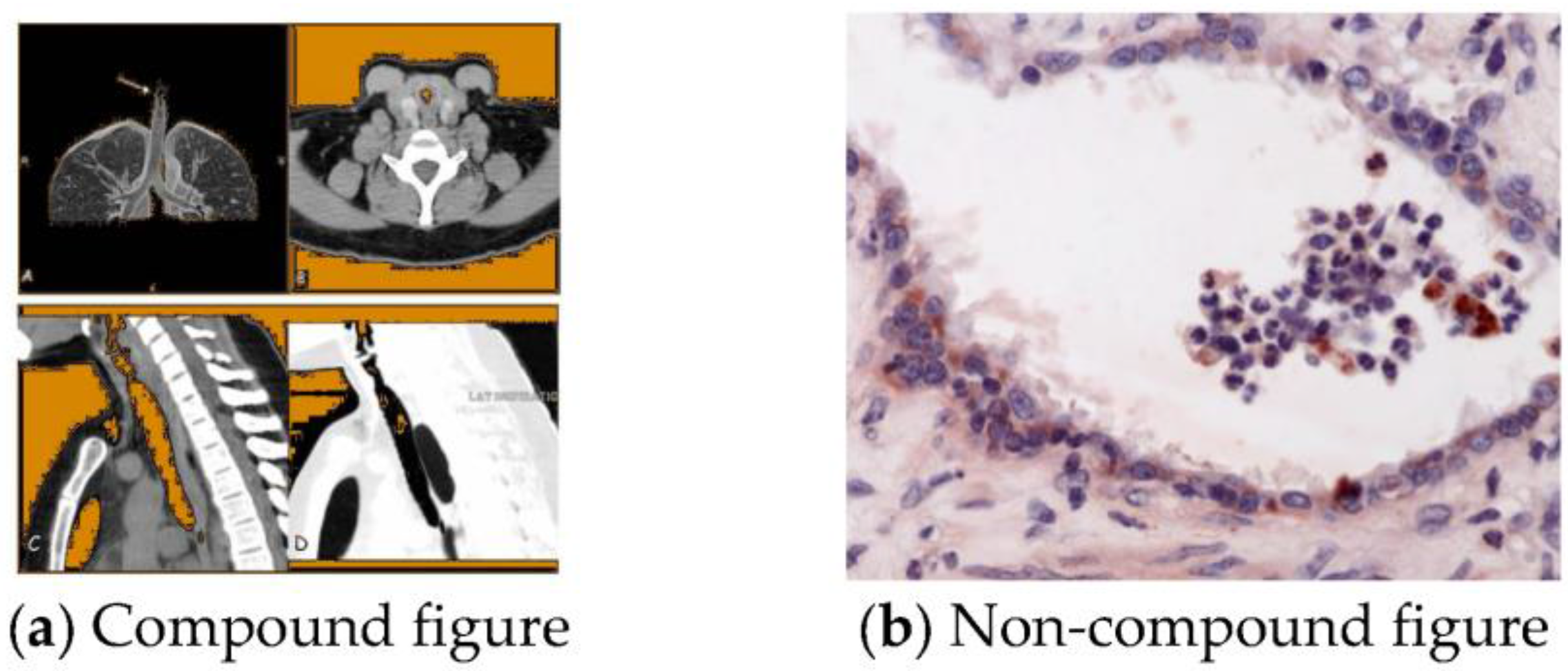
:1. Introduction
2. Methods
2.1. Textual Methods
2.1.1. Textual Convolutional Neural Networks
2.1.2. Textual Long Short-Term Memory Network
2.1.3. Textual Gated Recurrent Unit Network
2.1.4. Textual Rule Model of Delimiter
2.2. Visual Methods
2.2.1. Visual Convolutional Neural Networks
2.2.2. Visual Rule Model of Border
2.3. Mixed Method
3. Experiments
3.1. Dataset
3.2. Baselines
3.2.1. ImageCLEF2015
3.2.2. ImageCLEF2016
3.3. Experimental Results and Discussion
3.3.1. Textual Results
3.3.2. Visual Results
3.3.3. Mixed Results
3.3.4. Running Time
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lu, Z. PubMed and beyond: A survey of web tools for searching biomedical literature. Database 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Müller, H.; Despont-Gros, C.; Hersh, W.; Jensen, J.; Lovis, C.; Geissbuhler, A. Health care professionals’ image use and search behavior. In Proceedings of the Medical Informatics Europe, Maastricht, The Netherlands, 27–30 August 2006. [Google Scholar]
- De Herrera, A.G.S.; Kalpathy-Cramer, J.; Fushman, D.D.; Antani, S.; Müller, H. Overview of the ImageCLEF 2013 medical tasks. In Proceedings of the Working Notes of CLEF, Valencia, Spain, 23–26 September 2013. [Google Scholar]
- Demner-Fushman, D.; Antani, S.; Simpson, M.; Thoma, G.R. Design and development of a multimodal biomedical information retrieval system. J. Comput. Sci. Eng. 2012, 6, 168–177. [Google Scholar] [CrossRef]
- De Herrera, A.G.S.; Müller, H.; Bromuri, S. Overview of the ImageCLEF 2015 medical classification task. In Proceedings of the Working Notes of CLEF, Toulouse, France, 8–11 September 2015. [Google Scholar]
- De Herrera, A.G.S.; Schaer, R.; Bromuri, S.; Müller, H. Overview of the ImageCLEF 2016 medical task. In Proceedings of the Working Notes of CLEF, Évora, Portugal, 5–8 September 2016. [Google Scholar]
- Plataki, M.; Tzortzaki, E.; Lambiri, I.; Giannikaki, E.; Ernst, A.; Siafakas, N.M. Severe airway stenosis associated with Crohn’s disease: Case report. BMC Pulm Med. 2006, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Theegarten, D.; Sachse, K.; Mentrup, B.; Fey, K.; Hotzel, H.; Anhenn, O. Chlamydophila spp. infection in horses with recurrent airway obstruction: Similarities to human chronic obstructive disease. BMC Respir. Res. 2008, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Pelka, O.; Friedrich, C.M. FHDO biomedical computer science group at medical classification task of ImageCLEF 2015. In Proceedings of the Working Notes of CLEF, Toulouse, France, 8–11 September 2015. [Google Scholar]
- Wang, X.; Jiang, X.; Kolagunda, A.; Shatkay, H.; Kambhamettu, C. CIS UDEL Working Notes on ImageCLEF 2015: Compound figure detection task. In Proceedings of the Working Notes of CLEF 2015, Toulouse, France, 8–11 September 2015. [Google Scholar]
- De Herrera, A.G.S.; Markonis, D.; Schaer, R.; Eggel, I.; Müller, H. The medGIFT Group in ImageCLEFmed 2013. In Proceedings of the Working Notes of CLEF, Valencia, Spain, 23–26 September 2013. [Google Scholar]
- Csurka, G.; Dance, C.; Fan, L.; Willamowski, J.; Bray, C. Visual Categorization with Bags of Keypoints. In Proceedings of the Workshop on Statistical Learning in Computer Vision, European Conference on Computer Vision, Prague, Czech Republic, 11–14 May 2004. [Google Scholar]
- De Herrera, A.G.S.; Markonis, D.; Müller, H. Bag-of-Colors for Biomedical Document Image Classification. Proceedings of Medical Content-Based Retrieval for Clinical Decision Support (MCBR-CDS), Nice, France, 1 October 2012. [Google Scholar]
- Zhou, X. Grid-Based Medical Image Retrieval Using Local Features. Ph.D. Thesis, University of Geneva, Geneva, Switzerland, November 2011. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015. [Google Scholar]
- Kim, Y. Convolutional neural networks for sentence classification. arXiv 2014. [Google Scholar]
- Collobert, R.; Weston, J.; Bottou, L.; Karlen, M.; Kavukcuoglu, K.; Kuksa, P. Natural language processing (almost) from scratch. J. Mach. Learn. Res. 2011, 12, 2493–2537. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Tai, K.S.; Socher, R.; Manning, C.D. Improved semantic representations from tree-structured long short-term memory networks. arXiv 2015. [Google Scholar]
- Gal, Y.A. Theoretically Grounded Application of Dropout in Recurrent Neural Networks. arXiv 2015. [Google Scholar]
- Cho, K.; Van Merriënboer, B.; Bahdanau, D.; Bengio, Y. On the Properties of Neural Machine Translation: Encoder–Decoder Approaches. arXiv 2014. [Google Scholar]
- Mikolov, T.; Dean, J. Distributed representations of words and phrases and their compositionality. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 5–10 December 2013. [Google Scholar]
- Manning, C.D.; Surdeanu, M.; Bauer, J.; Finkel, J.R.; Bethard, S.; McClosky, D. The stanford corenlp natural language processing toolkit. In Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics: System Demonstrations, Baltimore, MD, USA, 22–27 June 2014. [Google Scholar]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the 13th International Conference on Artificial Intelligence and Statistics (AISTATS), Sardinia, Italy, 13–15 May 2010. [Google Scholar]
- Yu, Y.; Lin, H.; Meng, J.; Zhao, Z. Visual and Textual Sentiment Analysis of a Microblog Using Deep Convolutional Neural Networks. Algorithms 2016, 9. [Google Scholar] [CrossRef]
- Tieleman, T.; Hinton, G. Divide the gradient by a running average of its recent magnitude. COURSERA: Neural networks for machine learning, Technical Report. Available online: https://zh.coursera.org/learn/neural-networks/lecture/YQHki/rmsprop-divide-the-gradient-by-a-running-average-of-its-recent-magnitude (accessed on 21 April 2017).
- Graves, A. Generating sequences with recurrent neural networks. arXiv 2013. [Google Scholar]
- Kingma, D.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014. [Google Scholar]
- Yu, Y.; Lin, H.; Meng, J.; Zhao, Z.; Li, Y.; Zuo, L. Modality classification for medical images using multiple deep convolutional neural networks. JCIS 2015, 11, 5403–5413. [Google Scholar]
- Wan, L.; Zeiler, M.; Zhang, S.; Cun, Y.L.; Fergus, R. Regularization of neural networks using dropconnect. In Proceedings of the 30th International Conference on Machine Learning (ICML-13), Atlanta, GA, USA, 16–21 June 2013. [Google Scholar]

| Models 2 | ImageCLEF2015 (%) | ImageCLEF2016 (%) | ||
|---|---|---|---|---|
| 10FCV | Evaluation | 10FCV | Evaluation | |
| Baseline_Text | - | 78.34 | - | 88.13 |
| TCNN1 | 86.10 ± 0.12 | 82.24 ± 0.16 1 | 85.18 ± 0.14 | 89.38 ± 0.25 |
| TCNN5 | 86.37 | 82.30 | 85.75 | 89.81 |
| TLSTM1 | 86.50 ± 0.57 | 81.09 ± 0.54 | 85.15 ± 0.19 | 87.69 ± 0.25 |
| TLSTM5 | 87.16 | 81.67 | 85.84 | 88.69 |
| TGRU1 | 87.18 ± 0.07 | 82.01 ± 0.20 | 85.10 ± 0.71 | 88.28 ± 0.72 |
| TGRU5 | 87.23 | 82.40 | 85.91 | 88.72 |
| LR + Delimiter Features | 82.36 | 81.42 | 81.82 | 83.91 |
| Rule_TCNN5 | 87.38 | 82.61 | 86.45 | 90.05 |
| Rule_TLSTM5 | 87.50 | 81.90 | 86.14 | 89.53 |
| Rule_TGRU5 | 87.23 | 83.24 | 86.38 | 89.53 |
| TCNN5 + TLSTM5 + TLSTM5 | 87.89 | 82.95 | 86.49 | 89.90 |
| Model of Delimiter | 88.02 | 83.24 | 86.62 | 90.25 |
| Models 1 | ImageCLEF2015 (%) | ImageCLEF2016 (%) | ||
|---|---|---|---|---|
| 10FCV | Test | 10FCV | Test | |
| Baseline_Figure | - | 82.82 | - | 92.01 |
| VCNN1 | 85.40 ± 0.12 | 80.83 ± 0.45 | 86.41 ± 0.43 | 89.99 ± 0.44 |
| VCNN5 | 88.27 | 84.24 | 89.50 | 92.33 |
| LR + Border Features | 70.36 | 72.98 | 71.76 | 77.60 |
| Model of Border | 89.05 | 86.28 | 90.22 | 93.66 |
| Models | ImageCLEF2015 (%) | ImageCLEF2016 (%) | ||
|---|---|---|---|---|
| 10FCV | Test | 10FCV | Test | |
| Baseline_Mixed | - | 85.39 | - | 92.70 |
| TCNN5 + VCNN5 | 91.30 | 87.93 | 89.88 | 96.33 |
| TLSTM5 + VCNN5 | 91.57 | 87.47 | 90.21 | 96.18 |
| TGRU5 + VCNN5 | 90.26 | 88.35 | 90.30 | 96.12 |
| Rule-based mixed model | 90.85 | 87.52 | 89.91 | 96.18 |
| Mixed model (without rules) 1 | 91.40 | 88.07 | 90.24 | 96.24 |
| Models | ImageCLEF2015 (%) | ImageCLEF2016 (%) | ||||
|---|---|---|---|---|---|---|
| Accuracy 1 | Precision | Recall | Accuracy | Precision | Recall | |
| TCNN5 | 93.46 | 82.27 | 89.06 | 96.88 | 93.59 | 86.43 |
| TLSTM5 | 93.73 | 79.54 | 92.87 | 95.29 | 91.21 | 86.71 |
| TGRU5 | 92.40 | 83.17 | 87.91 | 95.36 | 91.79 | 86.10 |
| LR + Delimiter Features | 94.24 | 94.24 | 72.90 | 97.49 | 97.49 | 71.04 |
| Models | ImageCLEF2015 | ImageCLEF2016 | ||||
|---|---|---|---|---|---|---|
| Accuracy 1 | Precision | Recall | Accuracy | Precision | Recall | |
| VCNN5 | 90.76 | 84.16 | 90.22 | 95.32 | 91.63 | 95.58 |
| LR + Border Features | 92.86 | 92.86 | 56.94 | 95.58 | 95.58 | 59.91 |
| Models | ImageCLEF2015 (%) | ImageCLEF2016 (%) |
|---|---|---|
| TCNN5 + VCNN5 | 95.20 | 97.95 |
| TLSTM5 + VCNN5 | 95.20 | 97.80 |
| TGRU5 + VCNN5 | 95.14 | 97.87 |
| LR + Delimiter Features | 94.24 | 97.49 |
| Models | ImageCLEF2015 (%) | ImageCLEF2016 (%) |
|---|---|---|
| TCNN5 + VCNN5 | 93.47 | 97.35 |
| TLSTM5 + VCNN5 | 93.88 | 97.53 |
| TGRU5 + VCNN5 | 93.42 | 97.17 |
| LR + Border Features | 92.55 | 95.58 |
| Models | Training (ms) | Test (ms) |
|---|---|---|
| TCNN1 | 1.4 | 0.3 |
| TLSTM1 | 18.1 | 2.8 |
| TGRU1 | 11.8 | 3.1 |
| VCNN1 | 1.9 | 0.4 |
| Model of Delimiter | 0.0029 | 0.0017 |
| Model of Border | 0.0020 | 0.0021 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Y.; Lin, H.; Meng, J.; Wei, X.; Zhao, Z. Assembling Deep Neural Networks for Medical Compound Figure Detection. Information 2017, 8, 48. https://doi.org/10.3390/info8020048
Yu Y, Lin H, Meng J, Wei X, Zhao Z. Assembling Deep Neural Networks for Medical Compound Figure Detection. Information. 2017; 8(2):48. https://doi.org/10.3390/info8020048
Chicago/Turabian StyleYu, Yuhai, Hongfei Lin, Jiana Meng, Xiaocong Wei, and Zhehuan Zhao. 2017. "Assembling Deep Neural Networks for Medical Compound Figure Detection" Information 8, no. 2: 48. https://doi.org/10.3390/info8020048






